Abstract
Monoacylglycerol lipases (MGLs) catalyse the hydrolysis of monoacylglycerol into free fatty acid and glycerol. MGLs have been identified throughout all genera of life and have adopted different substrate specificities depending on their physiological role. In humans, MGL plays an integral part in lipid metabolism affecting energy homeostasis, signalling processes and cancer cell progression. In bacteria, MGLs degrade short-chain monoacylglycerols which are otherwise toxic to the organism. We report the crystal structures of MGL from the bacterium Bacillus sp. H257 (bMGL) in its free form at 1.2 Å and in complex with phenylmethylsulfonyl fluoride at 1.8 Å resolution. In both structures, bMGL adopts an α/β hydrolase fold with a cap in an open conformation. Access to the active site residues, which were unambiguously identified from the protein structure, is facilitated by two different channels. The larger channel constitutes the highly hydrophobic substrate binding pocket with enough room to accommodate monoacylglycerol. The other channel is rather small and resembles the proposed glycerol exit hole in human MGL. Molecular dynamics simulation of bMGL yielded open and closed states of the entrance channel and the glycerol exit hole. Despite differences in the number of residues, secondary structure elements, and low sequence identity in the cap region, this first structure of a bacterial MGL reveals striking structural conservation of the overall cap architecture in comparison with human MGL. Thus it provides insight into the structural conservation of the cap amongst MGLs throughout evolution and provides a framework for rationalising substrate specificities in each organism.
Abbreviations: MGL, monoacylglycerol lipase; bMGL, monoacylglycerol lipase from Bacillus sp. H257; hMGL, human monoacylglycerol lipase; PMSF, phenylmethylsulfonyl fluoride; SAXS, small angle X-ray scattering; RMSD, root mean square deviation; SDS-PAGE, sodium dodecyl sulphate polyacrylamide gel electrophoresis
Keywords: Monoacylglycerol lipase, Open conformation, Evolutionary conservation, X-ray crystallography, Molecular dynamics simulation, Small-angle X-ray scattering
Highlights
► First crystal structure of monoacylglycerol lipase from bacterial species. ► Small angle X-ray scattering shows that the protein is a monomer in solution. ► A large hydrophobic channel enables access of the substrate to the active site. ► Molecular dynamic simulations reveal open and closed states of the cap region. ► The cap architecture is conserved on a structural but not on a sequence level.
1. Introduction
Monoacylglycerol lipases (MGLs) were first identified in rat adipose tissue and catalyse the breakdown of monoacylglycerol (MG), a metabolic extracellular and intracellular intermediate, into free fatty acid (FA) and glycerol [1]. In mammals, extracellular sources of MG are dietary lipids which get hydrolysed in the intestine and in plasma. Intracellular MGs are derived from the hydrolysis of diacylglycerols during the degradation of triacylglycerols and glycerophospholipids [2]. MGL has also been shown to hydrolyse 2-arachidonoylglycerol, an important signalling molecule in the endocannabinoid metabolism of the central nervous system [3]. The endocannabinoid system regulates various physiological processes such as appetite, inflammation, pain sensation, and memory [4–7]. Moreover, a role of MGL in the progression of melanoma, human breast, ovarian and colorectal cancer has been identified [8,9]. Inhibition of MGL activity in these cancers shows a marked suppression of cell proliferation which also renders MGL a promising novel drug target for cancer treatment. Hence, MGL and its inhibition are considered to be of high therapeutic relevance [10,11]. In bacteria, MGs have been reported to be highly toxic. Consequently, MGs are often used as antimicrobial agents in the food industry, therapeutic, and pharmaceutical applications [12–14]. The antimicrobial activities have been shown to vary with chain length and saturation of MGs; saturated MGs with 10 or 12 carbon atoms in the chain were shown to be most effective [13,15]. Thus, MGL is crucial for bacterial survival.
MGLs are conserved across species with varying substrate specificities. Orthologs from mouse [16–18] and bacteria (e.g. Pseudomonas sp. LP7315 [19,20], Mycobacterium tuberculosis [21], Mycobacterium smegmatis [22], and Bacillus sp. H257 [23,24]) have been characterised biochemically. Recently, YJU3p has been identified as the functional ortholog of mammalian MGL in yeast [25]. Although YJU3p shares only 24% sequence identity with human MGL (hMGL), it was shown to degrade long-chain MGs with high specific activity exhibiting Vmax and Km values similar to mammalian MGL [25]. The 250 amino acid MGL ortholog from the moderately thermophilic Bacillus sp. H257 (also termed MGLP, UniProtKB accession code P82597) shares only 17% sequence identity with hMGL. bMGL is very specific for MG, especially 1-monolauroylglycerol, and does not show any activity towards di- and triacylglycerols [24]. The specificity and high catalytic rates for MG can be used as a tool for the quantification of MG in biological samples [26].
Members of the MGL family serve as an interesting model for evolutionary studies due to the fact that these lipases from humans to bacteria hydrolyse MG (albeit with quite different specificities with respect to the fatty acid chain length) and that they share very low sequence identity. Recently, 3D structures of human MGL (hMGL) in free form and in complex with inhibitors have been determined [27–29]. In this study, we report the crystal structures of MGL from Bacillus sp. H257 (bMGL) in its free form and in complex with the irreversible inhibitor phenylmethanesulfonyl fluoride (PMSF) at 1.2 Å and 1.8 Å resolution, respectively. bMGL adopts an α/β hydrolase fold and shows an open lid conformation in free as well as in complexed form. We unambiguously identified the catalytic aspartate and histidine residues, which complete the catalytic triad along with the previously identified serine. The structure presents an opportunity to gain a better understanding of the reaction mechanism, the substrate specificity and the structural conservation of MGLs. Unexpectedly, we uncovered that the overall architecture of MGLs is conserved from bacteria to mammals, despite the differences in the number of residues within the cap region, their low sequence identity, and a different composition of secondary structure elements.
2. Material and methods
2.1. Cloning, expression, and purification of MGL
The synthetic gene of bMGL cloned in pUC57 (ATG Biosynthetics, Merzhausen, Germany) was excised with the restriction enzymes NdeI and XhoI and ligated into vector pET28a(+) (Novagen, Merck, Whitehouse Station, USA) to generate an N-terminal (His)6 tagged protein using standard procedures. E. coli BL21 (DE3) cells harbouring pET28a(+)-bMGL were grown in Luria–Bertani broth at 37 °C from an overnight seed culture till they reached mid-log phase. Gene expression was induced using 0.5 mM IPTG at 37 °C for 4 h. The cells were harvested and lysed by sonication in buffer A (20 mM Tris–HCl pH 7.5, 100 mM NaCl). The lysate was centrifuged at 22,000 ×g for 30 min, and the soluble fraction was loaded onto a Ni-NTA agarose resin column (Qiagen, Hilden, Germany). The protein was eluted with 20 mM Tris–HCl pH 7.5, 100 mM NaCl and 250 mM imidazole, and was dialyzed against the same buffer without imidazole. Subsequently the protein was concentrated and loaded onto a Superdex 200 column (GE Healthcare) in buffer A at a flow rate of 0.5 mL/min. The purity of the protein was examined using SDS‐PAGE, and the protein concentration was determined by UV spectroscopy.
2.2. Small angle X-ray scattering analysis (SAXS)
Data collection for the SAXS studies was performed using a wavelength of 1.5 Å and a MAR Image Plate detector [345 mm] at beamline X33 at the European Molecular Biology Laboratory (EMBL), Hamburg, Germany. The distance between the sample and the detector was 2.7 m. The protein sample was measured at three different concentrations, namely 10.2, 5.2, and 1.1 mg/mL in buffer A. Bovine serum albumin (BSA) at a concentration of 4.3 mg/mL was used as standard solution. Data analysis was performed using the programme PRIMUS; scattering from the buffer was subtracted as background from the protein measurements [30]. Low and high concentration data were merged for data analysis. The evaluation of the radius of gyration (RG) and the forward scattering intensity (I(0)) was performed using the Guinier approximation [31]. The pair distribution function was calculated with GNOM [32,33]. Calculation of the theoretical scattering curve from the atomic structure and the subsequent calculation of the pair distribution function were performed with CRYSOL and GNOM, respectively [32–34].
2.3. Monoacylglycerol hydrolase activity assay
Monoacylglycerol hydrolase activity of bMGL was assayed using a protocol described previously [25]. The assay was also used to test the inhibition of bMGL by JZL184 [11], a specific inhibitor reported for hMGL, and PMSF, a commonly known serine hydrolase inhibitor. A concentration of 0.89 nM of bMGL, 10 μM of JZL184 and PMSF concentrations ranging from 40 μM to 200 μM were tested for the inhibition of bMGL. MG hydrolysis was measured after incubation for 20 min at 37 °C in the presence of inhibitor.
2.4. Crystallisation and data collection
Crystallisation trials were performed using the sitting drop vapour diffusion method in a 96-well plate at 20 °C by mixing equal volumes (0.5 μL) of ~ 20 mg/mL bMGL in buffer A and reservoir solutions, respectively. Well diffracting crystals were obtained without cleavage of the His-tag. Crystals were obtained with conditions from a commercial screen (Morpheus, Molecular Dimensions, Suffolk, UK) with the reservoir solution containing 0.1 M MES/imidazole pH 6.5, 12.5% w/v PEG 1000, 12.5% w/v PEG 3350, 12.5% v/v MPD, and 0.02 M of monosaccharides (d-glucose, d-mannose, d-galactose, l-fructose, d-xylose, and N-acetyl-d-glucosamine). No additional cryo-protectant was necessary for flash cooling the crystals in liquid nitrogen. A dataset was collected to 1.2 Å resolution at the X06DA PXIII beamline of the Swiss Light Source at the Paul Scherrer Institute (Villigen, Switzerland). Data collection statistics are listed in Table 1. Crystals of bMGL were soaked with a solution of 250 mM of the irreversible inhibitor PMSF for 90 min. A dataset was collected to 1.8 Å resolution at the X13 beamline, EMBL outstation, DESY, Hamburg, Germany.
Table 1.
Statistics of X-ray data collection and structure refinement.
| Contents | bMGL | bMGL–PMSF |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.0 | 0.81 |
| Resolution (Å) | 17.8–1.2 | 26.6–1.8 |
| Space group | P21 | P21 |
| Unit-cell parameters a, b, c (Å) β (°) |
38.15, 71.21, 43.66 111.7 |
38.05, 70.69, 43.44 111.7 |
| Total number of reflections | 249649 | 55676 |
| Unique reflections | 67545 | 17878 |
| Rsym | 0.065 (0.34) | 0.057 (0.19) |
| Completeness (%) | 99.8 (99.8) | 96.2 (79.63) |
| Mean I/σ(I) | 10.7 (3.4) | 11.1 (4.9) |
| Multiplicity | 3.7 (3.6) | 3.1 (2.8) |
| Refinement statistics | ||
| No. of protein atoms | 1934 | 1902 |
| No. of ligand and/or heteroatoms | 8 ((4R)-2-methylpentane-2,4-diol) | 18 (PMSF and (4R)-2-methylpentane-2,4-diol) |
| No. of solvent molecules | 348 | 202 |
| Rcryst | 13.5% | 15.8% |
| Rfree | 15.8% | 19.4% |
| Model geometry | ||
| RMSD bonds (Å) | 0.007 | 0.013 |
| RMSD angles (°) | 1.271 | 1.051 |
| Ramachandran distribution | ||
| Most favoured (%) | 97.2 | 97.6 |
| Additionally allowed (%) | 2.8 | 2.4 |
| Outliers (%) | 0.0 | 0.0 |
Crystal parameters and data collection statistics are derived from SCALA [36]. To calculate Rfree, 5% of the reflections were excluded from the refinement. Rsym is defined as Rsym = ΣhklΣi | Ii(hkl) − 〈I(hkl)〉 | /ΣhklΣiIi(hkl). Data in parentheses correspond to the highest resolution shells.
2.5. Structure determination and refinement
The uncomplexed bMGL dataset (1.2 Å) was indexed and integrated using iMosflm [35] and scaled using Scala [36]. Molecular replacement was carried out using the Balbes server [37]. A carboxylesterase structure (PDB ID: 1R1D) from Bacillus stearothermophilus exhibiting 34% sequence identity to bMGL was used as the template. Model building was performed using Arp/warp [38]. The model was subjected to rigid body and restrained refinement cycles using the programme REFMAC5 followed by several iterative rounds of refinement using PHENIX [39,40]. There, water molecules were added and the weights for the X-ray/stereochemistry and X-ray/ADP were optimised resulting in the lowest Rfree value. B-factors of the atoms were refined anisotropically. COOT was used to manually adjust and monitor the structure and the solvent, resulting in a final model with an Rfactor of 13.5% and an Rfree of 15.8% [41]. The MolProbity server was used to validate the model [42]. All amino acids were found in the allowed regions of the Ramachandran plot. In the final structure, no electron density was observed for residues Gly135, Gly136, Glu137, and Gly250. No electron density was observed for the N-terminal His-tag, the thrombin cleavage sequence and the linker region introduced from the vector.
X-ray diffraction data of the MGL–PMSF complex were indexed and integrated using iMosflm and scaled using Scala [35,36]. Rigid body refinement was carried out with PHENIX [40]. Refinement and validation were further carried out as described above. Already at the initial stages of model building and refinement, additional density for PMSF was clearly visible in the Fo–Fc map. The Ramachandran plot indicated 100% of the residues in the allowed regions. Figures depicting the protein structure were generated using PyMol (DeLano Scientific) unless stated otherwise [43].
2.6. Molecular dynamics simulations and access path analysis
The three missing residues, Gly135, Gly136, and Glu137, in the crystal structure of bMGL were modelled and optimised in YASARA (Fig. S6) [44]. pKa values and protonation states of the titratable amino acids were calculated at pH 7 using TITRA employing the Tanford–Kirkwood sphere model [45]. The GROMACS 4.5.3 software package was used to perform molecular dynamics simulations and equilibrations [46]. The structure was solvated with water inside a cubic box with a length of 7.5 nm, and minimised and equilibrated for 0.1 ns (NPT and NVT, respectively), with positions restraint on all heavy atoms followed by five unrestrained simulations of about 100 ns with explicit solvent employing OPLS all-atom force field and the TIP3P water model [47,48]. The backbone RMSD was monitored to ensure complete equilibration of the protein model. All calculations were carried out with a 2-fs time step, and long-range electrostatic interactions were computed using the particle mesh Ewald method. All bonds in the system were constrained using the LINCS algorithm. The neighbour list search was updated every 5 steps within a 1.0 nm cut-off. Van der Waals interactions were computed with a cut-off of 1.4 nm. The isotropic Parrinello–Rahman protocol was used for pressure (1 bar), and the velocity-rescaling thermostat was used for temperature coupling. The components of the system were separately coupled at 300 K with a coupling constant of 0.1 ps. Periodic boundary conditions were applied in all three dimensions. Calculations were performed on 64-bit Linux 48-core nodes of an AMD Magny-Cours cluster. Data analysis and image rendering were carried out with standard tools provided within GROMACS and YASARA, respectively [44,46]. The MOLE software package was used for analysing the access paths to the active site [49]. Standard VDW radii were applied for all calculations.
Accession numbers: The coordinates and structure factors were deposited in the Protein Data Bank (PDB ID: 3RM3 and 3RLI3RM33RLI).
3. Results
3.1. MGL from Bacillus sp. H257 is a monomer in solution adopting an α/β hydrolase fold with a partly flexible cap region
The structure of bMGL (Fig. 1A) was determined from a monoclinic crystal (space group P21) at a resolution of 1.2Å. Statistics of the data collection and structure refinement are shown in Table 1. One molecule of bMGL with approximate dimensions of 48Å×32Å×44Å was present in the asymmetric unit. The protein core adopts an α/β hydrolase fold that deviates slightly from the canonical α/β hydrolase fold (Fig. S1). The canonical fold is characterised by a central β-sheet, i.e. a β-hairpin followed by a parallel six-stranded sheet, flanked by α-helices on both sides. The central β-sheet of bMGL, on the other hand, lacks the first strand of the β-hairpin and thus consists of only seven β-strands. Similar deviations from the canonical α/β hydrolase fold are not uncommon and include enzymes missing the complete β-hairpin [50,51].
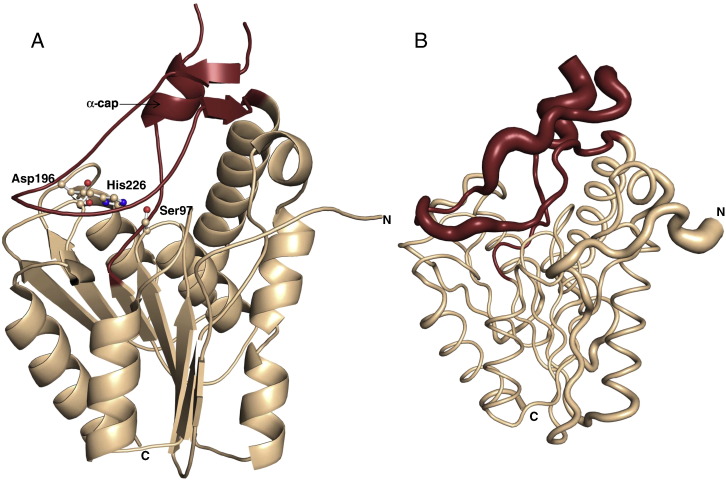
Fig. 1.
Overall structure of monoacylglycerol lipase (MGL) from Bacillus sp. H257. The α/β hydrolase core is coloured wheat brown, the cap region (Ile119–Thr164) is represented in ruby red. A) The catalytic residues Ser97, Asp196, and His226 are represented as ball and sticks. The small α-helix in the cap region is labelled as α-cap; the letters N and C represent the respective termini of the protein. B) Tube representation of bMGL in the same orientation with the flexibility of individual residues indicated based on B-factor values.
A striking feature in the structure of bMGL is the 45 amino acid cap region (Ile119 to Thr164), a structural moiety observed in many α/β hydrolases. It protrudes from the loop connecting β-strand 5 and α-helix 4 of the core structure. The region consists of a small α-helix (α-cap: Ala127-Ala130) and two short antiparallel β-strands (Tyr141-Asp143; Lys161-Pro163) connected by linker regions. A plot of Cα atom B-factors against the sequence indicates that a large part of the cap region is flexible and dynamic (Fig. 1B, Fig. S2). This is also underlined by a lack of electron density for residues Gly135, Gly136, and Glu137.
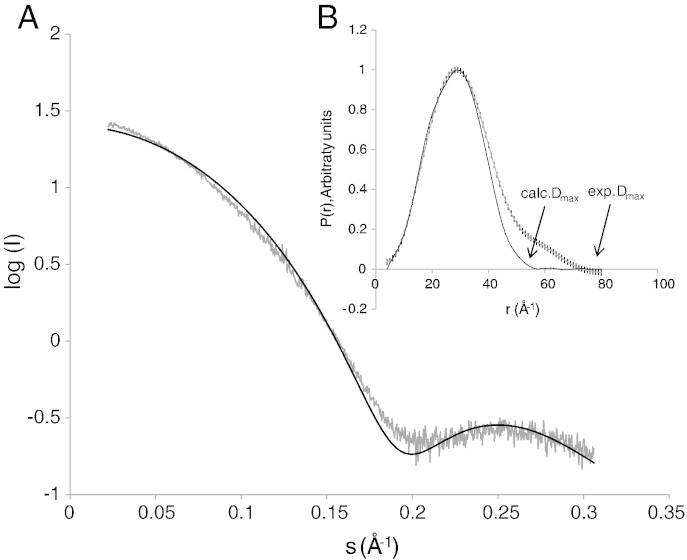
The monomeric state of the protein was confirmed using small angle X-ray scattering (SAXS). The molecular weight calculated from SAXS data is 28.7kDa, which matches the theoretical molecular weight (29.2kDa) for the monomeric protein. The scattering curve and the distance distribution function (Fig. 2A, B) calculated for the atomic model fit the experimental data almost perfectly. The deviation in the maximum distance (Dmax) between the calculated and the experimental data could be due to differences in the hydration shell or the fact that the N-terminal His-tag present in the protein is missing in the X-ray structure used for calculation. The radii of gyration were calculated using GNOM and are in very good agreement resulting in 19.2±0.0Å and 19.9±0.4Å for the theoretical and experimental data, respectively [33].
Fig. 2.
Small angle X-ray scattering (SAXS) measurement of bMGL in solution. A) The theoretical scattering curve of the atomic model (black) shows a good fit to the experimental data (grey). B) Pair distribution functions estimated from the SAXS data (grey) and calculated for the crystal structure (black).
3.2. Active site architecture, the substrate-binding pocket, and exit of hydrolytic products
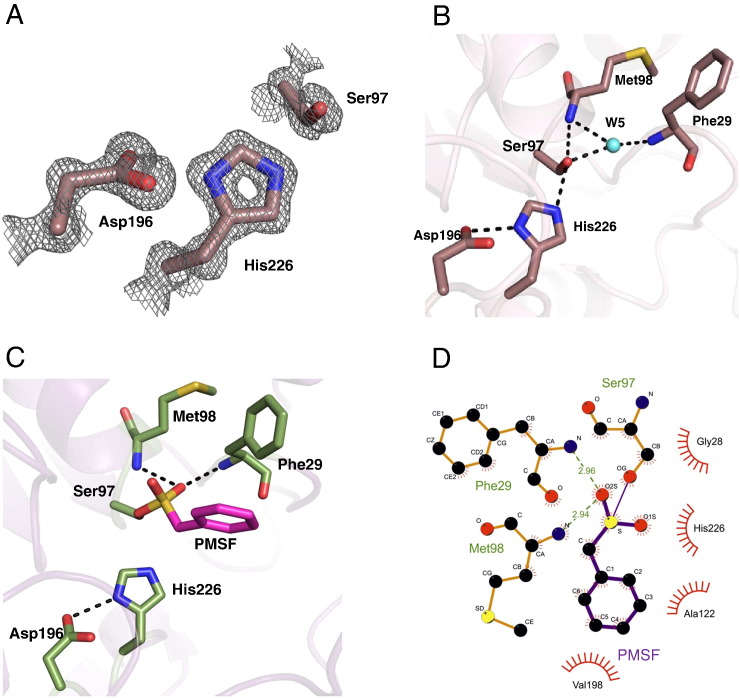
The crystal structure of bMGL shows the enzyme in an open conformation (Fig. 3A, B) as observed for hMGL [27,28]. The catalytic residues are buried at the bottom of a ~22Å long channel, which leads from the surface to the active site and thus keeps the active centre away from a polar environment. The rim at the entrance of the channel has a diameter of ~12Å and is lined by hydrophobic side chains mostly from the cap (Ile125, Ile128, Leu142, and Ile145) and Val198 from the core domain. The entrance is built mainly from hydrophobic side chains of the α/β hydrolase core (Phe29, Met98, Ala122, Leu167, Leu170, and Val198; Fig. S3). The lipophilic potential of bMGL was analysed using the Vasco software [52]. The results of the analysis are represented in colours ranging from red to blue, with red representing hydrophobic regions and blue representing hydrophilic regions of the protein. The residues lining the binding pocket leading to the active site can be observed as a red patch, indicating the rich hydrophobic nature of the pocket (Fig. 3B). The bottom of the substrate-binding pocket has a more polar nature with contributions from the catalytic residues Ser97, His226, and main chain atoms of residues from the core domain (e.g. Gly28, Phe29, Gly31, Thr32, Ser35, Met98, and Asp148; Fig. S3). Hydrophobic patches at the bottom of the substrate-binding site are formed by the side chains of residues Leu96, Ala122, Ala158, and Leu170. Based on sequence alignment, Ser97 had been predicted earlier to be the nucleophile of the active site [24]. From the crystal structure, we identified Asp196 and His226 as the other two residues completing the catalytic triad (Fig. 4A). Met98 and Phe29 form the oxyanion hole, which stabilises the tetrahedral intermediate formed in the course of the MG hydrolysis reaction. Hydrogen bonds play an important role in stabilising the integrity of the active site (Fig. 4B). The hydrogen atom of the backbone NH groups of Met98 and Phe29 form polar contacts with a water molecule (termed W5). In the uncomplexed enzyme, this water molecule also forms a hydrogen bond with the nucleophile Ser97. A hydrogen bond is formed between OG of Ser97 and NE2 of His226. Similarly, ND1 of His226 and OD2 of Asp196 share a hydrogen bond. All these hydrogen bonds are involved in stabilising the active conformation of the catalytic centre.
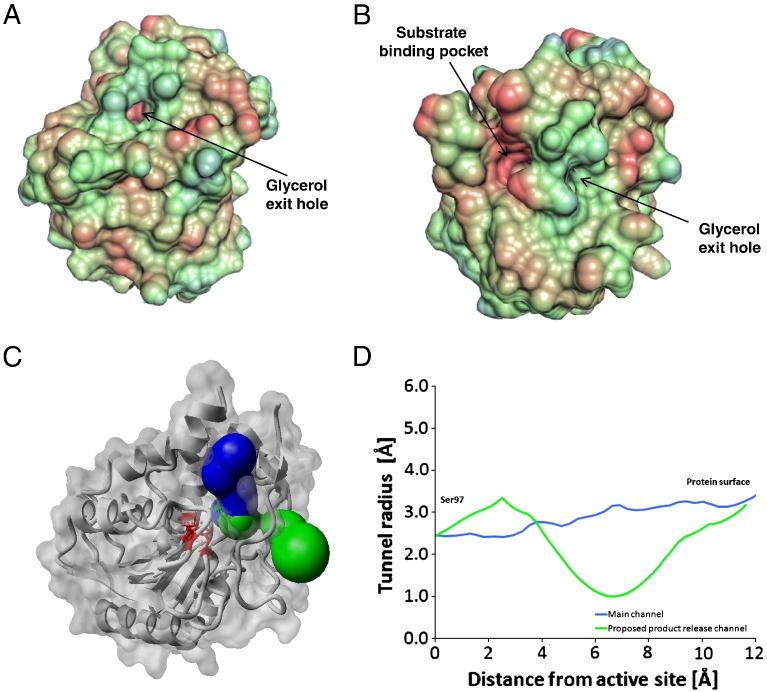
Fig. 3.
Surface and access paths to the catalytic site of bMGL. A and B) Surface representations of the uncomplexed bMGL structure showing the substrate-binding pocket and the glycerol exit hole in two different orientations. The colouring scheme represents the lipophilic potential: Regions coloured red are hydrophobic, blue are hydrophilic. A) Orientation of the protein as in Fig. 1. B) Orientation of the protein after rotation of 55 and 87° along X and Y axes, respectively, for better visualisation of the substrate binding pocket. C) Access paths to the active site of bMGL. The main active site channel is shown in blue, and the proposed glycerol exit hole is shown in green. Residues shown as red sticks are part of the catalytic triad (orientation of the molecule similar to Fig. 3B). D) Radii profile of the access paths of bMGL starting from the active site (Ser97) towards the protein surface.
Fig. 4.
Active site architecture of bMGL. A) The 2Fo–Fc sigma weighted electron density map (grey) is contoured at 1σ around the catalytic triad residues Ser97, Asp196 and His226 (violet sticks). B) The catalytic triad environment in the presence of a water molecule in the uncomplexed structure. H-bonds are depicted as dashed lines. The catalytic triad and oxyanion hole residues are shown in violet. The water molecule (W5) mediating the polar contacts is depicted in cyan. C) Environment of the catalytic triad in the bMGL–PMSF complex structure. H-bonds are depicted as dashed lines. The catalytic triad residues and oxyanion hole forming residues are shown in green. The phenyl group of the PMSF molecule covalently bound to Ser97 is represented in magenta. D) Depiction of atomic details of the interaction between bMGL and PMSF using LigPlot + [60].
A pore is located in the cap region of the protein lined by main-chain atoms of residues Ser147, Ile145, Ala158, and Thr159 and corresponds to the proposed exit hole for glycerol to leave the active site in human MGL (Fig. 3) [27]. Compared to the main entrance leading towards the active site which has a minimal radius of 3.1 Å, the exit hole radius is limited to approx. 1 Å (Fig. 3D). Therefore, only small molecules are expected to pass through this pore. However, due to the fact that it is located within the most flexible region of the protein, the pore geometry is expected to change during substrate binding and/or cap movement.
3.3. bMGL inhibition and bMGL–PMSF complex
Inhibition studies were performed on bMGL using JZL184, a compound reported to specifically inhibit hMGL and some commonly known serine hydrolase inhibitors, including PMSF [11]. The compound JZL184 did not inhibit bMGL (data not shown), whereas PMSF inhibited bMGL completely at a concentration of 200 μM (Fig. S4). Covalently bound PMSF did not change the overall structure of bMGL. The RMSD value for 243 Cα atoms based on an alignment of free and complex structure is 0.13 Å as calculated with Pymol [43]. Binding of PMSF resulted in the displacement of water molecule W5 observed in the uncomplexed structure (Fig. 4C). Hydrogen bonds mediated by W5 were replaced by equivalent interactions of the O2S oxygen atom of the sulphonic acid moiety in the PMSF molecule. The phenyl moiety of the bound inhibitor is projecting towards the entrance channel and forms hydrophobic contacts with Phe29, Leu167, Leu170 and Val198. The atomic details of the PMSF interactions at the active site are shown in Fig. 4D. No electron density was observed for 6 residues: Met132 to Glu137 (Fig. S5A). The minimal radius of the proposed glycerol exit channel in the bMGL-PMSF complex is larger (1.9 Å) than in the uncomplexed bMGL structure (Fig. 3D and S5B).
3.4. Molecular dynamics simulation shows open and closed states of the entrance channel and the glycerol exit hole
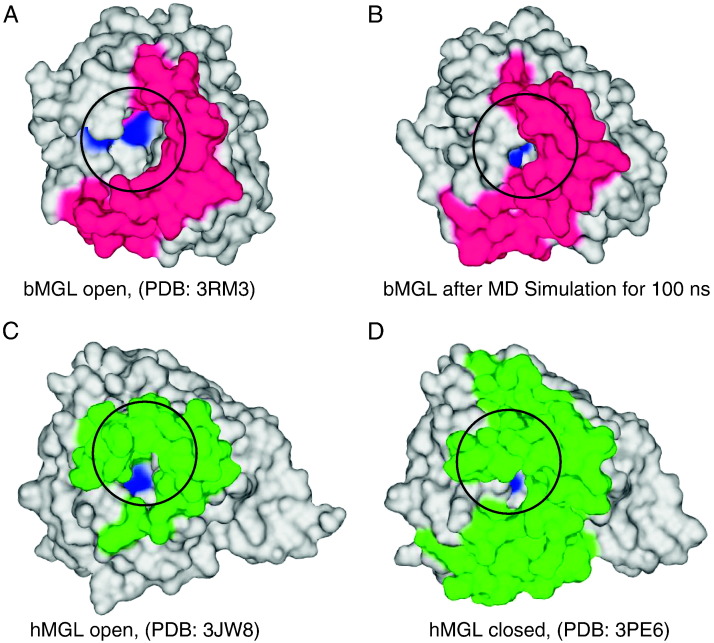
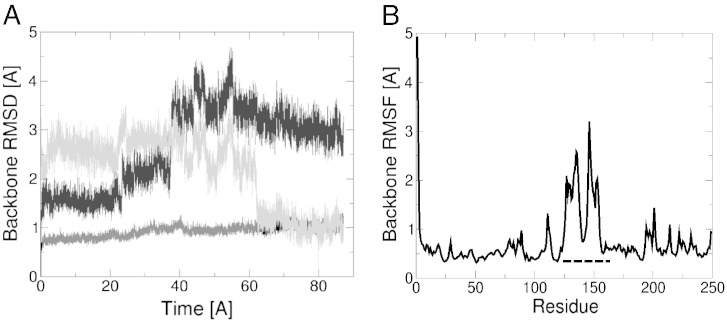
Residues missing within the cap region of the experimentally determined structure of bMGL were added by modelling (Fig. S6). Five 100ns molecular dynamics simulations of bMGL H257 solvated in explicit water starting from the open conformation were carried out to study the behaviour of bMGL in free solution. In three of five cases, the entrance to the active site remained open (Fig. 5A) similar to hMGL in the open conformation (Fig. 5C). In two other cases, the entrance was covered by the cap protecting the entrance to the active site. This yielded a closed conformation of bMGL similar to the recently described closed conformation of hMGL (Fig. 5B, D) [29]. The core region of bMGL in open and closed conformation showed a root mean square deviation (RMSD) around 1 Å relative to the structure in open conformation equilibrated in solution. The cap region showed an increasing RMSD up to 4.5 Å, indicating a significant structural rearrangement (Fig. 6A). In contrast to a complete unfolding of the cap, the observed decrease and increase of the initial cap RMSD (relative to the open conformation) and the final cap RMSD (relative to the closed confirmation) respectively, is a strong indication for a directed cap movement from the open to the closed state [53]. The flexibility of bMGL in solution was calculated using the root mean square fluctuations (RMSF) per residue (Fig. 6B). In addition to the N- and C-terminal regions, the cap region from Ile119 to Thr164 had highest flexibility of the protein in good agreement with the crystallographic B-factors (Fig. 1B, Fig. S2). The short helix α-cap of bMGL undergoes a partial unfolding during the closing event with movements up to 7 Å, as observed during simulations.
Fig. 5.
Surface representation of open and closed conformations of human and bacterial MGL. A) Crystal structure of bMGL in the open conformation. B) bMGL structure in the closed conformation after a 100 ns unrestrained molecular dynamics simulation in explicit water. C) Crystal structure of hMGL in the open conformation and D) hMGL structure in the closed conformation with inhibitor bound. Active site residues are shown in blue, cap regions are shown in magenta and green for bMGL and hMGL respectively. The circles mark the entrance to the binding pocket.
Fig. 6.
Flexibility of individual residues in bMGL. A) Backbone root mean square deviation of bMGL H257 during a 90 ns molecular dynamics simulation. The backbone RMSD of the cap region relative to the open conformation is shown in black and relative to the closed confirmation in light grey, the core region of the protein is shown in dark grey (bottom line). B) Backbone root means square fluctuations (RMSF) per residue of bMGL during a 90 ns molecular dynamics simulation. The cap region from Ile119 to Thr164 is indicated with a dotted line.
4. Discussion
4.1. Structural comparison of bacterial and human MGL
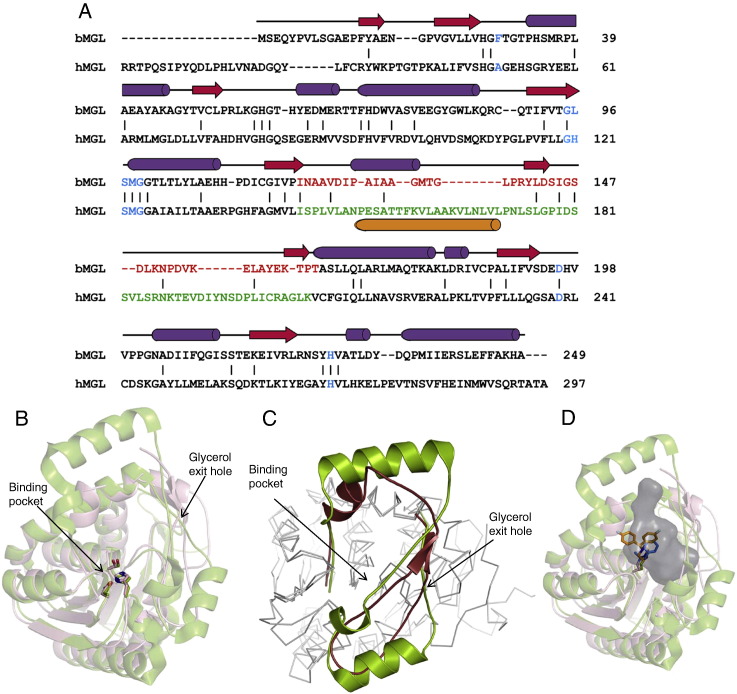
We performed a structural comparison of bMGL (250 aa) and hMGL (303 aa, PDB ID: 3HJU) which both possess an α/β hydrolase fold but share only 17% sequence identity. As shown in Fig. 7A, there is a 14-residue difference in the N-terminus of bMGL compared to hMGL in agreement with the fewer number of β-strands in the core domain of bMGL. It should be noted that two 3D structures of hMGL in open conformation were determined from an isoform with 313 amino acids (PDB ID: 3JWE, 3JW8), i.e. a 10 amino acids insertion at the N-terminus. Hence, there is a discrepancy of 10 amino acids in residue numbering between these and two other hMGL structures in open conformation (PDB ID: 3HJU) and closed conformation (PDB ID: 3PE6). Residues defining the catalytic centre of the proteins are well conserved. These include conservation of the canonical GXSXG consensus sequence (GHSMG and GLSMG in hMGL and bMGL, respectively) and of the catalytic aspartate and histidine residues. In addition, the structural positions of oxyanion hole forming residues (Ala51 and Met123in hMGL, Phe29 and Met98 in bMGL) are also conserved in both enzymes (Fig. 7A and B). The RMSD for Cα atoms of 206 amino acids of the α/β hydrolase core domain of bMGL (corresponding to Met1 to Pro118, and Ala165 to Gly250) in comparison with hMGL is 1.1 Å.
Fig. 7.
Structural comparison of bacterial and human MGL. A) Structure based sequence alignment of bMGL and hMGL (PDB ID: 3HJU) using Dali [58]. Secondary structure elements correspond to those observed in bMGL. β-strands and α-helices are depicted as red arrows and purple cylinders, respectively. Residues in blue correspond to the consensus G-X-S-X-G motif, the catalytic Asp and His, and the oxyanion hole forming residues. Residues within the cap regions of bMGL and hMGL are coloured in red and green, respectively. The proposed membrane binding helix α4 in the cap region of hMGL is shown as orange cylinder. B) Conservation of the catalytic triad evidenced by a structural superimposition. The structure of bMGL is shown as wheat brown cartoon and that of hMGL (PDB ID: 3JW8) as green cartoon. bMGL catalytic residues (Ser97-Asp196-His226) and hMGL (PDB ID: 3JW8) catalytic residues (Ser132-Asp249-His279) are shown as sticks in atomic colours. The substrate binding pocket and the proposed glycerol exit hole are indicated by arrows. C) Structural superposition of the cap region from bMGL (ruby red) and hMGL (green; PDB ID: 3JW8) shows a conserved architecture. D) bMGL (wheat brown) complexed to PMSF (blue) superimposed onto hMGL (green; PDB ID: 3JWE) complexed to SAR629 (orange). The cavity of bMGL as calculated by Casox is represented as a grey surface.
Comparison of the cap regions of hMGL with bMGL shows intriguing similarities despite a sequence identity of only 13% for that region (Fig. 7A, C). The 62-residue cap of hMGL has been described as U-shaped structure and comprises of three helices connected by loop regions [28]. The 45-amino acid cap of bMGL, however, harbours a short helical turn and a short two-stranded antiparallel β-sheet connected by linkers. Despite these differences, both proteins exhibit a similar overall shape and arrangement in the cap region in the open conformation [27,28]. Starting from β-strand 5 of bMGL, a loop and a short helical region traverse on top of the α/β hydrolase core. For the following three residues (Gly135 to Glu137) no electron density was observed (see Figs. 1A, 7A). However, comparison of hMGL and bMGL shows that these missing residues in the cap region of bMGL would fit in a similar position as the first cap helix of hMGL (Fig. 7C). The rest of the bMGL cap extends as a ~ 30 Å long loop completely crossing the α/β hydrolase core back and forth before re-joining the α-helix 4 of the core domain. Interestingly, amino acids at the N-terminal (residues Tyr141-Asp143) and C-terminal ends (Lys161-Pro163) of this long loop form a short antiparallel β-sheet, thus bringing these parts in very close spatial proximity. The corresponding cap region of hMGL (Val170-Lys206) forms a loop of almost identical overall shape, which also crosses the α/β hydrolase core twice. In human MGL, two α helices (Asp180-Leu184 and Lys188-Ser196) are formed within this large loop and can thus accommodate the ‘additional residues’ of its amino acid sequence compared to that of bMGL. To date, the only 3D structures of MGLs known are from human and Bacillus sp. H257. Interestingly, the bMGL structure presented here reveals that both lipases share the same cap architecture in the open conformation. MGL structures from various other organisms will need to be determined in order to discern whether this cap arrangement is a typical feature of MGLs and whether it has been conserved throughout evolution.
For human MGL, it has been proposed that the first helix of the cap region (α4) helps the protein attach itself to the membrane of neuronal cells [28]. Thereby, access to the substrate 2-AG is enabled, which is derived from the degradation of membrane phospholipids [28]. In bMGL, this region corresponds to a loop region, including the unobserved residues of the cap. Interestingly, electron density for α4 is also not observed in one of the monomers of the crystallographic dimer of hMGL complexed with the inhibitor SAR629 [27]. This suggests that this region is also flexible in human MGL. The proposed membrane-binding helix of hMGL is amphipathic, whereas the corresponding stretch in bMGL is composed of mainly small and polar residues. It could be that the predominantly observed membrane and lipid-droplet associated localisation of hMGL is influenced by this helix, whereas the hydrophilic character of bMGL is inferred by the lack of this corresponding amphipathic helix. These characteristics suggest that the restraints on the evolution of the cap region in MGLs are exerted largely by the overall cap architecture and by their respective physiological role.
Recently, the 3D structure of hMGL in complex with a novel inhibitor termed ‘Compound 1’ ((2-cyclohexyl-1,3-benzoxazol-6-yl){3-[4-(pyrimidin-2-yl)piperazin-1-yl]azetidin-1-yl}methanone) was solved (PDB ID: 3PE6) [29]. This structure shows a rearrangement of the cap region resulting in a closed conformation of the enzyme (Fig. 5D), with the inhibitor spanning the entire substrate-binding pocket [29]. Helix α4 is shown to undergo an 8 Å movement through an ‘inward coiling’ action and substantially occlude the entrance to the active site. The proposed glycerol exit hole is also closed in this conformation [29]. It is unknown at this stage whether the inhibitor compound 1 triggered the conformational rearrangement of the cap, especially since hMGL in complex with the inhibitor SAR629 shows an open conformation [27]. No correlation between the closing of the cap and the observed open or closed states of the proposed glycerol exit hole could be observed in our molecular dynamics calculations of bMGL: In one of the simulations, the glycerol exit hole stayed open, whereas in other cases, the exit hole closed independent of the open or closed state of the main entrance of the binding pocket (Fig. S7). Due to the fact that the transition between two stable conformations in free solution is a very rare event, no statistical evaluation of the equilibrium constant between the open and the closed state can be conducted in this study. However, the observation of two independent closing events within a total of 500 ns of simulation clearly demonstrates that stable open and closed conformations of bMGL can exist and that the transition between these two states might be possible under physiological conditions even in the absence of inhibitors.
The substrate specificity of α/β hydrolases is largely determined by the cap region of the respective enzyme [50]. bMGL has been reported to exert highest activity towards monolauroylglycerol (C12:0) (121.1 μmol of released glycerol/min ∗ mg protein) and less activity towards longer fatty acid chains as substrate e.g. monooleoylglycerol (C18:1) (69.0 μmol of released glycerol/min ∗ mg protein) [24]. The specific activity of mouse MGL towards monooleoylglycerol has been reported to be ~ 360 μmol of released FA/min ∗ mg protein [16]. In accordance with the different substrate preferences (long-chain MG vs. more water-soluble, short-chain MGs), helix α4 and the corresponding more hydrophilic region of bMGL could also aid in co-localising MGL at sites of high substrate availability. Experimental 3D structures with bound substrates of different chain lengths, which could give unambiguous insight into residues participating in substrate selectivity, are currently unknown. For hMGL, docking calculations have been performed to understand the binding of 2-arachidonoylglycerol at the active site [27,29]. Apart from the catalytically important residues, Tyr204 in the cap region has been identified to stabilise the glycerol moiety of MG bound to the active site. Structural comparison shows that Glu156 could perform a similar role in bMGL.
Analysis of the binding pocket reveals differences between the structures of bMGL and hMGL. The active site of bMGL is at the bottom of a channel buried between the cap and the core region about 20 Å away from solvent. JZL184 is a specific inhibitor for hMGL and is chemical similar to SAR629, the inhibitor in the complexed structure of hMGL in an open conformation [27]. JZL184 did not inhibit bMGL in our inhibition studies. Structural analysis of the substrate-binding pockets of bMGL and hMGL using the programme Casox suggests that hMGL possesses a larger binding cavity than bMGL (Fig. 7D) [52]. JZL184 might not be able to enter the narrower cavity of bMGL or bMGL might not be able to accommodate the two aromatic substituents of the inhibitor JZL184 in the same conformation as observed for SAR629 in the human structure. This provides a rationale for the inability of JZL184 to inhibit bMGL. The binding pocket of bMGL appears to be favourable for the binding of smaller MGs, whereas in the human enzyme, a rather large pocket would enable binding of longer MGs, which constitute the majority of available MG substrates.
hMGL has been shown to be inhibited by compounds which interact with cysteine residues [54]; hMGL has four cysteines, two of which (Cys201 and Cys242) are close to the catalytic centre and have been shown to interact with the reported isothiazolinone-based compound. bMGL also contains four cysteines, however, none of those have a positional conservation with hMGL and are spatially very distant from the site of catalysis.
4.2. Structural basis for thermostability of bMGL
bMGL was described as a moderately thermophilic protein and was biochemically characterised at 65 °C. We compared both structures to discover cues describing the structural basis of thermostability based on amino acid composition, salt bridges and cation-π interactions [55]. The bMGL structure shows that 16/250 residues (6.4%) form surface salt bridges whereas in hMGL (303aa isoform) 17/303 residues (5.6%) form salt bridges [56]. Two pairs of residues are involved in forming cation-π interaction in both bMGL (Arg87:Trp83 and Lys216:Phe208) and hMGL (Arg196:Tyr278 and Arg303:Trp299) [57]. Accordingly, bMGL has one cation-π interaction for every 125 residues whereas hMGL possesses one per 156 residues. An increase in the number of Arg, Glu, Pro, and Tyr residues accompanied by decrease in Ala, Asn, Gln, Leu, Ser, and Thr is also reported to contribute to the thermostability of proteins [55]. bMGL indeed shows a modest increase in the number of Glu and Tyr residues accompanied by a decrease of Asn, Gln, Leu and Ser residues. Taken together, all these individual factors could contribute to the increased thermostability of bMGL compared to hMGL.
4.3. Evolutionary conservation of MGL
The functional conservation combined with the variation in substrate specificity and low sequence identity across species makes MGL an excellent model for studying protein evolution through 3D structures. The 3D structures of hMGL and bMGL exhibit a high conservation of the fold despite very low sequence identity. A Dali search using the bMGL structure identified many other α/β hydrolases with the same core structure [58]. Somewhat similar cap architectures were observed in the feruloyl esterase (PDB ID: 2WTN; Z-score: 24.5, RMSD: 2.6 Å) and two other proteins (PDB ID: 3LLC; Z-score: 21.8, RMSD: 2.6 Å and 2HDW; Z-score: 21.6, RMSD: 3.0 Å) which are not yet biochemically characterised [59]. A multiple sequence alignment of MGLs from various species also shows that this enzyme class has a low overall degree of sequence conservation apart from the signature lipase motifs and the catalytic residues (Fig. S8). Interestingly, this analysis shows that hMGL shares a higher sequence identity with some MGL from bacteria (e.g. M. tuberculosis) compared to MGLs from plants and yeast. While the yeast ortholog YJU3p shares a mere 24% sequence identity with hMGL, reports suggest that the gene product of Rv0183 (33% sequence identity to hMGL) of the M. tuberculosis H37Rv strain is the closest known unicellular ortholog of hMGL. The mycobacterial enzyme is reported to be responsible for the degradation of cellular lipids of infected humans and has been shown to possess unaltered specificity towards both, 1-monolauroylglycerol (C12:0) and 1-monooleoyl glycerol (C18:1) [21]. In light of the here presented 3D structure of bMGL and the observed conservation of the cap architecture between human and bacterial MGLs, it is tempting to speculate a very similar structural architecture of the cap of the mycobacterial enzyme with secondary structure elements closely resembling those of human MGL.
Acknowledgements
This work was supported by the doctoral school “DK Molecular Enzymology” (W901-B12), by project (FWF P21296) and the SFB LIPOTOX (F30), which are funded by the Austrian Science Fund (FWF). We thank Dr. Georg Steinkellner for conducting the Casox and Vasco analysis. We are grateful for the assistance at the Membrane Protein Laboratory, Imperial College London (Wellcome/MPL grant: 079209/Z/06/Z) particularly to Dr. Momi Iwata's help in obtaining the initial crystals. Additionally, this work has been supported by the Austrian BMWFJ, BMVIT, SFG, Standortagentur Tirol and ZIT through the Austrian FFG-COMET‐Funding Program. The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbalip.2012.04.006.
Appendix A. Supplementary data
Supplementary materials.
References
- 1.Tornqvist H., Belfrage P. Purification and some properties of a monoacylglycerol-hydrolyzing enzyme of rat adipose tissue. J. Biol. Chem. 1976;251:813–819. [PubMed] [Google Scholar]
- 2.Fredrikson G., Tornqvist H., Belfrage P. Hormone-sensitive lipase and monoacylglycerol lipase are both required for complete degradation of adipocyte triacylglycerol. Biochim. Biophys. Acta. 1986;876:288–293. doi: 10.1016/0005-2760(86)90286-9. [DOI] [PubMed] [Google Scholar]
- 3.Dinh T.P., Kathuria S., Piomelli D. RNA interference suggests a primary role for monoacylglycerol lipase in the degradation of the endocannabinoid 2-arachidonoylglycerol. Mol. Pharmacol. 2004;66:1260–1264. doi: 10.1124/mol.104.002071. [DOI] [PubMed] [Google Scholar]
- 4.Di Marzo V., Goparaju S.K., Wang L., Liu J., Batkai S., Jarai Z., Fezza F., Miura G.I., Palmiter R.D., Sugiura T., Kunos G. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature. 2001;410:822–825. doi: 10.1038/35071088. [DOI] [PubMed] [Google Scholar]
- 5.Hohmann A.G., Suplita R.L., Bolton N.M., Neely M.H., Fegley D., Mangieri R., Krey J.F., Walker J.M., Holmes P.V., Crystal J.D., Duranti A., Tontini A., Mor M., Tarzia G., Piomelli D. An endocannabinoid mechanism for stress-induced analgesia. Nature. 2005;435:1108–1112. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- 6.Holt S., Comelli F., Costa B., Fowler C.J. Inhibitors of fatty acid amide hydrolase reduce carrageenan-induced hind paw inflammation in pentobarbital-treated mice: comparison with indomethacin and possible involvement of cannabinoid receptors. Br. J. Pharmacol. 2005;146:467–476. doi: 10.1038/sj.bjp.0706348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsicano G., Wotjak C.T., Azad S.C., Bisogno T., Rammes G., Cascio M.G., Hermann H., Tang J., Hofmann C., Zieglgansberger W., Di Marzo V., Lutz B. The endogenous cannabinoid system controls extinction of aversive memories. Nature. 2002;418:530–534. doi: 10.1038/nature00839. [DOI] [PubMed] [Google Scholar]
- 8.Nomura D.K., Long J.Z., Niessen S., Hoover H.S., Ng S.W., Cravatt B.F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye L., Zhang B., Seviour E.G., Tao K.X., Liu X.H., Ling Y., Chen J.Y., Wang G.B. Monoacylglycerol lipase (MAGL) knockdown inhibits tumor cells growth in colorectal cancer. Cancer Lett. 2011;307:6–17. doi: 10.1016/j.canlet.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nat. Rev. Drug Discov. 2008;7:438–455. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- 11.Long J.Z., Li W., Booker L., Burston J.J., Kinsey S.G., Schlosburg J.E., Pavon F.J., Serrano A.M., Selley D.E., Parsons L.H., Lichtman A.H., Cravatt B.F. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley A.J., Kabara J.J. Antimicrobial action of esters of polyhydric alcohols. Antimicrob. Agents Chemother. 1973;4:501–506. doi: 10.1128/aac.4.5.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabara J.J., Vrable R. Antimicrobial lipids: natural and synthetic fatty acids and monoglycerides. Lipids. 1977;12:753–759. doi: 10.1007/BF02570908. [DOI] [PubMed] [Google Scholar]
- 14.Thormar H., Hilmarsson H. The role of microbicidal lipids in host defense against pathogens and their potential as therapeutic agents. Chem. Phys. Lipids. 2007;150:1–11. doi: 10.1016/j.chemphyslip.2007.06.220. [DOI] [PubMed] [Google Scholar]
- 15.Kabara J.J., Lynch P., Krohn K., Schemmel R. The Pharmacological Effect of Lipids. The American Oil Chemists' Society; IL: 1978. pp. 25–36. [Google Scholar]
- 16.Karlsson M., Tornqvist H., Holm C. Expression, purification, and characterization of histidine-tagged mouse monoglyceride lipase from baculovirus-infected insect cells. Protein Expr. Purif. 2000;18:286–292. doi: 10.1006/prep.1999.1194. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson M., Contreras J.A., Hellman U., Tornqvist H., Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson M., Reue K., Xia Y.R., Lusis A.J., Langin D., Tornqvist H., Holm C. Exon-intron organization and chromosomal localization of the mouse monoglyceride lipase gene. Gene. 2001;272:11–18. doi: 10.1016/s0378-1119(01)00559-5. [DOI] [PubMed] [Google Scholar]
- 19.Sakiyama T., Yoshimi T., Miyake A., Umeoka M., Tanaka A., Ozaki S., Nakanishi K. Purification and characterization of a monoacylglycerol lipase from Pseudomonas sp. LP7315. J. Biosci. Bioeng. 2001;91:27–32. doi: 10.1263/jbb.91.27. [DOI] [PubMed] [Google Scholar]
- 20.Sakiyama T., Yoshimi T., Tanaka A., Ozaki S., Nakanishi K. Analysis of monoglyceride synthetic reaction in a solvent-free two-phase system catalyzed by a monoacylglycerol lipase from Pseudomonas sp. LP7315. J. Biosci. Bioeng. 2001;91:88–90. [PubMed] [Google Scholar]
- 21.Cotes K., Dhouib R., Douchet I., Chahinian H., de Caro A., Carriere F., Canaan S. Characterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem. J. 2007;408:417–427. doi: 10.1042/BJ20070745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhouib R., Laval F., Carriere F., Daffe M., Canaan S. A monoacylglycerol lipase from Mycobacterium smegmatis involved in bacterial cell interaction. J. Bacteriol. 2010;192:4776–4785. doi: 10.1128/JB.00261-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura S., Kitaura S. Purification and characterization of a monoacylglycerol lipase from the moderately thermophilic Bacillus sp. H-257. J. Biochem. 2000;127:419–425. doi: 10.1093/oxfordjournals.jbchem.a022623. [DOI] [PubMed] [Google Scholar]
- 24.Kitaura S., Suzuki K., Imamura S. Monoacylglycerol lipase from moderately thermophilic Bacillus sp. strain H-257: molecular cloning, sequencing, and expression in Escherichia coli of the gene. J. Biochem. 2001;129:397–402. doi: 10.1093/oxfordjournals.jbchem.a002870. [DOI] [PubMed] [Google Scholar]
- 25.Heier C., Taschler U., Rengachari S., Oberer M., Wolinski H., Natter K., Kohlwein S.D., Leber R., Zimmermann R. Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010;1801:1063–1071. doi: 10.1016/j.bbalip.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamura S., Takahasi M., Misaki H., Matsuura K. vol. 5162201. Toyo Jozo Co., Ltd. (Shizuoka, JP); United States: 1992. (Analytical Method Making Use of Monoglyceride Lipase). [Google Scholar]
- 27.Bertrand T., Auge F., Houtmann J., Rak A., Vallee F., Mikol V., Berne P.F., Michot N., Cheuret D., Hoornaert C., Mathieu M. Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 2010;396:663–673. doi: 10.1016/j.jmb.2009.11.060. [DOI] [PubMed] [Google Scholar]
- 28.Labar G., Bauvois C., Borel F., Ferrer J.L., Wouters J., Lambert D.M. Crystal structure of the human monoacylglycerol lipase, a key factor in endocannabinoid signaling. Chembiochem. 2010;11:218–227. doi: 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 29.Schalk-Hihi C., Schubert C., Alexander R., Bayoumy S., Clemente J.C., Deckman I., DesJarlais R.L., Dzordzorme K.C., Flores C.M., Grasberger B., Kranz J.K., Lewandowski F., Liu L., Ma H., Maguire D., Macielag M.J., McDonnell M.E., Mezzasalma Haarlander T., Miller R., Milligan C., Reynolds C., Kuo L.C. Crystal structure of a soluble form of human monoglyceride lipase in complex with an inhibitor at 1.35 A resolution. Protein Sci. 2011;20:670–683. doi: 10.1002/pro.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 31.Guinier A., Fournet G. Small-angle scattering of X-rays. Acta Crystallogr. 1956;9:839. [Google Scholar]
- 32.Semenyuk A.V., Svergun D.I. GNOM — a program package for small-angle scattering data processing. J. Appl. Crystallogr. 1991:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 34.Svergun D.I., Barberato C., Koch M.H.J. CRYSOL — a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 35.Leslie A.G.W. No. 26. 1992. pp. 27–33. (Recent Changes to the MOSFLM Package for Processing Film and Image Plate Data Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography). [Google Scholar]
- 36.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 37.Long F., Vagin A.A., Young P., Murshudov G.N. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrakis A., Morris R., Lamzin V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Echols N., Headd J.J., Hung L.W., Jain S., Kapral G.J., Grosse Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R.D., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. (see also http://www.pymol.org/citing)
- 44.Krieger E., Koraimann G., Vriend G. Increasing the precision of comparative models with YASARA NOVA — a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 45.Petersen M.T., Martel P., Petersen E.I., Drablos F., Petersen S.B. Surface and electrostatics of cutinases. Methods Enzymol. 1997;284:130–154. doi: 10.1016/s0076-6879(97)84009-8. [DOI] [PubMed] [Google Scholar]
- 46.Hess B., Kutzner B., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 47.Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 48.Price D.J., Brooks C.L., III A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 49.Petrek M., Kosinova P., Koca J., Otyepka M. MOLE: a Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15:1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Carr P.D., Ollis D.L. Alpha/beta hydrolase fold: an update. Protein Pept. Lett. 2009;16:1137–1148. doi: 10.2174/092986609789071298. [DOI] [PubMed] [Google Scholar]
- 51.van Pouderoyen G., Eggert T., Jaeger K.E., Dijkstra B.W. The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme. J. Mol. Biol. 2001;309:215–226. doi: 10.1006/jmbi.2001.4659. [DOI] [PubMed] [Google Scholar]
- 52.Steinkellner G., Rader R., Thallinger G.G., Kratky C., Gruber K. VASCo: computation and visualization of annotated protein surface contacts. BMC Bioinformatics. 2009;10:32. doi: 10.1186/1471-2105-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rehm S., Trodler P., Pleiss J. Solvent-induced lid opening in lipases: a molecular dynamics study. Protein Sci. 2010;19:2122–2130. doi: 10.1002/pro.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King A.R., Lodola A., Carmi C., Fu J., Mor M., Piomelli D. A critical cysteine residue in monoacylglycerol lipase is targeted by a new class of isothiazolinone-based enzyme inhibitors. Br. J. Pharmacol. 2009;157:974–983. doi: 10.1111/j.1476-5381.2009.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakravarty S., Varadarajan R. Elucidation of factors responsible for enhanced thermal stability of proteins: a structural genomics based study. Biochemistry. 2002;41:8152–8161. doi: 10.1021/bi025523t. [DOI] [PubMed] [Google Scholar]
- 56.Sarakatsannis J.N., Duan Y. Statistical characterization of salt bridges in proteins. Proteins. 2005;60:732–739. doi: 10.1002/prot.20549. [DOI] [PubMed] [Google Scholar]
- 57.Gallivan J.P., Dougherty D.A. Cation-π interactions in structural biology. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldstone D.C., Villas-Boas S.G., Till M., Kelly W.J., Attwood G.T., Arcus V.L. Structural and functional characterization of a promiscuous feruloyl esterase (Est1E) from the rumen bacterium Butyrivibrio proteoclasticus. Proteins. 2010;78:1457–1469. doi: 10.1002/prot.22662. [DOI] [PubMed] [Google Scholar]
- 60.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.