Abstract
Pluripotency is a property that early embryonic cells possess over a considerable developmental time span. Accordingly, pluripotent cell lines can be established from the pre-implantation or post-implantation mouse embryo as embryonic stem (ES) or epiblast stem (EpiSC) cell lines, respectively. Maintenance of the pluripotent phenotype depends on the function of specific transcription factors (TFs) operating within a pluripotency gene regulatory network (PGRN). As cells move from an ES cell to an EpiSC state, the PGRN changes with expression of some TFs reduced (e.g. Nanog) or eliminated (e.g. Esrrb). Re-expressing such TFs can move cells back to an earlier developmental identity and is being applied to attempt establishment of human cell lines with the properties of mouse ES cells.
Current Opinion in Genetics & Development 2013, 23:504–511
This review comes from a themed issue on Cell reprogramming
Edited by Huck Hui Ng and Patrick Tam
For a complete overview see the Issue and the Editorial
Available online 7th August 2013
0959-437X/$ – see front matter, © 2013 The Authors. Published by Elsevier Ltd. All rights reserved.
Introduction
The formation of epiblast cells within the inner cell mass (ICM) of pre-implantation mammalian embryos marks the establishment of pluripotency [1]. The resulting pluripotent cells are the cells from which all specialised cells that make up the developing embryo and indeed all tissues of the adult organism trace their origins. Despite the transient requirement for such cells, pluripotency is a capacity that lasts for several days spanning implantation and that can be propagated indefinitely in vitro by the establishment of pluripotent cell lines. Although they share the functional capacity for multilineage differentiation, pluripotent cell lines show differences in their properties. Not only are there differences between the growth factor requirements of pluripotent cell lines with an established pre-implantation (embryonic stem; ES) or post-implantation (so-called epiblast stem; EpiSC) identity but these cells also differ in the TFs that impinge upon the PGRN [2,3]. In this review we discuss recent insights into the operation of the PGRN with a particular focus on Nanog. We discuss how changes in the network can alter cell state as cells move from a pre-implantation to post-implantation identity and beyond, as well as when cells are reprogrammed to an ES cell state.
Factors regulating embryonic pluripotency
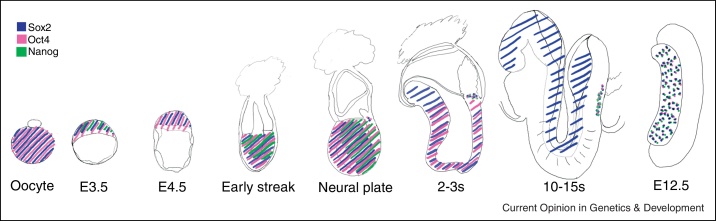
The ICM of early embryos is characterised by expression of three fundamental regulators of pluripotency: the TFs Oct4, Sox2 and Nanog (Figure 1). The requirement of these factors for specification of pluripotency in vivo and maintenance in vitro and their expression kinetics during pre-implantation development have been reviewed recently [4] and will not be recounted in detail here. However, it is worth noting that at E3.5 Nanog expression becomes heterogeneous in the ICM [5]. This is critical to the choice between maintaining pluripotency or differentiating into primitive endoderm. Cells retaining Nanog proceed to complete transcriptional and epigenetic resetting including reactivation of the inactive paternally inherited X chromosome in females [6]. A recent study has shown that in contrast to other pluripotency TFs, Nanog is initially transcribed in a random mono-allelic manner with a switch to bi-allelic expression occurring at the late blastocyst stage around E4.25 [7]. Why Nanog expression should be controlled in this particularly interesting way rather than by simply increasing the transcription of both alleles is an interesting question for the future.
Figure 1.
Nanog, Oct4 and Sox2 expression dynamics during murine development. Whereas Oct4 and Sox2 mRNAs are inherited maternally, Nanog mRNA is first detected in blastomeres of the 8 cell stage embryo [62••]. At E3.5 Nanog is expressed heterogeneously in the ICM, with segregation of Nanog positive and negative cells leading to formation of the epiblast and hypoblast at E4.5. Around implantation Nanog is downregulated before being re-expressed in the posterior epiblast. Nanog remains regionally expressed but progressively declines until being lost at the onset of somitogenesis. Oct4 and Sox2 also become regionally expressed post-implantation, with Sox2 higher in the anterior epiblast and Oct4 becoming progressively posterior. In PGCs (that retain Oct4 expression throughout development) Sox2 becomes detectable at neural plate stage, with Nanog upregulated by the 2–3s stage. Nanog and Sox2 are expressed by PGCs until E12.5 (an E12.5 genital ridge is represented) before becoming undetectable at E15.5 and E14.5 in males and females respectively. Oct4 expression is also lost at E14.5 as female germ cells enter meiosis but persists longer in male PGCs (E, embryonic day; s, number of somites).
Nanog expression is down-regulated in the epiblast before implantation [8], becoming re-activated in the posterior post-implantation epiblast [9••] (Figure 1). Subsequently, Nanog and Oct4 become undetectable when embryos have developed two or 15 somites, respectively. In contrast, Sox2 expression continues but becomes restricted to the neuroectoderm and caudal neural plate. Loss of pluripotency occurs at the onset of somitogenesis preceding the total elimination of Oct4 [9••]. Before this, Nanog expression in the epiblast becomes restricted at a time when cell fate becomes regionalized [9••,10]. Although the ability to express Nanog marks post-implantation epiblast cells as pluripotent, Nanog is strictly dispensable for pluripotency [9••]. Despite the fact that cell fate, morphogens and TFs are regionalized in gastrulating embryos, cells with demonstrable pluripotency persist throughout the epiblast [9••]. Therefore, before somitogenesis, the epiblast exists in a pre-commitment state, characterized by reduced, but reversible PGRN activity. Downregulation of Oct4 below a threshold level required to maintain the PGRN leads to the extinction of pluripotency through chromatin closure at key regulatory elements, such as those at the Nanog and Oct4 loci. Following loss of pluripotency, re-elevating Oct4 expression restores chromatin accessibility at regulatory elements and can rescue pluripotency for several days before DNA methylation changes preclude effective Oct4 action [9••].
Relationship to the germline
The pre-implantation PGRN becomes reactivated in primordial germ cells (PGCs) before epigenetic reprogramming occurs. PGC development has recently been reviewed [11]. Intriguingly, some of the same genes required to specify pre-implantation pluripotency are crucial for PGC development. Oct4 is essential to prevent apoptosis of PGCs after E9.5 [12] and Nanog is required for PGC development beyond E11.5 [13,14]. The recent development of protocols to efficiently generate PGCs from ES cells will enable the contribution of additional pluripotency factors to germ cell development to be systematically tested [15•]. How the activity of a gene regulatory network can on the one hand direct robust pluripotent identity while on the other be associated with a unipotent cell identity is a tantalising issue. Recently, the textbook example of reprogramming of unipotent PGCs to a pluripotent identity has been achieved using MEK/GSK3β inhibitors in place of FGF/SCF alongside co-culture with fibroblasts supplemented with LIF [16]. The precise steps involved in this conversion are not elucidated but perhaps altering the concentration of a single pluripotency TF may suffice.
Capturing pluripotency in vitro: ES cells and EpiSC
Pluripotent cells from the pre-implantation mouse embryo can be captured in vitro as ES cell lines. These cells can differentiate into each of the three primary germ layers and, when introduced into the pre-implantation embryo, can also colonise the germline. ES cells broadly maintain the molecular traits of the ICM, including expression of crucial pluripotency regulators [17] and the presence of two active X chromosomes in female cells. Despite this, ES cells differ from ICM cells most notably by having higher expression of genes involved in epigenetic silencing [17]. ES cells cultured in LIF/FCS show heterogeneous expression of several pluripotency TFs including Nanog, Rex-1, Stella, Klf4 and Tbx3 [4,18]. Nanog protein autorepresses Nanog gene transcription [19,20] thereby contributing to heterogeneity [19]. Surprisingly, ES cells with a reduced level of Oct4 do not exhibit such heterogeneity, instead showing relatively uniform, high expression of Nanog and other TFs [21••].
Post-implantation epiblast cells can also be established in vitro as EpiSC lines [2,3] but these differ from ES cells by requiring Activin/FGF rather than LIF/BMP for maintenance. EpiSCs can also be obtained by explanting pre-implantation mouse embryos in Activin/FGF instead of LIF/BMP [22••]. This indicates that environmental signals determine the cell type captured in vitro, an observation that extends to reprogramming experiments [23••]. In accordance with a post-implantation identity, EpiSC lines derived from female embryos have one inactive X chromosome [24]. EpiSCs are pluripotent, as demonstrated by their teratocarcinoma forming capacity and their ability to differentiate in vitro not only into somatic cells but also into germ cells [23••,25]. Despite this, questions remained about the developmental relevance of EpiSCs since they lack the efficient capacity of ES cells to resume development following introduction into blastocysts [2]. Using an Oct4-DE:GFP reporter that is predominantly active in the pre-implantation epiblast, EpiSCs from early post-implantation embryos show heterogeneous reporter expression with the minor GFP+ population possessing an increased ability to contribute to chimaeras following blastocyst injection [26]. In addition, overexpression of E-cadherin in EpiSCs also increases the proportion of chimaeras following blastocyst injection [27]. Excitingly, EpiSCs have recently been shown to participate efficiently in development when introduced into the appropriate environment of the post-implantation embryo, before but not following the loss of embryonic pluripotency [28•]. Underscoring the importance of this finding, ES cells lack this ability [28•]. These findings demonstrate that the ability to reveal the developmental competence of a cell line depends not only upon the intrinsic state of the cells but also requires a match between the developmental stage of the ‘captured’ cell line and the host environment: chimaera formation can occur when functionally equivalent cultured cells and host tissue are juxtaposed.
Learning from in vitro studies: modelling the implantation transition in vitro
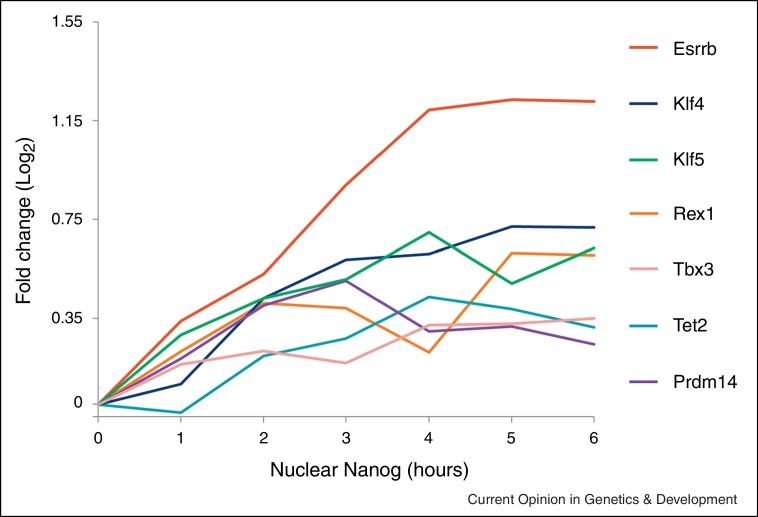
TFs expressed in the Nanog positive epiblast cells of the ICM, such as Esrrb, Klf4, Klf5, Rex1 and Tbx3 are undetectable in the E7.5 epiblast [29–32]. The requirement for pluripotency TFs during implantation is relatively difficult to study due to the inaccessibility of the peri-implantation embryo. However, the derivation of EpiSC by passaging of ES cells in serum free medium supplemented with Activin/FGF [24] provides a tractable model for studying the transition in pluripotent states that occurs at implantation. Several of the pluripotency TFs downregulated following implantation are also downregulated in EpiSCs [4]. Among the core pluripotency factors, Oct4 expression is maintained, whereas Sox2 and Nanog expression are reduced [4,24,26]. In ES cells, Nanog directs transcription of a cohort of regulators of pre-implantation pluripotency that are differentially expressed between ES cells and EpiSCs, including Esrrb, Klf4, Klf5, Rex1 and Tbx3 [33••] (Figure 2). Therefore, these genes may be co-ordinately down-regulated in response to declining Nanog levels at the peri-implantation stage. Perhaps reduced concentrations of Nanog and/or Nanog target gene(s) may be required to facilitate the pre-implantation to post-implantation pluripotency transition.
Figure 2.
Positively regulated transcriptional targets of Nanog. Expression changes of selected significantly upregulated genes identified by analysing the response to acute Nanog induction in a Nanog−/− ES cell line (data from supplementary information in [33••]).
Supporting the importance of Nanog downregulation at implantation, ES cells overexpressing Nanog resist differentiation into EpiSCs, and retain, albeit at reduced levels, expression of ES cell markers after repeated passaging in Activin/FGF [4]. Resistance to differentiation is directly correlated with the Nanog level [4], suggesting that high Nanog concentrations stabilise ES cell pluripotency even in the presence of differentiation signals such as retinoic acid, 3-methoxybenzamide or Activin/FGF [4,8].
New studies have identified two basic helix-loop-helix TFs that operate at this juncture. Tcf15 becomes expressed in the epiblast just before implantation and represses Nanog [34]. The second, Tfe3 relocalises from the nucleus to the cytoplasm during the implantation transition in vitro and in vivo. Preventing cytoplasmic relocalisation of Tfe3 blocks the loss of ES cell pluripotency [35].
Interestingly, Wnts have recently been demonstrated to sustain ES cells in culture when provided alongside LIF [36••]. Although this has been proposed to occur by blockade of the ES to EpiSC transition, the mechanisms involved are not fully resolved [37]. Gene repression by TCF3 appears to play a part and has been proposed to explain how GSK3β inhibition can promote ES cell self-renewal [38,39]. Interestingly, the Nanog target gene Esrrb is amongst the most functionally relevant targets of GSK3β inhibition [40•]. Recently, E-cadherin, which is physically linked to Wnt signalling via β-catenin, has been demonstrated to cooperate with LIFR/gp130 for LIF signalling [41], which could contribute to the Wnt mediated effect.
Learning from in vitro studies: the stalled reprogramming of Nanog-null cells.
Relative to the in vitro generation of EpiSC, reprogramming by enforced expression can provide complementary information on the role of TFs in promoting acquisition of pluripotency. Nanog is not in the original reprogramming factor cocktail [42]. However, Nanog is expressed late during reprogramming [43,44,45•] and is required to complete reprogramming [6]. Nanog−/− somatic cells can be reprogrammed to a state in which they acquire the morphology and growth factor dependence of ES cells [6]. However, as they neither activate endogenous pluripotency TF gene transcription, nor silence the reprogramming factor transgenes they are not fully reprogrammed [6]. This is interesting in light of recent data suggesting that pre-iPS cells may have high Oct4 transgene expression, which is incompatible with self-renewal of ES/iPS cells [46•,47]. Restoring Nanog expression to partially reprogrammed lines facilitates the transition to a fully reprogrammed state [6]. This raises the intriguing possibility that Nanog plays a critical role in imposing the transcriptional and epigenetic state required to silence transgene expression. Recent evidence provides some insight into the mechanisms by which Nanog may achieve reprogramming. Forced expression of the direct Nanog target gene Esrrb, in Nanog−/− pre-iPS cells triggers complete reprogramming when combined with 5’Azacytidine treatment [33••]. Furthermore, Nanog interacts with Tet1 ([48••] and our unpublished information) and induces Tet2 expression ([33••,48••] and Figure 2). Concomitant elevation of Tet1 and Nanog in Nanog −/− pre-iPSCs cooperatively enhances iPS cell generation [48••]. The overlap in chromatin binding between Tet1 and Nanog suggests that Nanog may bring Tet1 to the methylated regulatory regions of key pluripotency genes, thereby triggering hydroxymethylation, potential subsequent demethylation and activation of the PGRN. Therefore, Nanog might trigger completion of reprogramming both by directing transcription of key pluripotency regulators and by targeting silent PGRN loci for epigenetic reawakening.
Learning from in vitro studies: reprogramming EpiSCs to ES cells
The transition of EpiSCs to an ES-like state provides an additional approach to reveal the molecular requirements for attaining pre-implantation pluripotency. Overexpression of Nanog together with a change in culture conditions can drive reprogramming of EpiSC to ES-like cells [4,6]. This conversion is accompanied by acquisition of an ES cell gene expression profile and is marked by reactivation of the inactive X chromosome in female lines [6]. Although similar reprogramming capacities have been reported for other TFs including Esrrb [33••], Klfs [24,49], Nr5a2 [50], Stat3 [51] and, surprisingly, the germ cell marker Prdm14 [52•], the relative efficiency with which most of these factors reprogramme EpiSCs with respect to one another remains unresolved. Similarly to Esrrb, Klf4 and Klf5, Prdm14 is also a transcriptional target of Nanog ([33••] and Figure 2), and its ability to reprogramme EpiSC underscores the overlap in characteristics between migratory PGCs and ES cells.
Nanog was previously shown to be required for conversion of EpiSC to ES cells [6]. However, overexpression of the Nanog target Esrrb bypasses this requirement [33••], raising the possibility that the action of Nanog during reprogramming may be accounted for by Esrrb. Testing this notion by attempting to reprogramming Esrrb-null EpiSCs with Nanog should resolve this issue. Notably, while Esrrb requires 5′Azacytidine to complete reprogramming of Nanog−/− pre-iPS, reprogramming of Nanog−/− EpiSC is induced efficiently by Esrrb alone. Possibly EpiSCs have a closer methylation profile to ES cells than pre-iPS cells. In this regard, Nanog, Oct4 and Sox2 are expressed in EpiSCs and their promoters are unmethylated, while the pre-implantation markers Rex-1, Stella and Fbxo15 have methylated promoters in a fraction of the EpiSC population [9••,25,26]. This difference between the reprogramming of Nanog−/− pre-iPS and Nanog−/− EpiSCs highlights the dual activity that Nanog exerts during reprogramming, with only the transcriptional upregulation of silent target genes, and not the reversion of methylation marks being required for EpiSC reprogramming.
In contrast to human ES cells and EpiSC [2,51], mouse ES cells self-renew in response to LIF. Nanog was isolated on the basis of its ability to confer LIF independent self-renewal of mouse ES cells [8], an activity now shown to require the Nanog target gene Esrrb [33••]. Both Esrrb and the additional direct Nanog target gene Klf4 [33••], can confer LIF independence upon mouse ES cells [8,18,40•], though to varying degrees [33••]. It will be illuminating to determine more fully the epistatic relationship between TFs required to confer LIF independent self-renewal. Klf4 is also elevated in response to LIF [18] suggesting that the Nanog and the LIF-activated cascades may converge on a similar set of target genes to impose the pre-implantation PGRN configuration [53]. Supporting this interpretation, LIF/STAT3 signalling is limiting for EpiSC reprogramming [51], but Nanog expression can bypass this requirement [33••,54]. Intriguingly, hyper-activation of LIF signalling can even override the programmes induced by Activin and FGF in EpiSC, to promote the generation of chimaera-competent ES-like cells [55••]. In contrast, enforced Nanog expression in EpiSC lines cannot drive reprogramming without removal of Activin/FGF [6]. To determine whether extrinsic signals are dominant over intrinsic determinants in dictating pluripotent states, it will be important to test the ability of LIF hyper-activation to reprogramme Nanog−/− EpiSC and whether Nanog overexpression can reprogramme EpiSC cell lines cultured in N2B27/Activin/FGF supplemented with LIF.
Applying knowledge towards establishment of novel human ES cells
As mentioned above, human ES cells can be established from pre-implantation embryos under conditions used to establish mouse post-implantation (not pre-implantation) pluripotent cell lines [2,3]. This raises the question of whether an equivalent of the mouse pre-implantation pluripotent state exists in humans. Attempts have been made to generate human ES cells that possess desirable traits of mouse ES cells such as clonogenicity [56–58,59•]. LIF-dependent human ES cells were obtained using Oct4/Sox2/Nanog/lin28 [56,60] or using Nanog alongside Oct4/Sox2/Klf4/myc [57]. LIF-dependent human cells express pre-implantation markers, though to varying degrees [57,58,59•,60]. Studies using Oct4/Sox2/Klf4/myc without Nanog found that conversion of human ES cells to a LIF-dependent state was possible either in the presence of a compounds that boost Klf4 expression [58] or by including an Nr5a2 transgene [59•]: both of these TFs can reprogramme EpiSCs [24,50]. With one notable exception [59•], these cells remain dependent on continued transgene expression [57,58] or signal modulators [56,60]. Perhaps, in these latter cases self-renewal of converted human ES cells is not robustly sustained because LIF signalling cannot sufficiently activate the pre-implantation PGRN which requires further reinforcement from additional TFs.
Perspective
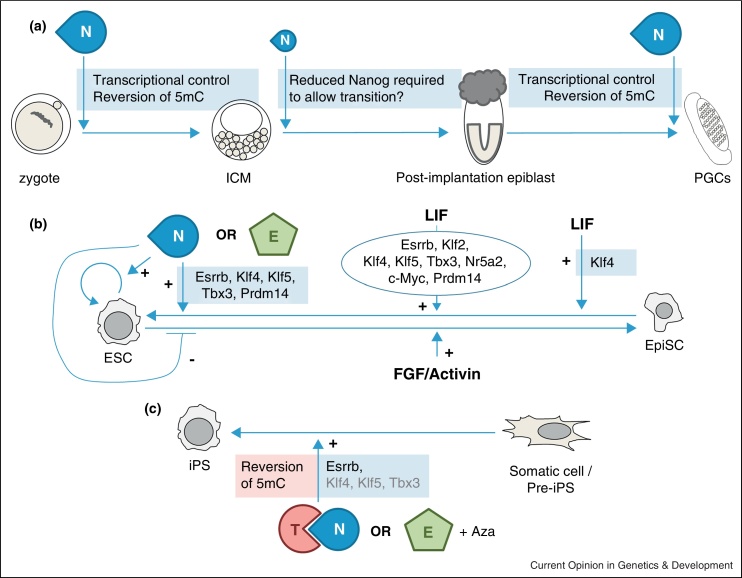
Nanog is crucial in driving establishment of pluripotency during specification of the pre-implantation epiblast and for maintenance of the specified PGC population later in development. By combining in vivo studies with results generated by in vitro reprogramming of Nanog−/− somatic cells, and the identification of Nanog transcriptional targets, at least two aspects of Nanog activity have emerged: first, Nanog regulates expression of other pre-implantation TFs, and thus stands close to the top of the transcriptional hierarchy governing the pre-implantation PGRN; second, Nanog interacts with a number of epigenetic factors [48••,61], targeting them to chromatin and possibly initiating reversion of repressive marks at silent genes during establishment of pluripotency (Figure 3). Further investigation of the protein partners and target genes of Nanog and other PGRN components will provide a fuller understanding of the relationships between distinct pluripotent cells and will continue to inform studies on pluripotency in human and other species.
Figure 3.
Schematic representation comparing the role of Nanog during development and reprogramming. Nanog interacts with other pluripotency TFs, as well as chromatin remodellers, such as NuRD and Tet1. By recruiting proteins to target gene chromatin Nanog can induce or repress mediators of self-renewal and differentiation. (a) Nanog is required twice during development: for the correct specification of a pluripotent epiblast in the blastocyst and for completion of germ cell development beyond E11.5. In both contexts Nanog may operate by modulating expression of its target genes and by counteracting accumulation of 5-methyl-cytosine at regulatory elements and consequent epigenetic silencing of key pluripotency genes. Nanog downregulation appears to be required in the epiblast to reduce expression of pluripotency TFs and allow dismantling of the pre-implantation PGRN. (b) Overexpression of Nanog induces LIF independent self-renewal and blocks ES cell differentiation. Nanog−/− EpiSC can be converted to a pre-implantation pluripotent state by complementation with Nanog or Esrrb. Conversion of wild-type EpiSC to ES cells can be achieved by overexpressing Nanog (possibly through transcriptional activation of the indicated TFs) or Esrrb. An alternative LIF-mediated conversion (which may operate through Klf4 induction) can be accelerated by additional enforced expression of the individual TFs indicated (oval). (c) Nanog expression is required for complete reprogramming of pre-iPSCs from somatic cells, potentially by inducing Esrrb expression and contributing to the reversion of 5-methyl-cytosine marks by Tet1 recruitment. Esrrb can substitute for Nanog during reprogramming of pre-iPSCs when used together with 5-azacytidine. N, Nanog; E, Esrrb; T, Tet1. Boxes associated with arrows indicate potential mechanisms of action.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We are grateful to Sally Lowell and Pablo Navarro for comments on the manuscript and to the Medical Research Council of the UK and CONACYT for support.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Nicola Festuccia, Email: nicola.festuccia@ed.ac.uk.
Ian Chambers, Email: ichambers@ed.ac.uk, I.Chambers@ed.ac.uk.
References
- 1.Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 3.Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 4.Osorno R., Chambers I. Transcription factor heterogeneity and epiblast pluripotency. Philos Trans R Soc Lond B Biol Sci. 2011;366:2230–2237. doi: 10.1098/rstb.2011.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyanari Y., Torres-Padilla M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- 8.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 9••.Osorno R., Tsakiridis A., Wong F., Cambray N., Economou C., Wilkie R., Blin G., Scotting P.J., Chambers I., Wilson V. The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development. 2012;139:2288–2298. doi: 10.1242/dev.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]; Examines the extinction of pluripotency relative to the kinetics of downregulation of Oct4, Sox2 and Nanog in the post-implantation epiblast. The time when the epiblast loses pluripotency is at the onset of somitogenesis. Enforcing Oct4 expression at later stages can resuscitate pluripotency.
- 10.Lawson K.A., Meneses J.J., Pedersen R.A. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 11.Magnusdottir E., Gillich A., Grabole N., Surani M.A. Combinatorial control of cell fate and reprogramming in the mammalian germline. Curr Opin Genet Dev. 2012;22:466–474. doi: 10.1016/j.gde.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomeli H., Nagy A., McLaughlin K.J., Scholer H.R. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi S., Kurimoto K., Yabuta Y., Sasaki H., Nakatsuji N., Saitou M., Tada T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development. 2009;136:4011–4020. doi: 10.1242/dev.041160. [DOI] [PubMed] [Google Scholar]
- 15•.Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]; Describes an efficient method for differentiating ES cells into PGCs. in vitro differentiation chronologically recapitulates in vivo development and yields bona fide germ cells able to generate functional gametes when grafted into neonatal testes.
- 16.Leitch H.G., Blair K., Mansfield W., Ayetey H., Humphreys P., Nichols J., Surani M.A., Smith A. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development. 2010;137:2279–2287. doi: 10.1242/dev.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2009;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 19.Navarro P., Festuccia N., Colby D., Gagliardi A., Mullin N.P., Zhang W., Karwacki-Neisius V., Osorno R., Kelly D., Robertson M. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31:4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fidalgo M., Faiola F., Pereira C.F., Ding J., Saunders A., Gingold J., Schaniel C., Lemischka I.R., Silva J.C., Wang J. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci U S A. 2012;109:16202–16207. doi: 10.1073/pnas.1208533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Karwacki-Neisius V., Goke J., Osorno R., Halbritter F., Ng J.H., Weisse A.Y., Wong F.C., Gagliardi A., Mullin N.P., Festuccia N. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12:531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that ES cells with heterozygous levels of Oct4 present homogeneous expression of Nanog and other TFs and have enhanced self-renewal. Oct4+/− ES cells also show important alterations in signalling including resistance to FGF-induced differentiation. Surprisingly, binding of Oct4 to enhancers of the PGRN is increased despite a lower Oct4 concentration.
- 22••.Najm F.J., Chenoweth J.G., Anderson P.D., Nadeau J.H., Redline R.W., McKay R.D., Tesar P.J. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell. 2011;8:318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that the type of pluripotent cell line formed from explanted pre-implantation mouse embryos depends on the culture environment; an observation with inference for human pluripotent cells.
- 23••.Han D.W., Greber B., Wu G., Tapia N., Arauzo-Bravo M.J., Ko K., Bernemann C., Stehling M., Scholer H.R. Direct reprogramming of fibroblasts into epiblast stem cells. Nat Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]; The authors report for the first time the direct conversion of differentiated cells into EpiSC. This occurs without cells going through an intermediate ES cell state and so, together with Ref. [22••], this study provides an important indication that the culture environment instructs the choice between a primed or naïve state during the derivation of pluripotent lines.
- 24.Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi K., Surani M.A. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136:3549–3556. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han D.W., Tapia N., Joo J.Y., Greber B., Arauzo-Bravo M.J., Bernemann C., Ko K., Wu G., Stehling M., Do J.T. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Ohtsuka S., Nishikawa-Torikai S., Niwa H. E-cadherin promotes incorporation of mouse epiblast stem cells into normal development. PLoS ONE. 2012;7:e45220. doi: 10.1371/journal.pone.0045220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Huang Y., Osorno R., Tsakiridis A., Wilson V. In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012;2:1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]; Demonstrates that EpiSC are functionally equivalent to the post-implantation epiblast. Grafting experiments show that EpiSC contribute efficiently to chimaeric embryos cultured in vitro, differentiating into derivatives of the three germ layers and PGCs.
- 29.Ema M., Mori D., Niwa H., Hasegawa Y., Yamanaka Y., Hitoshi S., Mimura J., Kawabe Y., Hosoya T., Morita M. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Chapman D.L., Garvey N., Hancock S., Alexiou M., Agulnik S.I., Gibson-Brown J.J., Cebra-Thomas J., Bollag R.J., Silver L.M., Papaioannou V.E. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 31.Pelton T.A., Sharma S., Schulz T.C., Rathjen J., Rathjen P.D. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- 32.Luo J., Sladek R., Bader J.A., Matthyssen A., Rossant J., Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 33••.Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S.R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uses Nanog-null cells containing an inducible Nanog protein to identify direct transcriptional targets of Nanog. The most prominent positively regulated TF is Esrrb. Esrrb can complement Nanog function in driving LIF-independent self-renewal and reprogramming EpiSC, NS and pre-iPS cells. Also shows that deletion of Esrrb in ES cells prevents Nanog from conferring cytokine independent self-renewal on transfected ES cells.
- 34.Davies O.R., Lin C.Y., Radzisheuskaya A., Zhou X., Taube J., Blin G., Waterhouse A., Smith A.J., Lowell S. Tcf15 primes pluripotent cells for differentiation. Cell Rep. 2013;3:472–484. doi: 10.1016/j.celrep.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that Wnt signals block ES cell differentiation into EpiSC and are essential for ES cell self-renewal. The combination of Wnt and LIF is sufficient to sustain undifferentiated ES cell cultures and allows the derivation of ES lines from non-permissive mouse strains. The authors also provide evidence of active Wnt signalling in the inner cell mass, becoming inactive post-implantation.
- 37.Niwa H. Wnt: what's needed to maintain pluripotency? Nat Cell Biol. 2011;13:1024–1026. doi: 10.1038/ncb2333. [DOI] [PubMed] [Google Scholar]
- 38.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the gsk3/tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies the transcriptional targets of GSK3β inhibition in ES cells: among the most responsive transcription factors is Esrrb. Esrrb is required to mediate the effects of GSK3β inhibition and cooperates with LIF in maintaining ES cell self-renewal. Esrrb−/− ES cells are derived and shown to retain the ability to contribute to chimaeras.
- 41.del Valle I., Rudloff S., Carles A., Li Y., Liszewska E., Vogt R., Kemler R. E-cadherin is required for the proper activation of the Lifr/Gp130 signaling pathway in mouse embryonic stem cells. Development. 2013;140:1684–1692. doi: 10.1242/dev.088690. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Analyses gene expression changes in single cells during the progression of reprogramming. Esrrb, Utf1, Lin28 and Dppa2 are among the last genes to be upregulated during reprogramming and are identified as good indicators of full reprogramming. This is also the first report that Nanog and Esrrb can efficiently replace the traditional OKMS combination in reprogramming cocktails.
- 46•.Radzisheuskaya A., Le Bin Chia G., Dos Santos R.L., Theunissen T.W., Castro L.F., Nichols J., Silva J.C. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that a reduced Oct4 level is required to enable completion of reprogramming.
- 47.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 48••.Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work extends the known list of Nanog-interacting proteins to include Tet1 and Tet2. Enforced expression of Nanog and Tet1 in combination enhances reprogramming efficiency. Nanog and Tet1 are shown to colocalise to the chromatin of pluripotency genes, possibly contributing to reversion of methylation during reprogramming.
- 49.Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Guo G., Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., van Oosten A.L., Theunissen T.W., Guo G., Silva J.C., Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Gillich A., Bao S., Grabole N., Hayashi K., Trotter M.W., Pasque V., Magnusdottir E., Surani M.A. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides the first report that a transcriptional repressor with a crucial role in germ cell development, Prdm14, can synergise with a pluripotency TF, Klf2, to induce reprogramming of EpiSC to pre-implantation pluripotency.
- 53.Bourillot P.Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N., Savatier P. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- 54.Theunissen T.W., van Oosten A.L., Castelo-Branco G., Hall J., Smith A., Silva J.C. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.van Oosten A.L., Costa Y., Smith A., Silva J.C. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hyperactivation of gp130 signalling is sufficient to drive reprogramming of pre-iPS and EpiSC to pre-implantation pluripotency. Importantly, the authors observe that elevated gp130 signalling in EpiSC results in upregulation of pre-implantation markers and chimaera competency, even in cells maintained in the presence of FGF and Activin.
- 56.Pomp O., Dreesen O., Leong D.F., Meller-Pomp O., Tan T.T., Zhou F., Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Buecker C., Chen H.H., Polo J.M., Daheron L., Bu L., Barakat T.S., Okwieka P., Porter A., Gribnau J., Hochedlinger K. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 2010;6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanna J., Cheng A.W., Saha K., Kim J., Lengner C.J., Soldner F., Cassady J.P., Muffat J., Carey B.W., Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Wang W., Yang J., Liu H., Lu D., Chen X., Zenonos Z., Campos L.S., Rad R., Guo G., Zhang S. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci U S A. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Characterises human cells resembling mouse pre-implantation pluripotent cells that are stable in the absence of transgene expression and that retain differentiation capacity.
- 60.Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 61.Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 62••.Guo G., Huss M., Tong G.Q., Wang C., Li Sun L., Clarke N.D., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]; During progression from the zygote to early blastocyst, the authors report a quantitative analysis of gene expression in single cells. This paper provides molecular insights into the mechanisms that drive lineage segregation and, for many factors, constitutes the only available report of expression pattern in a comprehensive collection of early developmental stages during the onset of zygotic transcription.