Abstract
CARD15/NOD2 encodes a protein involved in bacterial recognition by monocytes. Mutations in CARD15 have recently been found in patients with Crohn disease (CD), a chronic inflammatory condition of the digestive tract. Here, we report the mutational analyses of CARD15 in 453 patients with CD, including 166 sporadic and 287 familial cases, 159 patients with ulcerative colitis (UC), and 103 healthy control subjects. Of 67 sequence variations identified, 9 had an allele frequency >5% in patients with CD. Six of them were considered to be polymorphisms, and three (R702W, G908R, and 1007fs) were confirmed to be independently associated with susceptibility to CD. Also considered as potential disease-causing mutations (DCMs) were 27 rare additional mutations. The three main variants (R702W, G908R, and 1007fs) represented 32%, 18%, and 31%, respectively, of the total CD mutations, whereas the total of the 27 rare mutations represented 19% of DCMs. Altogether, 93% of the mutations were located in the distal third of the gene. No mutations were found to be associated with UC. In contrast, 50% of patients with CD carried at least one DCM, including 17% who had a double mutation. This observation confirmed the gene-dosage effect in CD. The patients with double-dose mutations were characterized by a younger age at onset (16.9 years vs. 19.8 years; P=.01), a more frequent stricturing phenotype (53% vs. 28%; P=.00003; odds ratio 2.92), and a less frequent colonic involvement (43% vs. 62%; P=.003; odds ratio 0.44) than were seen in those patients who had no mutation. The severity of the disease and extraintestinal manifestations were not different for any of the CARD15 genotypes. The proportion of familial and sporadic cases and the proportion of patients with smoking habits were similar in the groups of patients with CD with or without mutation. These findings provide tools for a DNA-based test of susceptibility and for genetic counseling in inflammatory bowel disease.
Introduction
Crohn disease (CD [MIM 266600]) and ulcerative colitis (UC [MIM 191390]) are classified as chronic idiopathic inflammatory bowel diseases (IBDs). These two disorders are usually easily differentiated. The inflammation in CD may involve any segment of the digestive tract, from the mouth to the anus, and may affect the mucosa and the deeper layers of the digestive wall. By contrast, UC lesions are continuous and are restricted to the mucosa of the large intestine, involving the rectum and a variable portion of the colon (Podolsky 1991). In the majority of cases, these characteristics permit a differential diagnosis between CD and UC in IBD, on the basis of clinical, endoscopic, and histological findings. However, in 10% of cases, the classification of the IBD remains difficult, and there is a need for additional diagnostic tools. Furthermore, even when a differential diagnosis is possible, the age at onset, the extent of the disease, extraintestinal manifestations, the natural course of the disease, and the response to treatments may define several clinical subgroups of patients. These phenotypic variations suggest several underlying etiological factors for IBD.
Despite numerous clinical and experimental studies, the etiopathogenesis of IBD has remained largely unknown. It is generally accepted that both CD and UC are multifactorial diseases caused by the interplay of genetic and environmental factors (Fiocchi 1998). Epidemiological studies have suggested that various exogenous factors, such as infectious agents and cigarette smoking, may contribute to the risk of being affected with IBD (Hugot et al. 1999). In addition to environmental factors, there is a strong genetic component in IBD, as is illustrated by the high level of concordance between identical twins combined with familial risk of IBD (for review, see Hugot et al. 1999). Linkage studies have revealed a number of putative IBD-susceptibility loci, suggesting that several genes are involved in predisposition to IBD (Hugot et al. 1996; Satsangi et al. 1996; Cho et al. 1998; Hampe et al. 1999; Ma et al. 1999; Duerr et al. 2000; Rioux et al. 2000; Vermeire et al. 2001; Cavanaugh and The IBD International Genetics Consortium 2001).
One of the genes causing susceptibility to CD was recently identified as the CARD15/NOD2 gene (Ogura et al. 2001a; Hugot et al. 2001). (The HUGO nomenclature committee recently proposed that the previous NOD2/IBD1 gene name be changed to CARD15.) CARD15 encodes a protein composed of two NH2-terminal caspase recruitment domains (CARDs), a nucleotide-binding domain (NBD), and 10 COOH-terminal leucine-rich repeats (LRR) (Ogura et al. 2001b). CARD15 is a member of the Ced4 superfamily, which includes APAF-1 (Zou et al. 1997) and NOD1/CARD4 (Bertin et al. 1999; Inohara et al. 1999). The LRRs are involved in the interaction with infecting bacterial lipopolysaccharides (LPS), whereas the CARDs enable the protein to induce apoptosis and the NF-κB signaling pathways (Inohara et al. 2001). Thus, CARD15 has been proposed to be an intracellular receptor for bacterial components in monocytes, where it is mainly expressed (Ogura et al. 2001b).
Three main mutations of CARD15, including a frameshift mutation encoding a truncated protein (1007fs), have been found in patients with CD but not in patients with UC (Hampe et al. 2001; Hugot et al. 2001; Ogura et al. 2001a). These mutations occur in the LRR domain or in its vicinity, suggesting that they alter the recognition of the bacterial LPS. This hypothesis has been supported by functional experiments performed by Ogura et al. (2001a), who demonstrated that the 1007fs mutation decreased the NF-κB activation by the LPS. In addition to the three main mutations, we were able to identify a large number of rare variants in the gene (Hugot et al. 2001).
Here, we report in detail the CARD15 mutations previously identified in series of 453 unrelated patients with CD, 159 patients with UC, and 103 healthy control subjects (Hugot et al. 2001). In addition, relationships between genotype and phenotype—including age at onset, disease location, severity of the disease, and complications—are examined in the group of patients with CD.
Subjects and Methods
Patients and Phenotype Recording
The patients in this study were recruited through a large European consortium involving clinicians from Belgium, Denmark, France, Germany, Ireland, Italy, Spain, and Sweden. Homogeneous diagnostic criteria based on clinical, endoscopic, radiological, and histological findings were used (Lennard-Jones 1989) and the same clinical questionnaire was completed for each patient. This questionnaire included: date of birth, sex, family history, age at onset, age at diagnosis, cigarette-smoking habits (smoker/ex-smoker or nonsmoker), disease location at onset, disease location at its maximal extent, granuloma, stenosis, transmural involvement, extradigestive symptoms, and therapeutic management. Stenosis and transmural involvement were defined by the occurrence, in the digestive tract, of at least one stricture or by the presence of fistula or abscess, respectively, as shown by radiological, endoscopic, or pathological examinations during the evolution of the disease. All patient data were recorded by a gastroenterologist.
A total of 453 unrelated patients with CD and 159 unrelated patients with UC were included in the study. Among patients with CD, 166 had sporadic disease, and 287 were derived from multiplex families. The group of patients with UC comprised 59 sporadic cases and 100 familial cases. The current average age at inclusion was 31.7 years (range 6–82 years) in the CD group and 34.5 years (range 6–85 years) in the UC group. Patients with indeterminate colitis (IC) were excluded, as well as patients from “mixed” families in which cases of CD and UC coexisted. As a control group, 103 unaffected white individuals, including CEPH French family members (n=26) and spouses of patients (n=77), were also analyzed. This study was approved by the relevant local ethics committees, and informed consent was obtained from each of the participants.
Strategy Outline for Mutation Analysis
The 11 constant exons and all of the intron-exon junctions of CARD15, corresponding to the previously published sequence (GenBank accession number AJ303140), were first screened by direct sequencing of DNA from 51 patients with CD, 10 patients with UC, and 103 control subjects. To increase the chance to detect sequence variations within this small group, patients with CD were chosen within families whose affected siblings were identical by descent at the IBD1 locus. A pair of primers was designed for each exon, except for exon 4, for which five overlapping fragments were amplified (table 1). Mutations were then searched for in the remaining patients. For the most polymorphic exons (number 2, 4, 6, 8, 9, and 11), the remaining patients with CD or UC were studied by direct sequencing of one of the DNA strands. In contrast, exons exhibiting little polymorphism in the initial screen (numbers 3, 5, 7, 10, and 12) were screened by a denaturing high-performance liquid chromatography (dHPLC) procedure. When a variation was observed in either the dHPLC profile or the first strand sequence, the relevant PCR product was further sequenced on both strands, to confirm the sequence variation.
Table 1.
Primers Used for Direct Sequencing and dHPLC Procedures Developed for CARD15 Screening[Note]
|
Primer |
|||
| ScreenedExon | Forward | Reverse | Size of PCR Product(bp) |
| 2 | ACCCTGCATCTGGCTTCTG | CCTTTCCTGAGAACTCTGTG | 555 |
| 3 | ACATTGCTCCATCAGCCTTC | GACTGCCCTTCCCTTTCTG | 201 |
| 4a | TGCCTCTTCTTCTGCCTTCC | AGTAGAGTCCGCACAGAGAG | 422 |
| 4b | TTTCTCTTTGTCTTCCCATTC | CCCTGTTCAGAGAAGCCC | 380 |
| 4c | GAAGTACATCCGCACCGAG | AGCCAAGAGAAATGTCATCAG | 446 |
| 4d | ATGTGCTGCTACGTGTTCTC | CAGACACCAGCGGGCACAG | 456 |
| 4e | ACCTTCAGATCACAGCAGCC | GCTCCCCCATACCTGAAC | 494 |
| 5 | TTGTCTTACTAGCTCCATTTTC | AGCCCATTGTCCACAGCC | 162 |
| 6 | CTGTTTGCATGATGGGGGG | GGGAGATCACAGCATTAGAG | 372 |
| 7 | CGTCCCGCTGCCCCTTTC | ACTCTCTCCCTGGCTTGTC | 435 |
| 7dHPLC | CCTGCCGCTGTGTTCTCTC | TCAAAAGTCCCAAGCCCCTC | 180 |
| 8 | AAGTCTGTAATGTAAAGCCAC | CCCAGCTCCTCCCTCTTC | 380 |
| 9 | CTTTCCCTGCTCTGACATAC | CCCCAGAGCAGAGAATCC | 156 |
| 10 | GCTGCAATGGAGAGTGGG | CTTTATTGGTTACCTTCACTTC | 654 |
| 10dHPLC | GTTCATCATCTTCCATAATCA | AAAGGCCAGCAATTATTGTC | 190 |
| 11 | CTCACCATTGTATCTTCTTTTC | GAATGTCAGAATCAGAAGGG | 228 |
| 12 | TAAAAACAGCCCTGACTTCC | AAACTCACAGCCTGCTCAC | 279 |
Note.— For exons 7 and 10, additional specific primers were designed for dHPLC analyses, as indicated. Exon 4 was screened by amplification of five overlapping DNA fragments.
Direct Sequencing Analysis
DNA was isolated from peripheral blood leukocytes by use of standard procedures. PCRs of CARD15 exons were performed in a 50-μl volume containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 250 μM dNTPs, a 0.50 μM concentration of each primer, 200 ng of genomic DNA, and 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Applied Biosystems). For all fragments, cycling was performed with an initial denaturation step of 10 min at 95°C to activate AmpliTaq Gold, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension step of 72°C for 10 min. Synthesis of appropriately sized PCR products was confirmed by agarose gel electrophoresis. PCR products used for sequencing analyses were purified according to a solid phase reversible immobilization (SPRI) protocol (Promega). Purified DNA products (5–10 ng) were used in a cycle-sequencing reaction using a Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems), and fragments were analyzed using an ABI 377 automated sequencer. Control subject and patient sequence data were aligned using the Sequence Navigator analysis software, version 1.0.1 (Perkin-Elmer Applied Biosystems) and were compared to the published CARD15 gene sequence. For homogeneity reasons, nucleotide numbering of CARD15 was based on the largest sequence published by Ogura et al. (2001b) (GenBank accession number AF178930), with the first base of the ATG start codon being nucleotide +1.
dHPLC Experiments
Exons 3, 5, 7, 10, and 12 were screened for variations by dHPLC (Liu et al. 1998). For exons 7 and 10, primers different from those used for DNA sequencing were chosen, to prevent intronic polymorphisms from interfering with dHPLC analysis (table 1). For all five fragments, PCR conditions were the same as described above.
Prior to dHPLC analysis, heteroduplex formation was induced by heat denaturation of PCR products at 98°C for 5 min, followed by a reannealing at 62°C for 30 min. Five microliters of these PCR products were then automatically loaded on the Hewlett-Packard ZORBAX Eclipse column (Varian Instruments) and were eluted on a linear acetonitrile gradient in a 0.1 M triethylamine acetate buffer (TEAA), pH 7.0, with a constant flow rate of 0.9 ml/min. The start and end points of the gradient were adjusted according to the size of the PCR products. Prior to sample analysis, restriction digests (pUC 18 HaeIII digest) were used at 50°C to test the dHPLC system and column performances and to give the reference by which to compare the size of DNA samples.
The denaturing run temperatures predicted by the Stanford dHPLC Melt program were tested by running a wild-type PCR product for each exon at the calculated run temperature ± 2°C. Experimental run temperatures were as follows: 59°C for exon 3; 59°C and 60°C for exon 5; 61°C and 62°C for exon 7; 57°C for exon 10; and 60°C and 61°C for exon 12.
Data were acquired using an ultraviolet detector at 260 nm (UV-Vis prostar 310, Varian Instruments), and heterozygous profiles were identified by visual inspection of the chromatograms on the basis of the appearance of superposed multiple eluting peaks. For each exon, heterozygous samples, when available, were used as internal positive controls.
Statistical Analysis
Statistical comparisons were performed using the STATVIEW version 4.51.1 package for Macintosh. χ2 tests and ANOVA were used to compare qualitative and quantitative variables between groups, respectively. Bonferroni correction for multiple comparisons was applied when necessary. A P value <.05 was considered significant. CIs for the odds ratios (OR) were calculated by the logit method. Multivariate analysis was performed using a logistic regression model to test the association between phenotype variables and genotypes.
Results
Mutational Spectrum
All coding exons of CARD15, including the intron-exon boundaries, were sequenced. In an initial screen—performed on a sample of 51 patients with CD, 10 patients with UC, and 103 healthy control subjects—we failed to detect frequent polymorphisms in exons 3, 5, 7, 10, and 12. We therefore developed a dHPLC procedure to screen these exons, whereas the remaining exons were studied by a direct sequence analysis strategy.
A total of 1,430 chromosomes from 453 patients with CD, 159 patients with UC, and 103 healthy control subjects were analyzed. We identified 67 sequence changes, corresponding to an average of two intragenic alterations per kilobase pair (table 2table 2table 2). The sequence variations were evenly distributed along the gene. However, because of the large difference in size between the exons, >50% (37/67) of these sequence changes occurred in exon 4, which contains 1,816 bp. In contrast, no variant was observed in the 70-bp coding region of exon 12.
Table 2.
CARD15 Variants Observed in 453 Patients with CD, 159 Patients with UC, and 103 Healthy Control Subjects[Note]
|
No (%) of Variant Alleles in Patient Group |
||||||
| Location andNucleotideChange | PeptideChange | PolymorphicMarkera | ProteinDomain | CD (n=906) | UC (n=318) | Control (n=206) |
| Exon 2: | ||||||
| 5′ UTR-37 T→G | Unknown | 2 | 1 | 0 | ||
| 5′ UTR-33 G→T | Unknown | 380 (42) | 79 (25) | 68 (33) | ||
| 5′ UTR-15 T→A | Unknown | 1 | 0 | 1 | ||
| 315 G→A | A105A | CARD1 | 1 | 0 | 0 | |
| 357 G→T | L119L | CARD1 | 1 | 0 | 0 | |
| 413 G→A | R138Q | CARD2 | 1 | 0 | 0 | |
| 418 G→A | A140T | CARD2 | 1 | 1 | 0 | |
| 469 T→C | W157R | CARD2 | 1 | 0 | 0 | |
| 534 C→G | S178S | CARD2 | 280 (31) | 135 (42) | 79 (38) | |
| Exon 3: | ||||||
| 566 C→T | T189M | CARD2 | 0 | 1 | 2 | |
| 633 C→T | A211A | CARD2 | 0 | 1 | 0 | |
| Exon 4: | ||||||
| 703 C→T | R235C | 1 | 0 | 0 | ||
| 743 T→G | L248R | 1 | 0 | 0 | ||
| 802 C→T | P268S | SNP5 | 375 (41) | 77 (24) | 57 (28) | |
| 866 A→G | N289S | NBD | 6 | 0 | 3 | |
| 871 G→A | D291N | NBD | 1 | 0 | 0 | |
| 881 C→G | T294S | NBD | 1 | 0 | 0 | |
| 902 C→T | A301V | NBD (P Loop) | 1 | 0 | 0 | |
| 931 C→T | R311W | NBD | 2 | 1 | 0 | |
| 1042 C→G | L348V | NBD | 1 | 0 | 0 | |
| 1055 A→G | H352R | NBD | 2 | 0 | 0 | |
| 1117 C→T | R373C | NBD | 1 | 0 | 0 | |
| 1241 A→G | N414S | NBD | 1 | 0 | 0 | |
| 1281 G→A | P427P | NBD | 2 | 0 | 0 | |
| 1292 C→T | S431L | NBD | 1 | 0 | 0 | |
| 1295 C→T | A432V | NBD | 1 | 0 | 0 | |
| 1321 G→A | E441K | NBD | 1 | 0 | 0 | |
| 1366 C→T | L456L | NBD | 0 | 0 | 1 | |
| 1377 C→T | R459R | SNP6 | NBD | 379 (42) | 76 (24) | 59 (29) |
| 1509 G→A | E503E | NBD | 0 | 1 | 0 | |
| 1581 C→G | P527P | NBD | 0 | 1 | 0 | |
| 1671 delCCTGGG | 558delLG | NBD | 1 | 0 | 0 | |
| 1761 T→G | R587R | SNP7 | 301 (33) | 142 (45) | 83 (40) | |
| 1788 G→A | T596T | 1 | 0 | 0 | ||
| 1833 C→T | A611A | 3 | 6 | 5 | ||
| 1834 G→A | A612T | 1 | 0 | 0 | ||
| 1835 C→T | A612V | 2 | 0 | 0 | ||
| 2050 C→T | R684W | 1 | 0 | 0 | ||
| 2104 C→T | R702W | SNP8 | 98 (11) | 10 (3) | 9 (4) | |
| 2107 C→T | R703C | 11 | 1 | 0 | ||
| 2137 C→T | R713C | 1 | 0 | 0 | ||
| 2174 C→G | A725G | 3 | 0 | 0 | ||
| 2220 C→T | I740I | 1 | 0 | 0 | ||
| 2264 C→T | A755V | LRR1 | 11 | 1 | 0 | |
| 2273 C→T | A758V | LRR1 | 1 | 0 | 0 | |
| 2332 G→A | E778K | LRR2 | 3 | 0 | 0 | |
| 2377 G→A | V793M | LRR2 | 4 | 0 | 0 | |
| IVS4: | ||||||
| IVS4+10 A→C | Unknown | 14 | 0 | 0 | ||
| Exon 5: | ||||||
| 2527 G→A | E843K | LRR4 | 1 | 0 | 0 | |
| Exon 6: | ||||||
| 2558 A→G | N853S | LRR5 | 2 | 0 | 0 | |
| 2587 A→G | M863V | LRR5 | 4 | 0 | 0 | |
| 2619 C→T | F873F | LRR5 | 1 | 0 | 0 | |
| IVS6: | ||||||
| IVS6+35 T→A | Unknown | 1 | 0 | 0 | ||
| IVS7: | ||||||
| IVS7-5 T→C | Unknown | 1 | 0 | 0 | ||
| Exon 7: | ||||||
| 2656 G→A | A885T | LRR6 | 0 | 1 | 0 | |
| Exon 8: | ||||||
| 2722 G→C | G908R | SNP12 | LRR6 | 55 (6) | 1 (.3) | 2 (1) |
| 2739 C→T | D913D | LRR7 | 1 | 0 | 0 | |
| 2753 C→A | A918D | LRR7 | 8 | 0 | 1 | |
| 2771 G→A | G924D | LRR7 | 1 | 0 | 0 | |
| Exon 9: | ||||||
| 2817 T→C | I939I | LRR8 | 1 | 0 | 0 | |
| 2863 G→A | V955I | LRR8 | 63 (7) | 31 (10) | 21 (10) | |
| Exon 10: | ||||||
| 2914 G→A | V972I | LRR9 | 1 | 0 | 0 | |
| 2925 C→T | L975L | LRR9 | 0 | 2 | 0 | |
| 2933 G→A | G978E | LRR9 | 1 | 0 | 0 | |
| Exon 11: | ||||||
| 3020insC | 1007fs | SNP13 | LRR10 | 96 (11) | 4 (1) | 4 (2) |
| IVS12: | ||||||
| IVS12-102 C→G | Unknown | 1 | 0 | 0 | ||
| Exon 12: | ||||||
| 3′ UTR +11 G→A | Unknown | 1 | 0 | 0 | ||
Note.— Variations were denoted according to the largest sequence reported by Ogura et al. (2001b). The A of the ATG of the initiator Met codon was denoted as “nucleotide +1.” Probable DCMs are indicated in boldface italics.
The correspondence with the polymorphic sites described elsewhere (Hugot et al. 2001) is indicated.
Table 2.
CARD15 Variants Observed in 453 Patients with CD, 159 Patients with UC, and 103 Healthy Control Subjects
|
No (%) of Variant Alleles in Patient Group |
||||||
| Location andNucleotideChange | PeptideChange | PolymorphicMarkera | ProteinDomain | CD (n=906) | UC (n=318) | Control (n=206) |
| Exon 2: | ||||||
| 5′ UTR-37 T→G | Unknown | 2 | 1 | 0 | ||
| 5′ UTR-33 G→T | Unknown | 380 (42) | 79 (25) | 68 (33) | ||
| 5′ UTR-15 T→A | Unknown | 1 | 0 | 1 | ||
| 315 G→A | A105A | CARD1 | 1 | 0 | 0 | |
| 357 G→T | L119L | CARD1 | 1 | 0 | 0 | |
| 413 G→A | R138Q | CARD2 | 1 | 0 | 0 | |
| 418 G→A | A140T | CARD2 | 1 | 1 | 0 | |
| 469 T→C | W157R | CARD2 | 1 | 0 | 0 | |
| 534 C→G | S178S | CARD2 | 280 (31) | 135 (42) | 79 (38) | |
| Exon 3: | ||||||
| 566 C→T | T189M | CARD2 | 0 | 1 | 2 | |
| 633 C→T | A211A | CARD2 | 0 | 1 | 0 | |
| Exon 4: | ||||||
| 703 C→T | R235C | 1 | 0 | 0 | ||
| 743 T→G | L248R | 1 | 0 | 0 | ||
| 802 C→T | P268S | SNP5 | 375 (41) | 77 (24) | 57 (28) | |
| 866 A→G | N289S | NBD | 6 | 0 | 3 | |
| 871 G→A | D291N | NBD | 1 | 0 | 0 | |
| 881 C→G | T294S | NBD | 1 | 0 | 0 | |
| 902 C→T | A301V | NBD (P Loop) | 1 | 0 | 0 | |
| 931 C→T | R311W | NBD | 2 | 1 | 0 | |
| 1042 C→G | L348V | NBD | 1 | 0 | 0 | |
| 1055 A→G | H352R | NBD | 2 | 0 | 0 | |
| 1117 C→T | R373C | NBD | 1 | 0 | 0 | |
| 1241 A→G | N414S | NBD | 1 | 0 | 0 | |
| 1281 G→A | P427P | NBD | 2 | 0 | 0 | |
| 1292 C→T | S431L | NBD | 1 | 0 | 0 | |
| 1295 C→T | A432V | NBD | 1 | 0 | 0 | |
| 1321 G→A | E441K | NBD | 1 | 0 | 0 | |
| 1366 C→T | L456L | NBD | 0 | 0 | 1 | |
| 1377 C→T | R459R | SNP6 | NBD | 379 (42) | 76 (24) | 59 (29) |
| 1509 G→A | E503E | NBD | 0 | 1 | 0 | |
| 1581 C→G | P527P | NBD | 0 | 1 | 0 | |
| 1671 delCCTGGG | 558delLG | NBD | 1 | 0 | 0 | |
| 1761 T→G | R587R | SNP7 | 301 (33) | 142 (45) | 83 (40) | |
| 1788 G→A | T596T | 1 | 0 | 0 | ||
| 1833 C→T | A611A | 3 | 6 | 5 | ||
| 1834 G→A | A612T | 1 | 0 | 0 | ||
| 1835 C→T | A612V | 2 | 0 | 0 | ||
| 2050 C→T | R684W | 1 | 0 | 0 | ||
| 2104 C→T | R702W | SNP8 | 98 (11) | 10 (3) | 9 (4) | |
| 2107 C→T | R703C | 11 | 1 | 0 | ||
| 2137 C→T | R713C | 1 | 0 | 0 | ||
| 2174 C→G | A725G | 3 | 0 | 0 | ||
| 2220 C→T | I740I | 1 | 0 | 0 | ||
| 2264 C→T | A755V | LRR1 | 11 | 1 | 0 | |
| 2273 C→T | A758V | LRR1 | 1 | 0 | 0 | |
| 2332 G→A | E778K | LRR2 | 3 | 0 | 0 | |
| 2377 G→A | V793M | LRR2 | 4 | 0 | 0 | |
| IVS4: | ||||||
| IVS4+10 A→C | Unknown | 14 | 0 | 0 | ||
| Exon 5: | ||||||
| 2527 G→A | E843K | LRR4 | 1 | 0 | 0 | |
| Exon 6: | ||||||
| 2558 A→G | N853S | LRR5 | 2 | 0 | 0 | |
| 2587 A→G | M863V | LRR5 | 4 | 0 | 0 | |
| 2619 C→T | F873F | LRR5 | 1 | 0 | 0 | |
| (continued) | ||||||
Table 2 (continued).
|
No (%) of Variant Alleles in Patient Group |
||||||
| Location andNucleotideChange | PeptideChange | PolymorphicMarkera | ProteinDomain | CD (n=906) | UC (n=318) | Control (n=206) |
| IVS6: | ||||||
| IVS6+35 T→A | Unknown | 1 | 0 | 0 | ||
| IVS7: | ||||||
| IVS7-5 T→C | Unknown | 1 | 0 | 0 | ||
| Exon 7: | ||||||
| 2656 G→A | A885T | LRR6 | 0 | 1 | 0 | |
| Exon 8: | ||||||
| 2722 G→C | G908R | SNP12 | LRR6 | 55 (6) | 1 (.3) | 2 (1) |
| 2739 C→T | D913D | LRR7 | 1 | 0 | 0 | |
| 2753 C→A | A918D | LRR7 | 8 | 0 | 1 | |
| 2771 G→A | G924D | LRR7 | 1 | 0 | 0 | |
| Exon 9: | ||||||
| 2817 T→C | I939I | LRR8 | 1 | 0 | 0 | |
| 2863 G→A | V955I | LRR8 | 63 (7) | 31 (10) | 21 (10) | |
| Exon 10: | ||||||
| 2914 G→A | V972I | LRR9 | 1 | 0 | 0 | |
| 2925 C→T | L975L | LRR9 | 0 | 2 | 0 | |
| 2933 G→A | G978E | LRR9 | 1 | 0 | 0 | |
| Exon 11: | ||||||
| 3020insC | 1007fs | SNP13 | LRR10 | 96 (11) | 4 (1) | 4 (2) |
| IVS12: | ||||||
| IVS12-102 C→G | Unknown | 1 | 0 | 0 | ||
| Exon 12: | ||||||
| 3′ UTR +11 G→A | Unknown | 1 | 0 | 0 | ||
Note.— Variations were denoted according to the largest sequence reported by Ogura et al. (2001b). The A of the ATG of the initiator Met codon was denoted as “nucleotide +1.” Probable DCMs are indicated in boldface italics.
The correspondence with the polymorphic sites described elsewhere (Hugot et al. 2001) is indicated.
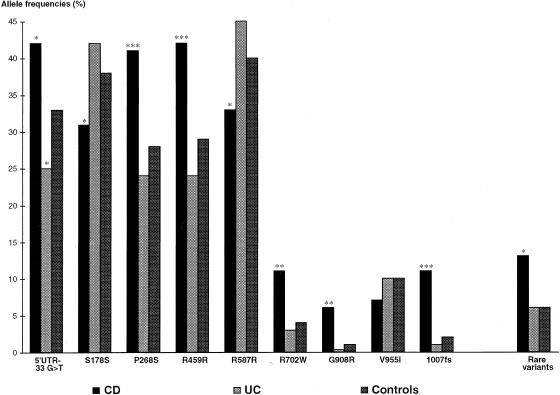
Among the 67 sequence alterations, 8 were detected in the noncoding regions of the gene (3′ and 5′ UTRs and intervening sequences; table 2table 2). Among these variants, the 5′ UTR-33 G→T alteration appeared to be more frequent in CD than in control subjects (P=.02; fig. 1) and the IVS4+10 A→C intronic change was observed in patients with CD only (n=14).
Figure 1.
Distribution of the nine most frequent variants and the group of rare sequence variants observed in CARD15 gene in CD, UC, and control populations. Allele frequencies of these variants in CD and UC populations were compared with those in the control group, and the level of significance (P value) is denoted here by asterisks: *, P<.05; **, P<.005; ***, P<.0005.
Except for the insertion frameshift mutation (1007fs) and the 6-bp deletion (558delLG), the remaining variants (57/59) resulted in a single-base-pair substitution (table 2table 2). Of these, 47 were due to transition-type changes, specifically the C→T transition (n=23). Of the 57 point mutations, 43 (75%) affected a CpG dinucleotide, whereas the average GC content was 54.8% (range 42.9%–64.3%) for the whole cDNA.
The definition of a “disease-causing” mutation (DCM) is problematic when functional assays to determine the phenotypic effects of specific variants have not been performed. To date, only the 1007fs variant has been shown to be defective in activating the NF-κB pathway (Ogura et al. 2001a). For the purposes of this study, we considered that CARD15 sequence alterations that led to a predicted premature truncation of the protein, small deletions or insertions and nonconservative missense mutations were considered as potential DCMs (n=31) (table 2table 2). Sequence alterations occurring in the noncoding sequence (n=8) and silent (n=17) or conservative (n=11) mutations were not considered pathological (table 2table 2).
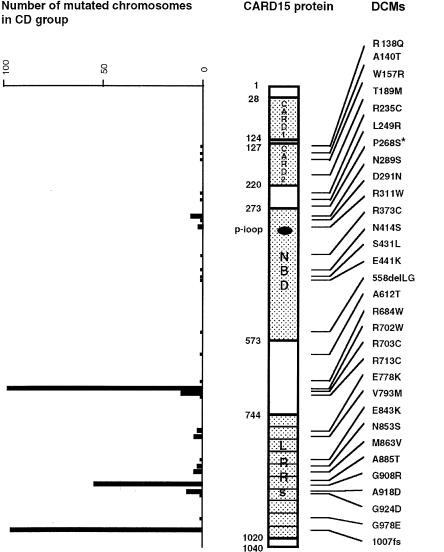
CARD15 encodes a protein that contains two NH2-terminal CARD domains, a centrally located NBD, and 10 COOH-terminal LRRs. Figure 2 shows the distribution of DCMs along the protein and the number of each identified DCM in the CD group. In the first CARD, no DCM was observed. In the second CARD, four DCMs (R138Q, A140T, W157R, and T189M) were identified, which may affect the putative α helices H1 (R138Q and A140T), H2 (W157R), and H4 (T189M). In the NBD, we observed eight DCMs and the conservative mutation (A301V), which is located in the P-loop box and thus may affect the function of the gene. Two frequent DCMs (R702W and R703C) were located between the NBD and the LRR domains. Finally, 11 DCMs were seen in the LRR domains, including the 2 frequent mutations, G908R and 1007fs. Along the protein, the proportion of DCMs per amino acid was 2% in the CARD domains, 2.7% in the NBD, and 4% in the LRRs (not significant). However, 93% of the mutated chromosomes carried a DCM located in the distal third of the gene (fig. 2).
Figure 2.
Distribution of the DCMs along the CARD15/NOD2 protein and number of mutated chromosomes observed among the patients with CD. The asterisk (*) denotes that the P268S mutation was not considered, because of its tight linkage disequilibrium with other variants (see text).
Distribution of CARD15 Variants in Patients with CD, Patients with UC, and Control Subjects
Nine sequence changes had an allele frequency of >5% in the CD group: 5′ UTR-33 G→T, S178S, P268S, R459R, R587R, R702W, G908R, V955I, and 1007fs (table 2table 2). Figure 1 shows the frequencies of these nine sequence changes in patients with CD, patients with UC, and control subjects. No difference in allele frequency was observed between UC and control subjects, except for the 5′ UTR-33 G→T variant (P<.05). However, this difference was not significant after the multiple comparisons were corrected for. In contrast, eight of the nine variants had a significantly higher frequency in the CD group compared with the control subjects, a result that could be due to linkage disequilibrium between the alleles and CD.
Of these nine polymorphisms, only four can be considered DCMs according to the proposed definition (see above). These sequence alterations were previously reported as SNP5 (P268S), SNP8 (R702W), SNP12 (G908R), and SNP13 (1007fs) (dbSNP accession numbers ss2978533, ss2978536, ss2978537, and ss2978539, respectively) (Hugot et al. 2001). As previously reported, the mutations R702W, G908R, and 1007fs were independently associated with the CD phenotype (P=.005, P=.003, and P=.00009, respectively).
The allele frequencies of the R702W, G908R, and 1007fs variants were, respectively, 0.09, 0.07, and 0.11 in cases of familial CD. In sporadic cases, the corresponding frequencies were 0.14, 0.04, and 0.09. Considering that these three main mutations are independent events (Hugot et al. 2001), the total frequency of the mutated chromosomes is equal to the sum of the three mutation frequencies. This sum (0.27) was identical in familial and sporadic cases, suggesting that the CARD15 mutations have the same prevalence in sporadic and familial cases. Similarly, the frequencies of the three main variants were identical in sporadic and familial cases of UC (data not shown).
The P268S variant was also associated with CD (P=.0003). However, previous studies demonstrated that this P268S variant occurs in phase with the R702W, G908R, and 1007fs mutations (Hugot et al. 2001) and that the association was related to this linkage disequilibrium. For this reason, we considered this variant further as a polymorphism. Similarly, the silent mutations S178S, R459R, and R587R and the 5′ UTR-33 G→T variant were found to be in linkage disequilibrium with the three main variants and were not considered further in the genotype-phenotype analyses. Lastly, the conservative variant V955I was not associated with CD.
Rare sequence variants were observed in patients with CD, patients with UC, and healthy control subjects (table 2table 2). These variations were more frequently encountered in CD (13% of CD chromosomes) than in control subjects (6% of the control chromosomes) (P<.01). This association was independent of the three main DCMs and the P268S variant, suggesting that at least some of these rare variants are involved in the disease predisposition (data not shown). However, for each variant, the number of patients carrying the alternative allele was too small to allow statistical calculations (table 2table 2). In contrast, the rare sequence variations were not associated with the UC phenotype (6% of the UC chromosomes; P=.76) (fig. 1).
In the study population, 229 patients with CD (51%) carried no DCMs in CARD15, 145 patients (32%) carried one DCM, and 79 patients (17%) had two mutated alleles, including 3 patients carrying three mutations. Previous studies have shown that these mutations generally occur on separate chromosomes, so that individuals in the latter group were further considered to have mutations on both homologous chromosomes (Hugot et al. 2001). This distribution was quite different from that observed in UC and control subjects, showing a large excess of individuals carrying two mutations among patients with CD (P<.0001) (table 3). It suggests a higher penetrance for susceptibility to CD among individuals with both mutated chromosomes. The small excess of CD patients carrying a single mutation shows that heterozygous individuals have a less marked risk of developing the disease.
Table 3.
Number of Different Genotypes Observed in CD/UC/Control Groups[Note]
|
No. of Genotypes Observed in CD/UC/Control Groups |
|||||
| Variant | R702W | G908R | 1007fs | Rare DCMs | Wild Type |
| R702W | 11/1/0 | 10/0/0 | 15/0/0 | 8/0/0 | 43/8/9 |
| G908R | 2/0/0 | 11/0/0 | 2/0/0 | 28/1/2 | |
| 1007fs | 11/0/0 | 7/0/0 | 40/4/4 | ||
| Rare DCMs | 2/0/0 | 34/5/6 | |||
| Wild type | 229/141/82 | ||||
Note.— Each cell of the table corresponds to the genotype combining the two genetic variants indicated in the corresponding row and column.
Genotype-Phenotype Correlations
We further examined the clinical manifestations in patients with CD according to their genotype. Because no major difference was noted in either the clinical presentation (data not shown) or allele frequencies (see above) between sporadic and familial CD groups, all the CD cases (sporadic or familial) were pooled for the genotype-phenotype analyses.
No phenotypic difference was observed between patients who were either (1) homozygous for one or (2) compound heterozygous for any of the R702W, G908R, and 1007fs mutations, suggesting that the respective roles of these mutations are comparable (data not shown). For DCMs, each private mutation was too rare to allow statistical analyses. For these reasons, we pooled frequent and rare mutations considered to be causative of CD (i.e., the 31 above-mentioned DCMs, except for the P268S variant; see above and table 2table 2), and we divided the 453 patients with CD into three distinct groups defined by the number of DCMs carried (table 4). The three groups correspond to normal homozygotes (G0), heterozygotes for a normal and mutated sequence (G1), and individuals with mutations in CARD15 on both homologous chromosomes (G2), respectively. The female/male ratio was slightly higher in G1 than in G0 (P=.05 after Bonferroni correction; not significant) and the average current age at inclusion was slightly lower in G2 than in G0 (29.1 years vs. 32.5 years; P=.04 after Bonferroni correction; not significant). This second observation may be explained by a recruitment bias related to the younger age at onset in G2 (see below). The percentages of familial and sporadic cases were identical in the three groups.
Table 4.
Genotype-Phenotype Relationships in Patients with CD[Note]
|
Value for No. of DCMs |
Comparison (P) |
||||||
| Detailed Phenotype | All (n = 453) | 0 (n=229) | 1 (n=145) | 2 (n=79) | 0 vs. 1 | 1 vs. 2 | 0 vs. 2 |
| Sex (M/F) | 45/55 | 48/52 | 38/62 | 49/51 | .05 | NS | NS |
| Age at inclusion (years) | 31.7 (6–82) | 32.5 (7–82) | 32.0 (6–65) | 29.1 (12–61) | NS | NS | .04 |
| Familial/sporadic | 63/37 | 60/40 | 68/32 | 64/36 | NS | NS | NS |
| Age at onset (years) | 19.4 (2–56) | 19.8 (3–56) | 20.0 (2–45) | 16.9 (4–40) | NS | .02 | .01 |
| Age at diagnosis (years) | 20.7 (3–61) | 21.3 (3–61) | 21.0 (3–54) | 18.4 (5–42) | NS | .05 | .03 |
| Transmural involvement | 41 | 38 | 44 | 46 | NS | NS | NS |
| Stenosis | 37 | 28 | 43 | 53 | .002 | NS | .00003a |
| Granuloma | 51 | 46 | 52 | 59 | NS | NS | .04 |
| Location at onset: | |||||||
| Ileon/right colon siteb | 87 | 86 | 87 | 92 | NS | NS | NS |
| Colorectal sitec | 59 | 62 | 61 | 43 | NS | .008 | .003 |
| Upper digestive tractd | 23 | 23 | 24 | 23 | NS | NS | NS |
| Perineal lesions | 31 | 28 | 35 | 30 | NS | NS | NS |
| Maximal extent: | |||||||
| Ileon/right colon siteb | 74 | 73 | 72 | 77 | NS | NS | NS |
| Colorectal sitec | 50 | 51 | 55 | 38 | NS | .01 | .04 |
| Upper digestive tractd | 19 | 19 | 20 | 20 | NS | NS | NS |
| Perineal lesions | 29 | 28 | 32 | 27 | NS | NS | NS |
| Extradigestive symptoms | 34 | 32 | 39 | 29 | NS | NS | NS |
| Eye | 9 | 7 | 12 | 5 | NS | NS | NS |
| Skin | 12 | 12 | 12 | 15 | NS | NS | NS |
| Joints | 25 | 23 | 30 | 22 | NS | NS | NS |
| Liver | 2 | 3 | 1 | 1 | NS | NS | NS |
| Medical management:e | |||||||
| Steroids | 85 | 90 | 81 | 81 | .01 | NS | .04 |
| Azathioprine/6-mercaptopurine | 41 | 46 | 36 | 36 | NS | NS | NS |
| Nutritional supportf | 39 | 36 | 40 | 43 | NS | NS | NS |
| Surgery | 55 | 55 | 52 | 63 | NS | NS | NS |
| Other immunosuppressor/anti-TNF antibodies | 7 | 6 | 8 | 6 | NS | NS | NS |
| Nonsmoker/smoker/ex-smoker | 60/27/13 | 60/26/14 | 58/30/11 | 63/27/11 | NS | NS | NS |
Note.— Patients with CD were categorized according to the number of DCMs they carried (0, 1, or 2), and the phenotypes were compared between groups. Quantitative traits are reported as mean (range). Qualitative traits are reported as % of patients. Significant P values of the tests are indicated.
P<.005 after Bonferroni correction for the multiple comparisons (n=78).
The ileocolonic site was defined by the involvement of the terminal ileum and/or the appendix and/or the right colon.
Colonic site was defined by the involvement of the transverse colon and/or the left colon and/or the rectum.
The upper digestive tract was defined by the involvement of at least one of the following sites: esophagus, stomach, duodenum, jejunum, and proximal ileum.
For medical management, patients were classified as positive if the corresponding treatment was used at least once during the evolution of the disease.
Nutritional support includes enteral and parenteral nutrition.
Patients carrying one or more CARD15 DCMs were characterized by the development of stenoses during the evolution of their disease, particularly in G2 patients, presumed to have mutations on both chromosomes (G2 vs. G0: 53% vs. 28%; P=.00003; OR 2.92; 95% CI 1.71–4.96). These comparisons remained significant after the Bonferroni correction for the multiple comparisons made (n=78). Also in G2, the involvement of the transverse colon, left colon, or rectum was significantly less common at the onset of the disease (G2 vs. G0: 43% vs. 62%; P=.003; OR 0.44; 95% CI 0.26–0.74) and during evolution (G2 vs. G0: 38% vs. 51%; P=.04; OR 0.57; 95% CI 0.33–0.96). Finally, the age at onset was, on average, slightly lower in G2 than in the two other groups (G2 vs. G0: 16.9 vs. 19.8; P=.01; and G2 vs. G1: 16.9 vs. 20.0; P=.02). As expected, concordant findings were observed regarding the age at diagnosis. It should be noted that the later reported associations were no longer significant after Bonferroni correction (n=78). Extraintestinal manifestations—including eye, skin, joint, and liver involvement—occurred at similar rates within the three groups.
Because some of the phenotypic criteria studied here may be linked (Polito et al. 1996), the relationships between all the phenotype variables and the genotype were explored using a stepwise logistic regression. Three explanatory variables remained independently associated with having two mutations, the presence of stenosis (P=.0007), the age at diagnosis (P=.0045), and the colorectal site as location at onset (P=.0094). After these three variables were taken into account, the development of granuloma also remained associated with the presence of two mutations, but the statistical test was of borderline significance (P=.07). No other variable adds information.
Regarding medical treatments, which were assumed to be related to the severity of the disease, there was a lower frequency of patients who had been treated at least once with steroids among those bearing one or more mutations when compared with patients having no mutation (G1 vs. G0: 81% vs. 90%; P=.01; G2 vs. G0: 81% vs. 90%; P=.04; OR 0.47; 95% CI 0.23–0.95). Surgical interventions tended to be more common in group 2, but this did not achieve statistical significance (G2 vs. G1: 63% vs. 52%; P=.13). However, no significant difference between groups was observed for the other treatments and for the cumulative number of medications used (data not shown). Finally, differences in smoking habits between the three groups were not statistically significant.
Because it is difficult to infer that the rare variants play a role in CD predisposition without any functional studies, we also performed the analyses after excluding patients carrying a private mutation. No major differences were observed although thresholds of significance were lower for the stricturing phenotype (G2 vs. G0: 57% vs. 28%; P=.00002; G1 vs. G0: 47% vs. 28%; P=.0004), the colorectal localization of the disease at onset (G2 vs. G0: 36% vs. 62%; P=.0003; G2 vs. G1: 36% vs. 59%; P=.006). These findings may suggest that some of the private mutations have a smaller biological effect than the three main variants.
A Genotyping Method for R702W, G908R, and 1007fs Variants
A screening method for the presence of the recurrent R702W, G908R, and 1007fs variants was developed in the laboratory. A brief description of the method is given below and in table 5, and more detailed explanations are available from the authors. The R702W mutation is detected by an allele-specific PCR assay. The mutated and wild-type alleles are independently amplified by use of two different sets of primers. A nonpolymorphic fragment is coamplified in the same tube as positive control of DNA amplification. The G908R mutation is detected by a restriction enzyme digestion of PCR-amplified DNA, the sequence variation creating a novel Hha1 restriction enzyme site. Finally, the 1-bp insertion variant (1007fs) genotyping is based on sizing of a labeled PCR product after electrophoresis in an acrylamide gel. Our method is based on fluorescently labeled primers and automated sequencers. These genotyping methods have proved, in our experience, to be robust, easy to perform, and cost effective (data not shown).
Table 5.
Genotyping Procedures for the R702W, G908R, and 1007fs CD-Associated Variants
| Mutation andGenotyping Method | Specific Primers | Size of the PCR Product(bp) |
| R702W; allele-specifica | SNP-specific primers: forward CCCACACTTAGCCTTGATG, reverse wild type ATCTGAGAAGGCCCTGCTCT, reverse mutated ATCTGAGAAGGCCCTGCTCC; constant primers: forward TGAGGCAAAACAACTGAGAC, reverse GCAGACATTGATTTTACACAG | 439 (specific); 102 (control) |
| G908R; PCR-RFLPb | Forward CCCAGCTCCTCCCTCTTC, reverse AAGTCTGTAATGTAAAGCCAC | Wild type: 380; mutated: 138+242 |
| 1007fs; PCR-product sizingc | Forward Fam-GAATGTCAGAATCAGAAGGG, reverse GCTCACCATTGTATCTTCTTTTC | Wild type: 230; mutated: 231 |
PCR was performed using 40 mM of each specific primer and 20 mM of each constant primer at an annealing tempertature of 58°C. The PCR product was then loaded onto a 2% agarose gel.
PCR products (annealing temperature 55°C) were digested by HhaI enzyme and were loaded onto a 2% agarose gel.
The fluorescently labeled PCR products (annealing temperature 55°C) were loaded onto a polyacrylamide gel and were run on a sequencer.
Discussion
The recent identification of CARD15 as a gene involved in susceptibility to CD (Hampe et al. 2001; Hugot et al. 2001; Ogura et al. 2001a) provides an opportunity to study the relationships between CARD15 genotype and phenotype in IBD. To address this question, we have performed a comprehensive mutational analysis of CARD15 in a European sample of 453 patients with CD, 159 individuals with UC, and 103 healthy control subjects.
Ogura et al. (2001b) reported two probable CARD15 isoforms, NOD2a and NOD2b, which are composed of 12 and 11 exons, respectively. The first alternative exon encodes a small 27–amino acid peptide without any known functional domain. We have not observed the NOD2a isoform in our studies (Hugot et al. 2001). The cDNA that we have proposed elsewhere (GenBank accession number AJ303140; Hugot et al. 2001) contains a larger 5′ UTR than the sequence of Ogura et al. (2001b). We have screened this 5′ UTR, the 11 common exons, and the intron-exon boundaries of the gene. However, to avoid heterogeneous annotations of mutations, our observations are reported using the nomenclature adopted by Ogura et al. (2001b) and, more recently, by Hampe at al. (2001), who also used the NOD2a isoform as the sequence reference (GenBank accession number AF178930; Ogura et al. 2001b).
The mutation-screening method used in the present study was not able to detect large DNA rearrangements, nor did the analysis include the promoter or other potential regulatory regions of the gene, all of which may influence the expression of the gene. Despite these limitations, we found a total of 67 different nucleotide alterations in the affected individuals and control subjects investigated. Of these alterations, 31 were classified as probable pathogenic mutations, including 29 missense mutations; a 1-bp insertion, leading to a frameshift that resulted in a truncated protein missing the last 33 amino acids; and an in-frame deletion of 6 bp. Of the 29 observed substitutions, 23 were C→T transitions found in CpG islands, which are known to be hotspots for mutations (Cooper et al. 1995).
The other sequence alterations included eight changes occurring in noncoding regions. Among these variants, the 5′ UTR-33 G→T alteration appeared to be more frequent in patients with CD than in control subjects (P=.02) and the IVS4+10 A→C intronic change was observed in patients with CD only (n=14). These findings suggest that these two variants may play a role in the CD predisposition. However, the consequences, at the RNA level, of these variations were not examined. Eleven conservative amino acid substitutions were also identified, and functional assays will be required to establish or discard their role in CD pathogenesis. Finally, 17 additional silent variations were found.
Most of the 67 CARD15 variants identified so far are rare. However, nine of them exhibited an allele frequency >0.05 in the group of patients with CD (5′ UTR-33 G→T, S178S, P268S, R459R, R587R, R702W, G908R, V955I, and 1007fs). Of these nine variants, we confirmed that R702W, G908R, and the frameshift mutation 1007fs (elsewhere referred to as “R675W,” “G881R,” and “980fs” [Hugot et al. 2001]) are significantly more frequent among patients with CD (allele frequencies of 11%, 6%, and 11%, respectively) than in healthy control subjects (4%, 1%, and 2%; P=.005, P=.003, and P=.00009, respectively). Similar results have been obtained for the 1007fs variant by other investigators (Hampe et al. 2001; Ogura et al. 2001a). The other frequent variations were found to be in linkage disequilibrium with alleles in this group, and we considered them to be polymorphisms. That assumption is even stronger for the P268S variation, which was not significantly associated with the disease when tested alone (Hugot et al. 2001). However, functional studies will be required before definitively ruling out a role for the nonsilent variations P268S, V955I, and the 5′ UTR-33 G→T variant.
The group of the 58 rare variants was also associated with the CD phenotype (P<.01), suggesting that at least some of them play a role in predisposition to CD. Twenty-seven of them encoded either nonconservative amino acid substitutions or a small deletion, including 20 mutations that were found exclusively or preferentially in patients with CD, which also supports an association with the CD phenotype. Because the number of patients carrying each mutation was too small, this question could not be resolved using statistical tests. On the other hand, it is important to note that stronger genotype-phenotype correlations were observed when the patients with CD carrying one or more private mutations were discarded from the analyses. The difference suggests that some of these private mutations may have a smaller biological effect than the three main variants. Functional studies are now required to further investigate this question.
CARD15 variations identified so far are evenly distributed along the entire coding sequence except in its 5′ portion encoding the first CARD domain. However, the mutations located in the third distal of the coding sequence are overrepresented in patients with CD (93%) and correspond to the vast majority of disease-associated mutations encountered so far. This finding suggests that the LRRs, which are known to interact with bacterial components, such as lipopolysaccharides (LPS), and their adjacent region have an important role in CD susceptibility. A reasonable hypothesis is that the mutations associated with CD disrupt proper NF-κB or apoptotic signaling, resulting in an inappropriate monocyte response to bacterial infection.
The present results confirm the gene-dosage effect reported elsewhere for the 1007fs (Hampe et al. 2001; Ogura et al. 2001a) and the R702W and G908R mutations (Hugot et al. 2001). These observations are now extended to include the rare DCMs, and they demonstrate that ∼50% of patients with CD carry at least one mutation (vs. 20% in control subjects). Of these, 17% (vs. 0% in control subjects) carry two mutations, which are assumed to affect separate homologues. The latter proportion is significantly higher than would be expected by chance (P<.003). This result suggests that a complete disruption of the NOD2-signaling pathway may be necessary, in many cases, for development of the disease.
No association was observed between any of the CARD15 variants and the UC phenotype. As previously mentioned, UC was not associated with the three main CD-associated variations R702W, G908R, and 1007fs. However, we also failed to identify any additional mutations specifically involved in predisposition to UC, and the overall rare variations were as frequent in the group of patients with UC as in the group of healthy control subjects (P=.76). These observations strongly suggest that the role of CARD15 in UC is minimal or nonexistent. This is consistent with previous reports of no linkage of the pericentromeric region of chromosome 16 to UC (Cavanaugh and The IBD International Genetics Consortium 2001).
The detailed analyses of the genotype/phenotype relationships did not allow us to find any specific group of clinical features that were associated with particular CARD15 mutations (data not shown). The effects of the vast majority of DCMs seem to be roughly comparable. To see whether the presence of one or more mutated copies of the CARD15 gene was associated with a specific phenotypic outcome, we categorized patients with CD into three classes, according to the number of carried mutations. Patients bearing one or more mutations developed significantly more stenoses of the digestive tract. However, on the whole, patients bearing only one mutation exhibited few other differences from those bearing none of the identified mutations.
On the other hand, patients carrying two mutations were characterized by a stricturing phenotype, a younger age at onset, a less frequent involvement of the left colon and rectum, and a more frequent formation of granulomas. These variables were found to be independently associated with having two CARD15 mutations. In other words and from the clinical point of view, the association of a young age at onset, a purely ileal involvement, and a stricturing phenotype characterizes patients with a double-dose mutation. In contrast, this complete phenotypic association is observed less often in patients with no or one mutation. Interestingly, these findings suggest that patients having two mutations resemble those initially reported by Crohn et al. (1932), who defined the disease as a “regional enteritis” phenotype.
These phenotype-genotype correlations confirm the gene-dosage effect at the subphenotype level. The younger age at onset is in accordance with the previously reported higher linkage score on chromosome 16 when patients diagnosed at an earlier age are analyzed (Brant et al. 2000). However, the most significant result of these genotype-phenotype analyses is that patients with CARD15 mutations have a stricturing phenotype more frequently than do patients carrying no mutations. This is particularly true for patients with mutations on both chromosomes, who have a threefold higher risk of developing a stenosis than do patients with no mutations. This difference may be useful in clinical classification and management of the patients. However, prospective clinical studies are now needed to confirm the prognostic value of CARD15 genotyping. No difference was observed among the three genotype groups for the severity of the disease (as estimated by the need for medication and surgery) or in the extradigestive manifestations.
The allele frequency of the three main variants is comparable in familial and sporadic cases, and the proportion of familial to sporadic cases is identical in the three groups of patients defined by the number of carried mutations. Therefore, mutations in CARD15 could not be invoked to explain the small clinical differences observed between these two categories of patients (Colombel et al. 1996; Lee and Lennard-Jones 1996; Polito et al. 1996). Finally, smoking habits were similar between groups, suggesting that additional independent genetic factors may interact with this environmental risk factor (Tobin et al. 1987; Cosnes et al. 1996).
This study also proposes a practical approach for CARD15 genotyping. Confirming that the variants R702W, G908R, and 1007fs are the most important ones for the phenotype classification and that they represent 81% of the mutated alleles, we propose that initial genotyping efforts should be focused on these three polymorphisms. A rapid and efficient procedure is reported here, based respectively on allele-specific PCR, PCR-RFLP, and PCR product–sizing methods. This procedure is easily implemented in laboratories involved in molecular biology. In case of a negative result, this study also provides the basis for the detection of rarer mutations, suggesting that the ends of exon 4, exon 8, and exon 6—which carry the other most frequent mutations—should be screened first.
In addition to endoscopic, radiological, and histological analyses, CARD15 genotyping may represent an additional tool for the clinician in his (her) clinical practice and research. This study helps to define the limits of the usefulness of CARD15 genotyping. However, additional clinical studies are now necessary for determination of which clinical situations CARD15-based tests may be the most relevant and cost effective for patients and researchers.
Acknowledgments
We are grateful to the patients and their families for their participation in this study. Drs. J. Balanzo, B. Bonaz, Y. Bouhnik, G. Cadiot, A. Cortot, S. Cucchiara, B. Crusius, J. J. Delchier, B. Duclos, J. L. Dupas, J. P. Galmiche, J. P. Gendre, D. Golfain, C. Grännö, D. Heresbach, A. Lachaux, H. Lautraite, C. Lenaerts, E. Lerebours, V. Levy, R. Löfberg, H. Malchow, P. Marteau, A. Morali, F. Pallone, S. Pena, A. Rotenberg, I. Rousseau, J. Schmitz, F. Shanahan, I. Sobhani, H. Svensson, A. Van Gossum, M. Van Winckel, and M. Veyrac, as well as gastroenterologists from Nord, Pas de Calais, Somme, and Seine Maritime, generously assisted in the recruitment of families for the study. We are grateful to Dr. C. Bellanné-Chantelot, J. C. Beaudoin, C. Billon, O. Bluteau, T. H. Bui, L. Cazes, C. Guidicelli, P. Legoix, M. Legrand, C. Massart, C. Merlin, P. Pasturaud, S. Martin-Blanc, A. Martins, J. M. Sebaoun, C. de Toma, M. Sahbatou, C. Vaury, and Dr. J. Zucman for their expert technical assistance. We thank Dr. L. Pascoe for critically reading the manuscript. This project received support from the following organizations: the European Union (BIOMED BMH4-97-2098), the Ministère de l'Enseignement National de la Recherche et de la Technologie, the Institut National de la Santé et de la Recherche Médicale, the Direction Générale de la Santé, Association François Aupetit, the Institut de Recherche sur les Maladies de l'Appareil Digestif, and the Swedish Society of Medicine.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Database of Single Nucleotide Polymorphisms, http://www.ncbi.nlm.nih.gov/SNP/index.html (for SNP5 [accession number ss2978533], SNP8 [accession number ss2978536], SNP12 [accession number ss2978537], and SNP13 [accession number ss2978539]
- dHPLC Melt Program, http://insertion.stanford.edu/melt.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for CARD15/NOD2 genomic sequence information [accession numbers AJ303140, AF178930, and NT_030834])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CD [MIM 266600] and UC [MIM 191390])
References
- Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GHW, Glucksmann MA, DiStefano PS (1999) Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-κB. J Biol Chem 274:12955–12958 [DOI] [PubMed] [Google Scholar]
- Brant SR, Panhuysen CIM, Bailey-Wilson JE, Rohal PM, Lee S, Mann J, Ravenhill G, Kirschner BS, Hanauer SB, Cho JH, Bayless TM (2000) Linkage heterogeneity for the IBD1 locus in Crohn’s disease pedigrees by disease onset and severity. Gastroenterology 119:1483–1490 [DOI] [PubMed] [Google Scholar]
- Cavanaugh J, the IBD International Genetics Consortium (2001) International collaboration provides convincing linkage replication in complex disease through analysis of a large pooled data set: Crohn disease and chromosome 16. Am J Hum Genet 68:1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Nicolae D, Gold LH, Fields CT, LaBuda MC, Rohal PM, Pickles MR, Qin L, Fu Y, Mann JS, Kirschner BS, Jabs EW, Weber J, Hanauer SB, Bayless TM, Brant SI (1998) Identification of novel susceptibility loci for inflammatory bowel disease on chromosome 1p, 3q, 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci USA 95:7502–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombel JF, Grandbastien B, Gower-Rousseau C, Plegat S, Evrard JP, Dupas JL, Gendre JP, Modigliani R, Bélaiche J, Hostein J, Hugot JP, Van Kruiningen H, Cortot A (1996) Clinical characteristics of Crohn’s disease in 72 families. Gastroenterology 111:604–607 [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Antonorakis SE (1995) The nature and mechanisms of human gene mutation. In: Scriver C, Beaudet AL, Sly WS, Valle D (eds) Metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 259–291 [Google Scholar]
- Cosnes J, Carbonel F, Beaugerie L, Le Quintrec Y, Gendre JP (1996) Effects of cigarette smoking on the long-term course of Crohn’s disease. Gastroenterology 110:424–431 [DOI] [PubMed] [Google Scholar]
- Crohn BB, Ginzburg L, Oppenheimer GD (1932) Regional ileitis: a pathologic and clinical entity. JAMA 99:1323–1329 [PubMed] [Google Scholar]
- Duerr RH, Barmada MM, Zhang L, Pfutzer R, Weeks DE (2000) High-density genome scan in Crohn’s disease shows confirmed linkage to chromosome 14q11-12. Am J Hum Genet 66:1857–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiocchi C (1998) Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology 115:182–205 [DOI] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJP, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJS, Bridger S, van Deventer S, Forbes A, Nikolaus S, Lennard-Jones JE, Foelsch UR, Krawczak M, Lewis C, Schreiber S, Mathew CG (2001) Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 357:1925–1928 [DOI] [PubMed] [Google Scholar]
- Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, MacPherson AJS, Cardon LR, Sakul H, Harris TJ, Buckler A, Hall J, Stokkers P, van Deventer SJ, Nurnberg P, Mirza MM, Lee JC, Lennard-Jones JE, Mathew CG, Curran ME (1999) A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet 64:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603 [DOI] [PubMed] [Google Scholar]
- Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Groupe d’Etude Thérapeutique des Affections Inflammatoires Digestives, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Lennard JE, Cortot A, Colombel JF, Thomas G (1996) Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 379:821–823 [DOI] [PubMed] [Google Scholar]
- Hugot JP, Zouali H, Lesage S, Thomas G (1999) Etiology of the inflammatory bowel diseases. Int J Colorect Dis 14:2–9 [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G (1999) Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J Biol Chem 274:14560–14567 [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Chen FF, Muto A, Nunez G (2001) Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem 276:2551–2554 [DOI] [PubMed] [Google Scholar]
- Lee JCW, Lennard-Jones JE (1996) Inflammatory bowel disease in 67 families each with three or more affected first-degree relatives. Gastroenterology 111:587–596 [DOI] [PubMed] [Google Scholar]
- Lennard-Jones JE (1989) Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl 170:2–6 [DOI] [PubMed] [Google Scholar]
- Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD (1998) Denaturing high-performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res 26:1396–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Ohmen JD, Li Z, Bentley LG, McElree C, Pressman S, Targan SR, Fischel-Ghodsian N, Rotter JI, Yang H (1999) A genome-wide search identifies potential new susceptibility loci for Crohn’s disease. Inflamm Bowel Dis 5:271–278 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DN, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH (2001a) A frameshift mutation in Nod2 associated with susceptibility to Crohn’s disease. Nature 411:603–606 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G (2001b) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J Biol Chem 276:4812–4818 [DOI] [PubMed] [Google Scholar]
- Podolsky DK (1991) Inflammatory bowel disease. N Engl J Med 325:928–937 [DOI] [PubMed] [Google Scholar]
- Polito JM, Childs B, Mellits ED, Tokayer AZ, Harris ML, Bayless TM (1996) Crohn’s disease: influence of age at diagnosis on site and clinical type of disease. Gastroenterology 111:580–586 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB, Bitton A, Williams CN, Greenberg GR, Cohen Z, Lander ES, Hudson TJ, Siminovitch KA (2000) Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet 66:1863–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger J, Lathrop GM, Bell JI, Jewell DP (1996) Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet 14:199–202 [DOI] [PubMed] [Google Scholar]
- Tobin MV, Logan RF, Langman MJ, McConnell RB, Gilmore IT (1987) Cigarette smoking and inflammatory bowel disease. Gastroenterology 93:316–321 [DOI] [PubMed] [Google Scholar]
- Vermeire S, Satsangi J, Peeters M, Parkes M, Jewell DP, Vlietinck R, Rutgeerts P (2001) Evidence for inflammatory bowel disease of a susceptibility locus on the X chromosome. Gastroenterology 120:834–840 [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X (1997) Apaf-1, a human protein homolog to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90:405–413 [DOI] [PubMed] [Google Scholar]