Abstract
Significance: Redox biology is a rapidly developing area of research due to the recent evidence for general importance of redox control for numerous cellular functions under both physiological and pathophysiological conditions. Understanding of redox homeostasis is particularly relevant to the understanding of the aging process. The link between reactive oxygen species (ROS) and accumulation of age-associated oxidative damage to macromolecules is well established, but remains controversial and applies only to a subset of experimental models. In addition, recent studies show that ROS may function as signaling molecules and that dysregulation of this process may also be linked to aging. Recent Advances: Many protein factors and pathways that control ROS production and scavenging, as well as those that regulate cellular redox homeostasis, have been identified. However, much less is known about the mechanisms by which redox signaling pathways influence longevity. In this review, we discuss recent advances in the understanding of the molecular basis for the role of redox signaling in aging. Critical Issues: Recent studies allowed identification of previously uncharacterized redox components and revealed complexity of redox signaling pathways. It would be important to identify functions of these components and elucidate how distinct redox pathways are integrated with each other to maintain homeostatic balance. Future Directions: Further characterization of processes that coordinate redox signaling, redox homeostasis, and stress response pathways should allow researchers to dissect how their dysregulation contributes to aging and pathogenesis of various age-related diseases, such as diabetes, cancer and neurodegeneration. Antioxid. Redox Signal. 19, 1362–1372.
Introduction
Reactive oxygen species (ROS) are a group of chemically reactive molecules derived from partial reduction of molecular oxygen. These reactive species are generated as by-products of cellular metabolism and include hydrogen peroxide (H2O2) and oxygen-containing free radicals, such as superoxide anion radical (O2•−) and hydroxyl radical (HO•). Due to their high reactivity, ROS are generally thought to be toxic to the cell and damage a variety of macromolecules, including nucleic acids, proteins, and lipids (67). Apart from their toxicity, ROS have been implicated as important signaling molecules (19, 60). Whereas the role of ROS in aging and longevity has been recognized for a long time, the precise mechanisms are still not fully clear. Oxidative stress and damage to macromolecules due to increased mitochondrial ROS are commonly considered as important contributors to aging and have been documented in a wide variety of species ranging from yeast to humans. Although the importance of ROS as signaling molecules has brought much attention in recent years, their signaling function (and dysfunction) in the aging process remain controversial. Considering that ROS may have a dual function in regulating longevity by both causing oxidative stress and functioning as important signaling molecules, elucidation of the role played by ROS in aging presents a challenge, as it is often difficult to differentiate between the two functions. Moreover, the specific effects of ROS on organism's lifespan largely depend on the chemical nature and function of the particular ROS signaling molecules. In this review, we focus on the understanding of ROS-based signaling in determining longevity, discuss ROS sources, properties and regulatory targets of ROS, and highlight recent advances on the mechanisms of redox sensing and regulation that might help uncover the specific roles of ROS in aging.
Sources and Biochemical Properties of ROS
There are many processes and reactions that result in the production of ROS within the cell. A major source of intracellular ROS in the aging process is superoxide anion radical O2•− produced by mitochondria (Fig. 1). During the process of cellular respiration, O2•− is continuously generated as a result of electron leak from the electron-transport chain. O2•− is produced mainly at complex I and complex III due to transfer of electrons to molecular oxygen. Superoxide radical that is produced by complex I is released into the mitochondrial matrix, whereas complex III forms O2•− both in the matrix and the inner mitochondrial space (9, 68). Another major source of superoxide is NADPH oxidases, a family of membrane-bound enzymes that catalyze controlled production of O2•− by coupling NADPH-derived electrons to oxygen. Superoxide can be also produced in the cell through one-electron transfer reactions catalyzed by a number of enzymes, including monoamine oxidase, cyclooxygenases, lipoxygenases, and components of the cytochrome P450 system.
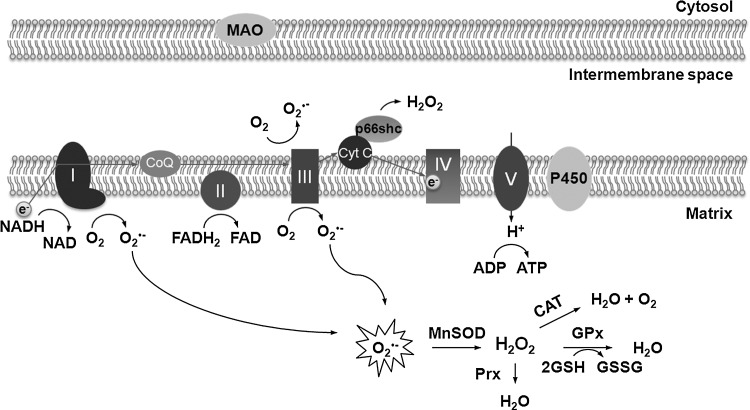
FIG. 1.
ROS production in mammalian mitochondria. Superoxide anion radical O2•− is a major source of intracellular ROS. It is formed mainly in mitochondria as a result of electron leakage from the electron transport chain at complex I and complex III. O2•− can be also produced by several enzymes that catalyze one-electron transfer reactions, such as monoamine oxidase (MAO) and the cytochrome P450 components. Superoxide is unstable and is dismutated spontaneously or by the activity of mitochondrial manganese superoxide dismutase (MnSOD), into H2O2. In addition to hydrogen peroxide produced by MnSOD, H2O2 can be also produced by p66sch protein through 2-electron transfer from cytochrome C (Cyt C). Once hydrogen peroxide is formed, it is further degraded by enzymes such as catalase (CAT) in peroxisomes and glutathione peroxidases (GPx) and peroxiredoxins (Prx) in various cellular compartments.
Superoxide radical is very unstable (half-life 10−6 s) and is rapidly dismutated to H2O2 and molecular oxygen by superoxide dismutases (SODs) (25). In mammalian cells, manganese superoxide dismutase (MnSOD) is present within mitochondrial matrix, whereas cytosolic and extracellular forms of SOD contain copper and zinc in their active site (CuZnSOD) (24). SOD accelerates the rate of spontaneous dismutation more than 1000-fold. If superoxide is not removed by SOD, it can react with iron–sulfur ([Fe–S]) clusters, releasing iron and compromising the functions of [Fe–S] cluster-containing proteins (34). Due to its high reactivity and inability to diffuse through membranes because of the negative charge, O2•− is considered as a poor signaling molecule (12).
In contrast to O2•−, H2O2 is less toxic and is better suited for signaling purposes, in part due to its longer half-life (cellular half-life ∼10−3 s) (25). In addition, it is an uncharged molecule and can diffuse within the cell by crossing membranes. Much of H2O2 is produced from superoxide radical by SODs, but it can be also produced by two-electron reactions. For example, p66Shc generates H2O2 by transferring electrons from the respiratory protein cytochrome C in mitochondria (28). Moreover, H2O2 is formed as a by-product of oxidative protein folding in the endoplasmic reticulum (ER) (71). Each disulfide bond produced by Ero1 protein and protein disulfide isomerase, which form the major pathway for disulfide bond formation in eukaryotes, is expected to result in the production of stoichiometric amounts of H2O2 in the ER lumen. In addition, H2O2 can be generated in the ER lumen by other systems. For example, an ER-localized NADPH oxidase-4 (Nox4) has been reported to produce H2O2 directly without generating O2•− (83). Once H2O2 is formed, it is rapidly eliminated by the activity of several classes of enzymes, such as catalase and thiol-dependent peroxidases, which are members of the peroxiredoxin (Prx) and glutathione peroxidase (GPx) families.
If not removed, H2O2 can be homolytically cleaved in the presence of redox-active Fe(II) iron (Fenton reaction) to form highly reactive hydroxyl radicals (HO•). Hydroxyl radicals are toxic and cause damage to different macromolecules, including nucleic acids, proteins, and lipids. They are also short-lived (half-life 10−9 s), which limits their use as signaling molecules (25).
Depending on biochemical properties of ROS signaling molecules, each of these species (O2•−, H2O2, and HO•) has a distinct set of cellular targets. As discussed above, O2•− targets mainly [Fe–S] cluster-containing proteins. One such example in Escherichia coli is a redox-sensitive transcription factor SoxR, which regulates transcription of genes involved in degradation of O2•− (e.g., mitochondrial MnSOD) and regeneration of [Fe–S] clusters in proteins (ferredoxin:NADPH oxidoreductase) (33). HO• has a very broad reactivity and can interact with a large number of biological molecules.
The major targets of H2O2 are two sulfur-containing amino acids, cysteine (Cys) and methionine (Met), which are the most susceptible amino acids to oxidation by this compound (43). Oxidation of both free Met and Met residues by H2O2 forms methionine sulfoxide (MetSO). As this modification can alter the structure and function of target proteins, organisms evolved a family of enzymes, referred to as methionine sulfoxide reductases (Msr), which can repair oxidatively-damaged Met. However, H2O2 reacts with Met residues very slowly (rate constant of 10−2 M−1•s−1), which makes Met an unfavorable target for the H2O2-receptor function (34).
Although oxidation of free Cys by H2O2 is also slow (2–20 M−1•s−1, at neutral pH) (79), the reactivity of Cys residues in proteins can be significantly increased by the protein environment. High reactivity of Cys and its ability to be reversibly modified by H2O2 make it ideally suited for redox signaling. The following unique features of Cys are often employed by major regulatory protein targets of H2O2. First, Cys thiols can covalently interact with other thiol groups and form intra- and intermolecular disulfide bonds, a reaction promoted by H2O2. Disulfide bond formation is reversible as disulfides can be reduced by either thioredoxin (TRX) or glutaredoxin (GRX) systems. Second, Cys (along with His) is often employed in metal-binding sites in proteins to coordinate metals such as Zn. Upon oxidation of Cys residues in metal-containing redox-active proteins, the metal ion is released, resulting in conformational changes that can alter protein function. Finally, the side chain of reactive Cys can be directly oxidized by H2O2 to form cysteine sulfenic acid (R-SOH) (Fig. 2). The sulfenic acid form is unstable and is either further oxidized to cysteine sulfinic acid (R-SO2H) or can react with an adjacent Cys-SH group, leading to disulfide bond formation. Alternatively, R-SOH can be stabilized by formation of sulfenyl amide in some proteins (e.g., in protein tyrosine phosphatase PTP1B) (63). Reversible oxidation of Cys thiols is known to play an important role in redox regulation via the formation of sulfenic acid intermediates.
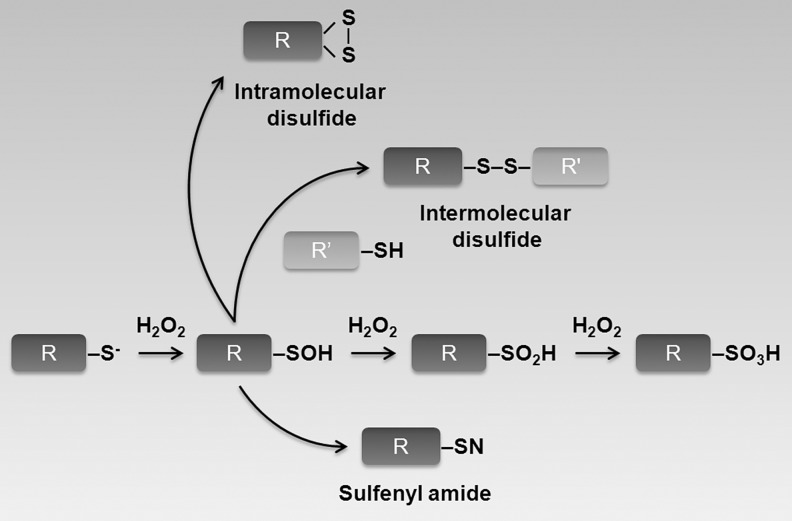
FIG. 2.
Redox-dependent cysteine modifications. During oxidative stress, reactive Cys residues can be oxidized by H2O2 to form cysteine sulfenic acid (R-SOH). R-SOH can be further oxidized by H2O2 to cysteine sulfinic acid (R-SO2H) and cysteine sulfonic acid (R-SO3H). Alternatively, R-SOH can covalently interact with adjacent Cys-SH groups and form disulfide bonds (either intra- or intermolecular), or can be converted to sulfenyl amide (R-SN).
Specificity of ROS Signaling
ROS-mediated signaling has been shown to regulate a number of different cellular processes, ranging from activation of stress response pathways to regulation of normal growth and development. Specific examples include regulation of protein tyrosine phosphatases (63), PTEN (40), and hypoxia-inducible factor (HIF-1a) (4, 30, 45), circadian rhythms (18), as well as regulation of autophagy (64) and inflammatory responses (5, 52). Moreover, ROS have been implicated in activation of the ataxia-telangiectasia (ATM) protein kinase, which is involved in stem cell self-renewal, in response to DNA double-strand breaks (29), among other ROS signaling targets. Although a role for intracellular ROS in signal transduction has been established, it is still controversial how the specificity of redox signaling by ROS is achieved. ROS have distinct chemical properties, which distinguish them from other signaling molecules. In contrast to other signaling molecules, which regulate functions of proteins through non-covalent binding of ligands to their cognate receptors, ROS often operate through chemical interactions with specific amino acid residues (e.g., Cys) in target proteins that lead to covalent modifications (53). Given a large number of Cys residues in proteins and high reactivity of ROS in general, a wide range of promiscuous targets could be affected by these highly reactive molecules. Nevertheless, experimental evidence from studies of known redox signaling pathways shows a high level of specificity in ROS signaling.
The signaling function of ROS is generally thought to result from direct oxidation of ROS-specific receptor proteins involved in various cellular signaling pathways (19, 46) and that high specificity and selectivity of this interaction is achieved by different reactivity of various Cys residues (82). A second mode of regulation involves a cascade of thiol oxidoreductases that transfer oxidizing equivalents from H2O2 to target proteins. One example of such system is the regulation by thiol-dependent peroxidases, which are a group of peroxide scavenging enzymes that have evolved high reactivity toward H2O2. In addition to degrading peroxides, these proteins can also oxidize protein thiols and form disulfides in other substrates. This capability is employed by the Gpx3-Yap1 redox relay in Saccharomyces cerevisiae to regulate transcriptional response to oxidative stress. Gpx3 serves as a sensor of H2O2 (15) and can activate Yap1 transcription factor by forming multiple (up to three) intramolecular disulfide bonds, which promote accumulation of Yap1 in the nucleus and Yap1-dependent gene activation (55). An additional thiol peroxidase, Tsa1, has also been reported to sense H2O2 and activate Yap1-mediated transcriptional response to oxidative stress in S. cerevisiae by forming disulfides in Yap1 (54, 62, 69). Moreover, Schizosaccharomyces pombe peroxiredoxin Tpx1 has been shown to activate Pap1 transcription factor and Sty1 stress-activated protein kinase in a similar fashion (3, 76, 77). These observations suggest a possibility that H2O2 can specifically oxidize a narrow group of proteins that have high reactivity to H2O2 (e.g., thiol peroxidases), which in turn transfer the signal to specific transcription activators (and other proteins) by forming disulfide bonds in these targets. Therefore, uncoupling H2O2 sensing function from transcriptional regulators in such redox relays can dramatically increase specificity of H2O2 signaling. How widespread this mechanism is in living organisms remains unclear. Recently, a study of a yeast strain lacking all thiol peroxidases demonstrated that, at least in this organism, thiol peroxidases are major targets of H2O2 signaling and mediate genome-wide regulation of gene expression in response to H2O2 (21). Yeast cells lacking all 8 thiol peroxidases were unable to sense and transcriptionally respond to H2O2, whereas other oxidant/stress signaling pathways were not affected. These data suggested that thiol peroxidases have dual function in redox regulation: they (i) protect cells from oxidative stress by scavenging toxic H2O2 molecules, and (ii) specifically transmit oxidative signals to activate transcription factors and regulatory proteins in response to H2O2 (Fig. 3). This model also suggests that thiol peroxidases are the main regulators of H2O2 response in yeast. Although direct oxidation of signaling and regulatory proteins by H2O2 also likely exists, it plays a secondary role (21). Whether thiol peroxidases regulate redox signaling by sensing H2O2 and activating cellular target proteins in other organisms, including mammals, awaits further investigation. In line with a possible involvement of thiol peroxidase-mediated signaling in regulation of metazoan aging are recent reports that show a dual role of Prxs in modulating stress resistance and longevity in C. elegans (56) as well as in stimulating survival of murine macrophages exposed to oxidized low density lipoprotein (oxLDL) (11). In addition to protecting cells from oxidative stress by degrading H2O2, in both studies Prxs have been shown to activate the p38 MAPK stress-related signaling pathway. Prx activity has been also implicated in regulating PTEN signaling by protecting this protein from oxidation-induced inactivation (8). However, it should be noted that it is often difficult to differentiate between the contribution that the signaling and antioxidant effects of thiol peroxidases make to their role in longevity.
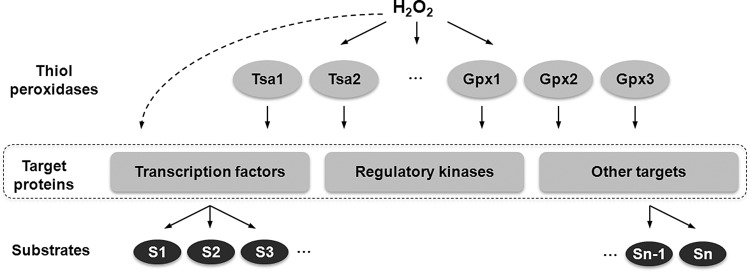
FIG. 3.
A model for the role of yeast thiol peroxidases in regulation of gene expression in response to H2O2. Due to their high specificity toward hydroperoxides, Tsa1, Tsa2, and other thiol peroxidases serve as the proximal H2O2 sensors. These proteins are initially oxidized by H2O2 and then oxidize regulatory and signaling proteins. Upon activation, regulatory proteins interact with their specific substrates (S1, S2, etc.), resulting in the transcriptional response and activation of various signaling pathways. Although this model does not exclude direct oxidation of transcription factors and other regulatory proteins by H2O2 (dashed arrow), at physiological H2O2 concentrations it likely plays a minimal role.
Another possible mechanism by which H2O2 specificity may be achieved is by controlling the concentration of H2O2 and co-localization of the target protein in vicinity of the source of H2O2 production. Local H2O2 concentration can be modulated by peroxiredoxins (Prxs), which are themselves subject to H2O2-mediated inactivation. In eukaryotic cells, catalytic Cys in Prx proteins can be overoxidized by H2O2 to a sulfinic acid (-SO2H) form, inactivating Prx (60, 82). This inhibition is reversible by ATP-dependent reduction of the sulfinic acid by the tightly regulated enzyme sulfiredoxin (Srx) (2, 80). According to the “floodgate hypothesis”, active Prxs normally keep H2O2 concentration low, thereby preventing cellular damage (82). However, during signal transduction, rapid production of H2O2 results in high local H2O2 concentrations, which could inactivate Prxs. Prx inactivation leads to further accumulation of H2O2 for signaling purposes, which could oxidize downstream target proteins in the specific region of the cell (31). This model is consistent with the recent observation of a transient tyrosine phosphorylation on membrane-associated peroxiredoxin 1 (Prx1) upon stimulation of cells by a growth factor, which leads to inactivation of Prx1 (81). This transient phosphorylation only occurs on membrane-associated, but not cytosolic Prx1 leading to localized accumulation of H2O2 needed for signaling. In addition, recent studies suggest that H2O2 not only can diffuse across plasma membrane, but may also be transported through membrane aquaporin channels (1, 48). Therefore, H2O2 channeling to specific cellular sites may provide additional mechanisms, by which high specificity of ROS signaling can be achieved. Finally, it is also possible that overall specificity of H2O2 signaling may result from a combination of the different mechanisms described above and other unidentified factors that may modulate signaling functions of ROS.
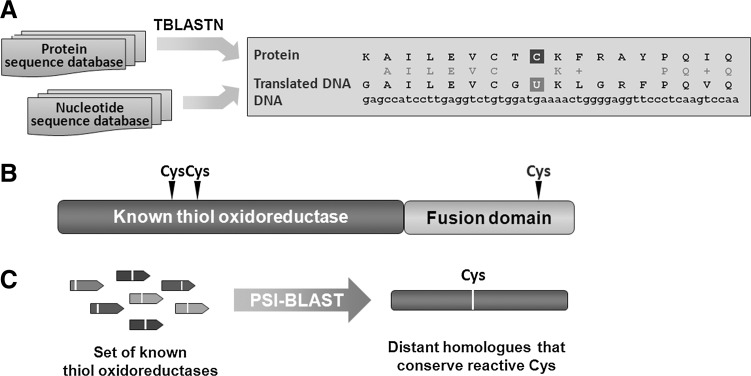
Genome-Wide Identification of Reactive Cys Residues
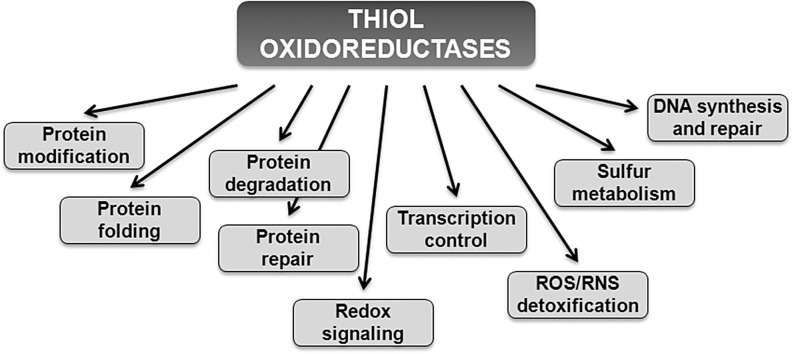
Reactive Cys residues in redox signaling proteins serve as major regulatory targets of H2O2. Many of these regulatory proteins belong to a class of enzymes known as thiol oxidoreductases, which control a variety of cellular functions (Fig. 4). These proteins utilize catalytic Cys residues for reduction or oxidation of their substrates and are ideally suited for the reaction with H2O2 (22). Although many thiol oxidoreductases have been characterized, identification of reactive Cys in proteins has been limited to experimental approaches that utilized individual proteins. Recent advances in the field were made by employing computational approaches (20, 23), which were used to identify reactive Cys residues on a genome-wide level (Fig. 5), as well as large-scale proteomic (mass spectrometry-based) approaches, including two-dimensional electrophoresis gel (2DE)-based fluorescent methods (10), OxICAT (42), and isoTOP-ABPP (isotopic Tandem Orthogonal Proteolysis—Activity-Based Protein Profiling) (78), which were used to identify Cys-containing proteins oxidized in vivo. In particular, computational studies revealed that thiol oxidoreductases are present in all living organisms and generally account for around 0.5%–1% of all proteins encoded in the genome. The abundance of thiol oxidoreductases in cells demonstrates a previously not appreciated complexity of redox regulation and suggests that thiol-based redox signaling is a widespread process across different domains of life. Rapid accumulation of publicly available genome sequences combined with comparative genomics analyses should allow detection of new thiol oxidoreductases and identification of redox-active Cys in these proteins. It would be also important to further characterize the function of thiol oxidoreductases and their contribution to redox regulation of cellular processes, including aging.
FIG. 4.
Functions of thiol oxidoreductases.
FIG. 5.
Computational methods for high-throughput identification of reactive Cys residues. (A) Redox-active Cys residues in thiol oxidoreductases are highly conserved, and in some organisms a functionally similar amino acid, selenocysteine (one letter designation is U), can replace the catalytic Cys. Detection of such Cys/selenocysteine pairs in homologous sequences using TBLASTN could predict the location of redox-active Cys residues in proteins. (B) Proteins containing redox-active Cys can sometimes form fusions with thiol oxidoreductases. Thus, catalytic Cys residues can be predicted by searching for conserved Cys in protein domains fused to known thiol oxidoreductases. (C) Distant homologs of known thiol oxidoreductases containing conserved catalytic redox-active Cys could also be identified in an iterative approach by PSI-BLAST.
Role of Redox Signaling in Aging
The “free radical theory” of aging postulates that aging results from accumulation of oxidative damage to macromolecules (32). However, organisms evolved specific stress response pathways to protect cells from oxidative stress. These pathways sense ROS and activate transcription factors that regulate expression of genes involved in oxidative stress defence. Although the sources of ROS and the mechanisms of ROS-inducible oxidative stress-protective responses are well defined, little is known about the role of redox signaling in aging.
Whereas young organisms are capable of effectively responding to environmental and internal stresses, such as hypoxia, altered metabolism, and oxidative stress, there is compelling evidence that these regulatory processes are altered during aging. It is thought that the inability of aged cells to respond effectively to cellular damage leads to decreased stress resistance and accumulation of “senescence factors,” such as DNA mutations, by-products of metabolism, oxidatively damaged proteins, and protein aggregates (13, 59, 66). Consistent with this view, genetic mutations that extend lifespan are associated with increased cellular stress resistance in variety of species, including yeast, worms, fruit flies, and mice (44).
Role of ROS-mediated signaling in regulating yeast stress resistance and lifespan
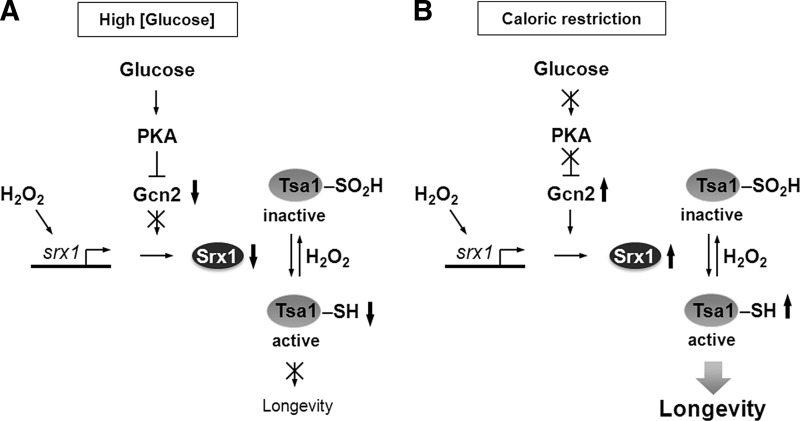
Emerging evidence from studies in model organisms revealed that ROS-mediated signaling plays an important and previously unappreciated role in regulating cellular stress resistance and organism's lifespan. One of the examples that illustrate importance of redox signaling in aging comes from the study of lifespan extension by caloric restriction (CR) in S. cerevisiae. Peroxiredoxin Tsa1 and its partner sulfiredoxin Srx1 have been shown to mediate extension of replicative lifespan and increased H2O2 resistance elicited by CR in yeast (49). During aging, Tsa1 is subject to H2O2-mediated inactivation by hyperoxidation, and CR enhances expression of Srx1, which is required for reactivation of Tsa1 contributing to increased resistance to oxidative stress and longevity (Fig. 6). These findings suggest that stress resistance associated with CR depends on ROS-mediated signaling pathways and that, during aging, hyperoxidation of Tsa1 can inhibit the ability of cells to activate its downstream regulatory targets and respond to oxidative stress. Considering that CR can extend lifespan in a wide variety of species and that peroxiredoxins (Prxs) have been conserved during evolution, it is possible that Prxs can also modulate longevity and contribute to the lifespan extension effect of CR in higher eukaryotes. Consistent with this possibility, a neuronal Prx was identified in D. melanogaster as a downstream target of the insulin signaling-regulated transcription factor FOXO, which is involved in the increased resistance to H2O2 and the lifespan extension effect of CR (41). It should be noted, however, that some organisms (e.g., C. elegans), lack any Srx gene or Srx-like enzyme activity and do not show an increase in Prx hyperoxidation with age (70). Thus, the induction of Srx by CR observed in yeast is unlikely to mediate CR‐induced increases in lifespan in these organisms. A detailed mechanism by which Prxs may contribute to longevity and the identity of transcription factors and regulatory proteins responsible for the Prx-dependent lifespan extension during CR remains to be established. Moreover, the exact signaling role of Prxs in the normal aging process, as well as whether Prx inactivation can be beneficial for signaling (see the “floodgate hypothesis” above) remain puzzling (31). In a recent report, Day et al. provided evidence that in S. pombe Prx inactivation by high H2O2 may enhance cell survival during oxidative stress by preserving the pool of reduced Trx (14). S. pombe peroxiredoxin Tpx1 and transcription factor Pap1 form a redox relay that regulates the transcriptional response to H2O2. In the absence of stress, Tpx1 is maintained in cells in the reduced state by the NADPH-dependent Trx system, with Trx1 serving as a primary reductant. During exposure to H2O2, Tpx1 activates Pap1 by forming disulfide bonds; however, high levels of H2O2 lead to overoxidation and inhibition of Tpx1 activity. Surprisingly, the authors found that hyperoxidation of Tpx1 increases the pool of reduced Trx1 and promotes survival of cells exposed to toxic levels of H2O2. They hypothesized that the beneficial effect of Tpx1 inactivation may result from an increased level of reduced Trx1, which can be used for repair of oxidatively damaged proteins by methionine sulfoxide reductases (Msr). Although this study proves that sensitivity of Tpx1 to hyperoxidation may provide a benefit during acute exposure to H2O2, it does not offer a clear explanation why sensitivity of Tpx1 to hyperoxidation has evolved (36) and whether preserving the pool of reduced Trx1 by Tpx1 inactivation may affect the lifespan of yeast cells.
FIG. 6.
Model for Tsa1-dependent lifespan extension by caloric restriction. (A) At high concentration, glucose stimulates the activity of cyclic AMP-dependent protein kinase A (PKA), leading to inhibition of translation of the SRX1 gene. As a result of decreased production of Srx1, Tsa1 is inactivated by H2O2-induced hyperoxidation. (B) During caloric restriction, low PKA activity stimulates Tsa1 activity by enhancing Gcn2-dependent translation of Srx1, which reactivates hyperoxidized Tsa1.
In addition, ROS signaling has been implicated in extending chronological lifespan caused by inactivation of the nutrient-responsive target of rapamycin (TOR) kinase in S. cerevisiae (57), which has been shown to be involved in the lifespan extension by CR. It was hypothesized that these effects can be attributed to “mitochondrial hormesis” or “mitohormesis” (61, 65), a process whereby elevated mitochondrial ROS generation leads to constitutive activation of antioxidant defense mechanisms and expression of survival genes. However, specific signaling pathways and downstream effectors that are activated by CR-induced ROS production remain obscure. More recently, both low-glucose media and reduced signaling of the nutrient-sensitive TOR and protein kinase A (PKA) pathways have been shown to extend the chronological lifespan of S. pombe through increased ROS production and activation of the stress-dependent Sty1 mitogen-activated protein (MAP) kinase (87). Sty1 kinase is involved in upregulation of stress-related genes and can be activated by multiple stresses, including H2O2. During stress, Sty1 is phosphorylated by Wis1 and translocates to the nucleus, where it activates its downstream transcription factor Atf1. It should be noted that among other stress-related genes, Atf1 regulates expression of Srx1 that is necessary for reactivation of Tpx1 in S. pombe. In turn, Tpx1 may also regulate Sty1 signaling by enhancing H2O2-induced Sty1 phosphorylation (76). Whether Tpx1 and Srx1 can mediate the CR-induced lifespan extension in S. pombe remains unknown.
Links between ROS-induced damage/ROS-signaling responses and aging in higher eukaryotes
Although a role for ROS in aging in higher eukaryotes remains controversial (50, 74), recent studies manipulating the expression of SOD and catalase, antioxidant genes involved in control of ROS levels, in nematodes provide additional support for the importance of ROS-mediated signaling in regulating aging. SOD and catalase are the major antioxidant enzymes that detoxify O2•− and H2O2, respectively. Whereas yeast and fruit flies have two SOD genes encoding cytoplasmic and mitochondrial enzymes, there are five SOD isozymes in C. elegans (two mitochondrial SODs, encoded by sod-2 and sod-3; two cytosolic SODs, encoded by sod-1 and sod-5; and one extracellular SOD, encoded by sod-4). There are also three different catalases, encoded by ctl-1, ctl-2, and ctl-3 genes. Several groups examined the consequences of eliminating expression of these enzymes or their overexpression in worms (reviewed in Refs. 26 and 74). According to the free radical theory of aging, which postulates that aging results from accumulation of molecular damage caused by ROS, a decreased lifespan would be expected, when ROS levels are elevated by deletion of genes such as SOD or catalase. On the other hand, overexpression of these ROS-detoxifying genes would be expected to extend lifespan. Indeed, previous studies in yeast and fruit flies have demonstrated that deletion of either mitochondrial or cytosolic SOD genes shortens lifespan, which could be interpreted as consistent with this theory. However, deletion of individual sod genes had little effect on lifespan in C. elegans, and in some cases (e.g., deletion of sod-2) it could even extend lifespan (16, 73). Furthermore, extended lifespan in sod-2 mutant worms was associated with increased levels of oxidative damage, suggesting that ROS toxicity does not play a major role in lifespan regulation in these animals (73). One possible explanation of why deletion of individual sod genes failed to shorten lifespan is compensation by additional sod genes. However, a recent report from the Hekimi laboratory demonstrates that worms lacking all five sod genes are viable and have normal lifespans, despite significantly increased sensitivity to multiple stresses (75). These observations indicate that oxidative damage caused by superoxide radical does not contribute to worm aging in all experimental conditions. It appears that SOD activity is dispensable and is required only when animals are exposed to acute stresses.
Consistent with these findings, lifespan extension from overexpression of sod-1, the major cytosolic Cu/ZnSOD, and sod-2, the major mitochondrial MnSOD, in C. elegans was not caused by reduced oxidative damage (6). Instead, SOD overexpression was associated with elevated levels of H2O2 and increased protein oxidation, suggesting a possible role of H2O2-mediated signaling in regulating lifespan in worms.
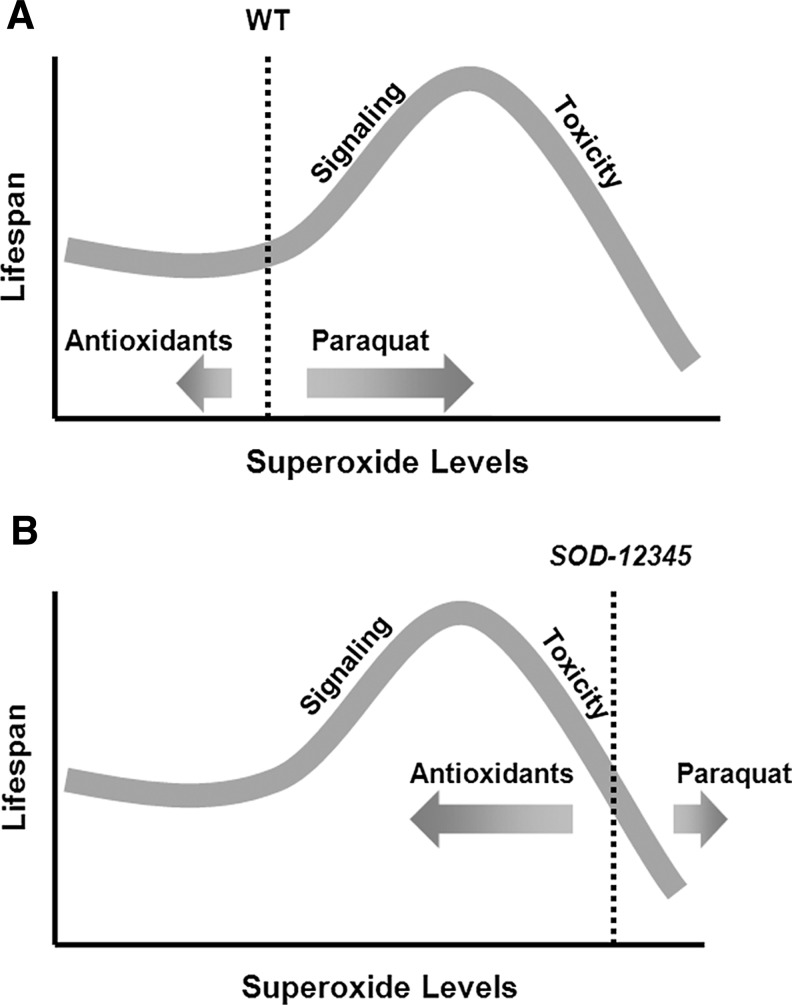
Further support for the role of ROS-mediated signaling in aging is provided by experiments in which worms were treated with increasing concentrations of paraquat, a superoxide generator, or vitamin C, which has a superoxide scavenging activity (75). This study demonstrated that increased superoxide levels, due to addition of low levels of paraquat, resulted in lifespan extension in wild-type worms, whereas at high doses of paraquat the lifespan was decreased. In contrast, treatment of wild-type worms with antioxidants had little effect on their lifespan. This biphasic pattern suggested a model in which C. elegans longevity is dependent on a balance between the prosurvival ROS-mediated signaling and ROS toxicity (Fig. 7). At low levels of superoxide (caused by a decrease in mitochondrial superoxide detoxification through the deletion of sod-2 or produced by the addition of low levels of paraquat), the prosurvival effect of ROS signaling on lifespan is much stronger than the ROS toxic effect leading to lifespan extension. But, at high concentrations, superoxide leads to a dose-dependent decrease in lifespan due to its toxicity. Consistent with this model, treatment of sod quintuple mutant with antioxidants increased lifespan by 15%–20%, whereas the addition of low levels of paraquat decreased lifespan, presumably because the basal superoxide level in this mutant exceeds optimum concentration required for ROS signaling (75). Together, these observations suggest that both lack of antioxidant genes and their overexpression can lead to complex effects due to a dual function of ROS in redox control. Thus, researchers have to be cautious when interpreting such studies (51).
FIG. 7.
Model for the effect of ROS on lifespan in wild-type and sod-12345 worms. (A) ROS may have a dual function in regulating longevity: (i) they can cause oxidative stress (toxicity), and (ii) activate prosurvival signaling. At low concentrations of superoxide, produced by addition of low levels of paraquat, ROS extend lifespan of wild-type worms in a dose-dependent manner, as the prosurvival signaling effect of ROS is greater than the toxic effect. At high concentrations, superoxide leads to a dose-dependent decrease in lifespan due to its toxicity. (B) ROS levels in sod-12345 worms exceed the optimum concentration required for signaling. Decreasing superoxide levels in these animals through addition of antioxidants leads to lifespan extension, whereas treatment with paraquat shortens the lifespan.
Lifespan extending effects of ROS in long-lived C. elegans, in which mitochondrial function is genetically disrupted
Impaired mitochondrial function has been implicated in the aging process in a number of different ways. Although mutations of some genes that encode subunits of mitochondrial electron transport chain, for example, gas-1 (37) and mev-1 (35), have been found to accelerate aging in worms due to an increase in ROS-induced oxidative stress, some mutations that affect mitochondrial function lead to lifespan extension (e.g., isp-1 and nuo-6) (85). It has been proposed that both isp-1 and nuo-6 increase C. elegans lifespan by lowering levels of ROS and oxidative damage to proteins. However, recent analysis of superoxide generation in these mutants revealed that isp-1 and nuo-6 worms have in fact increased rather than decreased ROS levels, and that superoxide is necessary and sufficient for the lifespan extension (84). These observations support a model wherein superoxide produced by mitochondria can act as a signaling molecule triggering specific, yet unidentified, signaling pathways that induce protective and repair mechanisms. The nature of the signaling pathways that may contribute to the ROS signaling-mediated lifespan extension remains largely unknown, and considering the large number of potential ROS targets (see above) it would not be surprising if multiple signaling pathways are involved. One potential mechanism that has been uncovered recently links mitochondrial ROS production to the induction of the mitochondrial unfolded protein response (UPRmt) (17). In this study, inhibition of mitochondrial activity in the intestine or neuronal cells, by tissue specific knockdown of a cytochrome C oxidase 1 subunit (cco-1), increased ROS levels and extended C. elegans lifespan. The authors also demonstrated that inhibition of mitochondrial function can promote longevity in a cell-nonautonomous manner, as knockdown of mitochondrial electron transport chain components in neurons was able to induce the UPRmt signaling pathway in the intestine and extend lifespan. Although the chemical nature of the signal that couples mitochondrial inhibition in neuronal cells to activation of the UPRmt in the intestine remains unknown, it is a possibility that ROS produced in mitochondria may act as a diffusible signaling molecule to activate the UPRmt-mediated transcriptional response. In this regard, it would be important to test whether tissue-specific upregulation of ROS levels in worms by manipulating the expression of antioxidant genes may also extend lifespan in a cell-nonautonomous manner.
Similar to yeast, glucose restriction extends lifespan in C. elegans through increased mitochondrial ROS generation and ROS-dependent induction of long-term stress resistance (65). Moreover, treatment of calorically restricted nematodes with different antioxidants prevented both stress resistance and the lifespan extension effect, suggesting that induction of stress resistance during CR is dependent on generation of ROS.
Together, the above examples in yeast and nematodes revealed that, while ROS can contribute to aging by damaging macromolecular structures, the generation of ROS at low levels can promote longevity by activating redox-regulated signaling pathways. It remains an open question whether increased ROS signaling can promote lifespan extension in mammals. In multicellular organisms, ROS-mediated signaling has been adapted for purposes other than oxidative stress defence (i.e., transcriptional activation of antioxidant response genes). Such nonstress-related ROS-mediated signaling is unique to eukaryotes and appears to regulate normal growth and development. The naked mole rat (Heterocephalus glaber), an exceptionally long-lived rodent (maximum lifespan exceeds 30 years), provides an example of how elevated ROS signaling may contribute to the increased longevity in mammals. In addition to delayed aging, naked mole rats show increased oxidative stress and ROS production at young age, yet their proteome is resistant to oxidative damage (58). Whether elevated ROS production contributes to the exceptional longevity of this organism or is just a correlative event remains unknown. Further elucidation of mechanisms underlying unique stress resistance and longevity of the naked mole rat should improve our knowledge of the role played by ROS-mediated signaling in aging, and availability of this organism's genome, which has been recently sequenced, should accelerate these studies (38).
Role of ROS-inducible pathways in regulating longevity in mammals
Several signaling pathways that have been implicated in regulating lifespan in mammals are subject to regulation by ROS, but the specific upstream ROS sensor proteins remain largely unknown. One of the pathways activated by ROS that might link ROS-mediated signaling with the aging process is the Keap1-Nrf2 pathway. Nrf2 transcription factor regulates the expression of phase 2 detoxification and antioxidant genes. In turn, activation of Nrf2 is repressed by an inhibitor protein named Keap1, which targets Nrf2 for proteasomal degradation through ubiquitination of its specific Lys residues (39). However, H2O2 and other oxidative stress and environmental cues inactivate Keap1 so that it can no longer promote Nrf2 degradation, leading to its nuclear accumulation. The ROS-sensing mechanism involves modification of specific cysteine residues in Keap1 and conformational changes that inactivate Keap1 activity (86). Recently, an Nrf2 homolog in worms, SKN-1, has been shown to mediate the lifespan extension effect of reduced insulin/IGF-1-like signaling in worms (72) linking ROS signaling with the aging process.
Future Directions
ROS, which have been long considered as toxic compounds causing oxidative stress, are now recognized as unique signaling messengers that are produced in a regulated manner and control a wide range of cellular processes, including but not limited to, cell proliferation, circadian rhythms, immune response, and aging. Intracellular levels of ROS are sensed by various cellular transducers, which relay the signal to activate downstream targets and signaling pathways. Dysregulation of ROS homeostasis and ROS signaling have been linked to the development of various age-related diseases, including diabetes (27), cancer (47), and neurodegeneration (7), but their exact roles in the aging process still remain puzzling.
In recent years, many of the mechanisms regulating the production of ROS and basic principles of ROS-based redox signaling have been identified. Despite this progress, studies on the role of ROS-mediated signaling in aging lag behind, in part due to insufficient information about the specific regulatory targets of ROS. Large-scale identification of reactive Cys residues and studies on thiol oxidoreductases, enzymes that utilize catalytic redox-active Cys residues, found that an increasing number of proteins are subject to redox regulation and revealed complexity of redox-mediated signaling pathways. It would be important to identify functions of these proteins under both physiological and pathophysiological conditions. Moreover, further characterization of these pathways and mechanisms that coordinate redox signaling may provide better understanding on how dysregulation of ROS-mediated redox signaling contributes to aging and allow development of new strategies to delay the aging process in humans.
Abbreviations Used
- CR
caloric restriction
- ER
endoplasmic reticulum
- Gpx
glutathione peroxidase
- Grx
glutaredoxin
- H2O2
hydrogen peroxide
- Hsp33
heat-shock protein 33
- Msr
methionine sulfoxide reductase
- Prx
peroxiredoxin
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- Srx
sulfiredoxin
- TOR
target of rapamycin
- Trx
thioredoxin
- UPRmt
mitochondrial unfolded protein response
Acknowledgments
This work was supported by National Institutes of Health grants AG021518 and AG038004 (to V.N.G.), and AG040191 (to V.M.L.). This research was conducted while V.M.L. was an Ellison Medical Foundation/AFAR Postdoctoral Fellow.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bienert GP. Moller AL. Kristiansen KA. Schulz A. Moller IM. Schjoerring JK. Jahn TP. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem. 2007;282:1183–1192. doi: 10.1074/jbc.M603761200. [DOI] [PubMed] [Google Scholar]
- 2.Biteau B. Labarre J. Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 3.Bozonet SM. Findlay VJ. Day AM. Cameron J. Veal EA. Morgan BA. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J Biol Chem. 2005;280:23319–23327. doi: 10.1074/jbc.M502757200. [DOI] [PubMed] [Google Scholar]
- 4.Brunelle JK. Bell EL. Quesada NM. Vercauteren K. Tiranti V. Zeviani M. Scarpulla RC. Chandel NS. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Bulua AC. Simon A. Maddipati R. Pelletier M. Park H. Kim KY. Sack MN. Kastner DL. Siegel RM. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) J Exp Med. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabreiro F. Ackerman D. Doonan R. Araiz C. Back P. Papp D. Braeckman BP. Gems D. Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic Biol Med. 2011;51:1575–1582. doi: 10.1016/j.freeradbiomed.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabrese V. Sultana R. Scapagnini G. Guagliano E. Sapienza M. Bella R. Kanski J. Pennisi G. Mancuso C. Stella AM. Butterfield DA. Nitrosative stress, cellular stress response, and thiol homeostasis in patients with Alzheimer's disease. Antioxid Redox Signal. 2006;8:1975–1986. doi: 10.1089/ars.2006.8.1975. [DOI] [PubMed] [Google Scholar]
- 8.Cao J. Schulte J. Knight A. Leslie NR. Zagozdzon A. Bronson R. Manevich Y. Beeson C. Neumann CA. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. Embo J. 2009;28:1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q. Vazquez EJ. Moghaddas S. Hoppel CL. Lesnefsky EJ. Production of reactive oxygen species by mitochondria: Central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 10.Chiappetta G. Ndiaye S. Igbaria A. Kumar C. Vinh J. Toledano MB. Proteome screens for Cys residues oxidation: The redoxome. Methods Enzymol. 2010;473:199–216. doi: 10.1016/S0076-6879(10)73010-X. [DOI] [PubMed] [Google Scholar]
- 11.Conway JP. Kinter M. Dual role of peroxiredoxin I in macrophage-derived foam cells. J Biol Chem. 2006;281:27991–28001. doi: 10.1074/jbc.M605026200. [DOI] [PubMed] [Google Scholar]
- 12.D'Autreaux B. Toledano MB. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 13.David DC. Ollikainen N. Trinidad JC. Cary MP. Burlingame AL. Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day AM. Brown JD. Taylor SR. Rand JD. Morgan BA. Veal EA. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Delaunay A. Pflieger D. Barrault MB. Vinh J. Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 16.Doonan R. McElwee JJ. Matthijssens F. Walker GA. Houthoofd K. Back P. Matscheski A. Vanfleteren JR. Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durieux J. Wolff S. Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RS. Green EW. Zhao Y. van Ooijen G. Olmedo M. Qin X. Xu Y. Pan M. Valekunja UK. Feeney KA. Maywood ES. Hastings MH. Baliga NS. Merrow M. Millar AJ. Johnson CH. Kyriacou CP. O'Neill JS. Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fomenko DE. Gladyshev VN. Comparative genomics of thiol oxidoreductases reveals widespread and essential functions of thiol-based redox control of cellular processes. Antioxid Redox Signal. 2012;16:193–201. doi: 10.1089/ars.2011.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fomenko DE. Koc A. Agisheva N. Jacobsen M. Kaya A. Malinouski M. Rutherford JC. Siu KL. Jin DY. Winge DR. Gladyshev VN. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc Natl Acad Sci USA. 2011;108:2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fomenko DE. Marino SM. Gladyshev VN. Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cells. 2008;26:228–235. [PMC free article] [PubMed] [Google Scholar]
- 23.Fomenko DE. Xing W. Adair BM. Thomas DJ. Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 24.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 25.Gadjev I. Stone JM. Gechev TS. Programmed cell death in plants: New insights into redox regulation and the role of hydrogen peroxide. In: Jeon KW, editor. International Review of Cell and Molecular Biology. San Diego, CA: Academic Press; 2008. pp. 87–144. [DOI] [PubMed] [Google Scholar]
- 26.Gems D. Doonan R. Antioxidant defense and aging in C. elegans: Is the oxidative damage theory of aging wrong? Cell Cycle. 2009;8:1681–1687. doi: 10.4161/cc.8.11.8595. [DOI] [PubMed] [Google Scholar]
- 27.Gil-del Valle L. de la CML. Toledo A. Vilaro N. Tapanes R. Otero MA. Altered redox status in patients with diabetes mellitus type I. Pharmacol Res. 2005;51:375–380. doi: 10.1016/j.phrs.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Giorgio M. Migliaccio E. Orsini F. Paolucci D. Moroni M. Contursi C. Pelliccia G. Luzi L. Minucci S. Marcaccio M. Pinton P. Rizzuto R. Bernardi P. Paolucci F. Pelicci PG. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Guo Z. Kozlov S. Lavin MF. Person MD. Paull TT. ATM activation by oxidative stress. Science. 2011;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 30.Guzy RD. Hoyos B. Robin E. Chen H. Liu L. Mansfield KD. Simon MC. Hammerling U. Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Hall A. Karplus PA. Poole LB. Typical 2-Cys peroxiredoxins—Structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 33.Hidalgo E. Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 35.Ishii N. Fujii M. Hartman PS. Tsuda M. Yasuda K. Senoo-Matsuda N. Yanase S. Ayusawa D. Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- 36.Karplus PA. Poole LB. Peroxiredoxins as molecular triage agents, sacrificing themselves to enhance cell survival during a peroxide attack. Mol Cell. 2012;45:275–278. doi: 10.1016/j.molcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kayser EB. Morgan PG. Hoppel CL. Sedensky MM. Mitochondrial expression and function of GAS-1 in Caenorhabditis elegans. J Biol Chem. 2001;276:20551–20558. doi: 10.1074/jbc.M011066200. [DOI] [PubMed] [Google Scholar]
- 38.Kim EB. Fang X. Fushan AA. Huang Z. Lobanov AV. Han L. Marino SM. Sun X. Turanov AA. Yang P. Yim SH. Zhao X. Kasaikina MV. Stoletzki N. Peng C. Polak P. Xiong Z. Kiezun A. Zhu Y. Chen Y. Kryukov GV. Zhang Q. Peshkin L. Yang L. Bronson RT. Buffenstein R. Wang B. Han C. Li Q. Chen L. Zhao W. Sunyaev SR. Park TJ. Zhang G. Wang J. Gladyshev VN. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi M. Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Kwon J. Lee SR. Yang KS. Ahn Y. Kim YJ. Stadtman ER. Rhee SG. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee KS. Iijima-Ando K. Iijima K. Lee WJ. Lee JH. Yu K. Lee DS. JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends life span. J Biol Chem. 2009;284:29454–29461. doi: 10.1074/jbc.M109.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leichert LI. Gehrke F. Gudiseva HV. Blackwell T. Ilbert M. Walker AK. Strahler JR. Andrews PC. Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine RL. Moskovitz J. Stadtman ER. Oxidation of methionine in proteins: Roles in antioxidant defense and cellular regulation. IUBMB Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- 44.Lithgow GJ. Miller RA. The determination of aging rate by coordinated resistance to multiple forms of stress. In: Wallace DC, editor. The Molecular Biology of Aging. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2008. pp. 427–481. [Google Scholar]
- 45.Mansfield KD. Guzy RD. Pan Y. Young RM. Cash TP. Schumacker PT. Simon MC. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martindale JL. Holbrook NJ. Cellular response to oxidative stress: Signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 47.Mates JM. Segura JA. Alonso FJ. Marquez J. Intracellular redox status and oxidative stress: Implications for cell proliferation, apoptosis, and carcinogenesis. Arch Toxicol. 2008;82:273–299. doi: 10.1007/s00204-008-0304-z. [DOI] [PubMed] [Google Scholar]
- 48.Miller EW. Dickinson BC. Chang CJ. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc Natl Acad Sci USA. 2010;107:15681–15686. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molin M. Yang J. Hanzen S. Toledano MB. Labarre J. Nystrom T. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell. 2011;43:823–833. doi: 10.1016/j.molcel.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 50.Muller FL. Lustgarten MS. Jang Y. Richardson A. Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 51.Murphy MP. Holmgren A. Larsson NG. Halliwell B. Chang CJ. Kalyanaraman B. Rhee SG. Thornalley PJ. Partridge L. Gems D. Nystrom T. Belousov V. Schumacker PT. Winterbourn CC. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011;13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakahira K. Haspel JA. Rathinam VA. Lee SJ. Dolinay T. Lam HC. Englert JA. Rabinovitch M. Cernadas M. Kim HP. Fitzgerald KA. Ryter SW. Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nathan C. Specificity of a third kind: Reactive oxygen and nitrogen intermediates in cell signaling. J Clin Invest. 2003;111:769–778. doi: 10.1172/JCI18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okazaki S. Naganuma A. Kuge S. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid Redox Signal. 2005;7:327–334. doi: 10.1089/ars.2005.7.327. [DOI] [PubMed] [Google Scholar]
- 55.Okazaki S. Tachibana T. Naganuma A. Mano N. Kuge S. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell. 2007;27:675–688. doi: 10.1016/j.molcel.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 56.Olahova M. Taylor SR. Khazaipoul S. Wang J. Morgan BA. Matsumoto K. Blackwell TK. Veal EA. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci USA. 2008;105:19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Y. Schroeder EA. Ocampo A. Barrientos A. Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perez VI. Buffenstein R. Masamsetti V. Leonard S. Salmon AB. Mele J. Andziak B. Yang T. Edrey Y. Friguet B. Ward W. Richardson A. Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poon HF. Vaishnav RA. Getchell TV. Getchell ML. Butterfield DA. Quantitative proteomics analysis of differential protein expression and oxidative modification of specific proteins in the brains of old mice. Neurobiol Aging. 2006;27:1010–1019. doi: 10.1016/j.neurobiolaging.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Rhee SG. Kang SW. Jeong W. Chang TS. Yang KS. Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 61.Ristow M. Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 62.Ross SJ. Findlay VJ. Malakasi P. Morgan BA. Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol Biol Cell. 2000;11:2631–2642. doi: 10.1091/mbc.11.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salmeen A. Andersen JN. Myers MP. Meng TC. Hinks JA. Tonks NK. Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 64.Scherz-Shouval R. Shvets E. Fass E. Shorer H. Gil L. Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schulz TJ. Zarse K. Voigt A. Urban N. Birringer M. Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 66.Squier TC. Oxidative stress and protein aggregation during biological aging. Exp Gerontol. 2001;36:1539–1550. doi: 10.1016/s0531-5565(01)00139-5. [DOI] [PubMed] [Google Scholar]
- 67.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 68.St-Pierre J. Buckingham JA. Roebuck SJ. Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 69.Tachibana T. Okazaki S. Murayama A. Naganuma A. Nomoto A. Kuge S. A major peroxiredoxin-induced activation of Yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J Biol Chem. 2009;284:4464–4472. doi: 10.1074/jbc.M807583200. [DOI] [PubMed] [Google Scholar]
- 70.Thamsen M. Kumsta C. Li F. Jakob U. Is overoxidation of peroxiredoxin physiologically significant? Antioxid Redox Signal. 2011;14:725–730. doi: 10.1089/ars.2010.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tu BP. Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 72.Tullet JM. Hertweck M. An JH. Baker J. Hwang JY. Liu S. Oliveira RP. Baumeister R. Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Raamsdonk JM. Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Raamsdonk JM. Hekimi S. Reactive oxygen species and aging in Caenorhabditis elegans: Causal or casual relationship? Antioxid Redox Signal. 2010;13:1911–1953. doi: 10.1089/ars.2010.3215. [DOI] [PubMed] [Google Scholar]
- 75.Van Raamsdonk JM. Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci USA. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veal EA. Findlay VJ. Day AM. Bozonet SM. Evans JM. Quinn J. Morgan BA. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol Cell. 2004;15:129–139. doi: 10.1016/j.molcel.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 77.Vivancos AP. Castillo EA. Biteau B. Nicot C. Ayte J. Toledano MB. Hidalgo E. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Natl Acad Sci USA. 2005;102:8875–8880. doi: 10.1073/pnas.0503251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weerapana E. Wang C. Simon GM. Richter F. Khare S. Dillon MB. Bachovchin DA. Mowen K. Baker D. Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winterbourn CC. Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 80.Woo HA. Chae HZ. Hwang SC. Yang KS. Kang SW. Kim K. Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 81.Woo HA. Yim SH. Shin DH. Kang D. Yu DY. Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 82.Wood ZA. Poole LB. Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 83.Wu RF. Ma Z. Liu Z. Terada LS. Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol Cell Biol. 2010;30:3553–3568. doi: 10.1128/MCB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang W. Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang W. Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 86.Zhang DD. Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zuin A. Carmona M. Morales-Ivorra I. Gabrielli N. Vivancos AP. Ayte J. Hidalgo E. Lifespan extension by calorie restriction relies on the Sty1 MAP kinase stress pathway. EMBO J. 2010;29:981–991. doi: 10.1038/emboj.2009.407. [DOI] [PMC free article] [PubMed] [Google Scholar]