Abstract
Nucleic acids transiently morph into alternative conformations that can be difficult to characterize at the atomic level by conventional methods because they exist for too little time and in too little abundance. We recently reported evidence for transient Hoogsteen base pairs in canonical B-DNA based on NMR carbon relaxation dispersion. While the carbon chemical shifts measured for the transient state were consistent with a syn orientation for the purine base, as expected for A(syn)•T(anti) and G(syn)•C+(anti) HG base pairing, HG type hydrogen bonding could only be inferred indirectly. Here, we develop two independent approaches for directly probing transient changes in N-H⋯N hydrogen bonds and apply them to the characterization of transient Hoogsteen type hydrogen bonds in canonical duplex DNA. The first approach takes advantage of the strong dependence of the imino nitrogen chemical shift on hydrogen bonding and involves measurement of R1ρ relaxation dispersion for the hydrogen-bond donor imino nitrogens in G and T residues. In the second approach, we assess the consequence of substituting the hydrogen-bond acceptor nitrogen (N7) with a carbon (C7H7) on both carbon and nitrogen relaxation dispersion data. Together, these data allow us to obtain direct evidence for transient Hoogsteen base pairs that are stabilized by N-H⋯N type hydrogen bonds in canonical duplex DNA. The methods introduced here greatly expand the utility of NMR in the structural characterization of transient states in nucleic acids.
Keywords: transient state, Hoogsteen base pair, 15N relaxation dispersion, 7-deazapurine, single-atom substitution
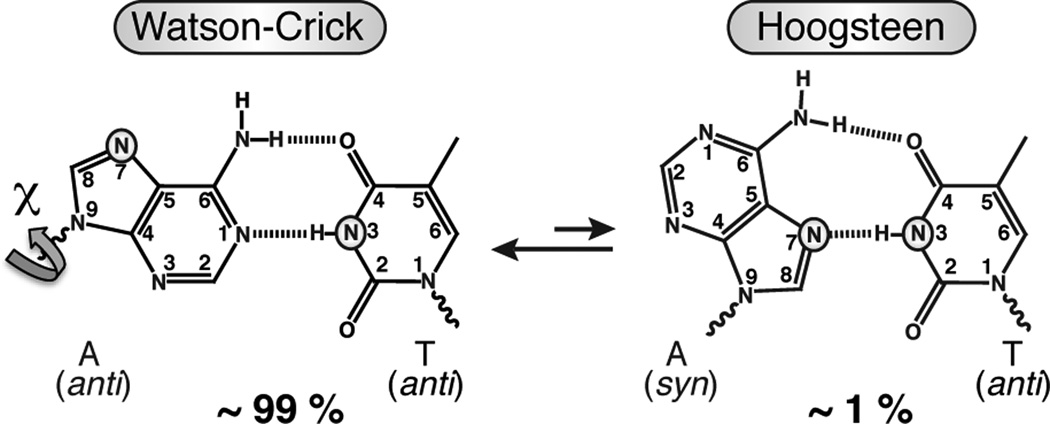
Recently, we presented evidence based on NMR carbon rotating-frame (R1ρ) relaxation dispersion data for low-populated (< 1 %) and short-lived (< 5 ms) Hoogsteen (HG) A•T and G•C+ base pairs at CA and TA steps of canonical duplex DNA (Figure 1).1 The observation of transient HG base pairs in naked duplex DNA together with prior high-resolution structural studies showing HG base pairs in duplex DNA, when damaged2 or in complex with proteins3–5 and small molecule6,7 ligands, suggests a potentially wider functional role for HG base pairs than previously thought. The carbon R1ρ relaxation dispersion experiments allowed the determination of base and sugar carbon chemical shifts for a transient state, which are downfield shifted relative to the WC base pair. While these downfield carbon chemical shifts were consistent with a syn purine base orientation, as expected for A(syn)•T(anti) and G(syn)•C+ (anti) HG base pairing, HG type hydrogen bonding (H-bonding) could only be inferred indirectly. Such evidence came from the measured enthalpy and entropy differences between the ground and transient state and the observation of pH dependent relaxation dispersion in G•C base pairs, including for pyrimidine C C6, which is consistent with creation of protonated G•C+ HG base pairs stabilized by two H-bonds. Here, we develop two independent approaches for more direct probing of transient changes in N-H⋯N H-bonds and apply them to the characterization of transient HG base pairing in canonical duplex DNA.
Figure 1.
Schematic of an equilibrium between ground state WC and transient state HG A•T base pair showing the relative populations obtained previously from NMR relaxation dispersion.1 The WC-HG transition for A•T and G•C base pairs requires an anti-to-syn rotation around the glycosidic bond (χ) that creates an N-H⋯N HG H-bond between T/C N3 and A/G N7 (highlighted in A•T).
It is well known that the NMR chemical shift for hydrogen bond donor nitrogens, such as imino nitrogens in nucleic acids (G N1 and T N3), are strongly dependent on detailed aspects of H-bonding.8 The measurement of imino 15N relaxation dispersion can, therefore, provide a basis for measuring H-bond dependent nitrogen chemical shifts in transient nucleic acid states and thereby allow for direct assessment of transient changes in H-bonding. Despite this unique utility, 15N relaxation dispersion data have never been reported for nucleic acids, even though the same experiments that are widely used in protein applications can be readily adapted for this purpose.9
We first examined what, if any, are the differences between the imino 15N chemical shifts when comparing WC versus HG base pairs in canonical duplex DNA. We stabilized an A•T HG base pair within duplex DNA by using N1-methyladenine, which is impaired from forming WC base pairing and which was previously shown to adopt an HG base pair inside an otherwise B-DNA duplex.1,2 In A•T base pairs, T N3 serves as an H-bond donor in both WC and HG base pairs (Figure 1). Despite being engaged in an N-H⋯N type H-bonding in the two cases, T N3 showed a sizeable upfield shift (−2.8 ppm, minus sign denotes upfield) in the HG versus WC geometry (Figure S1). We confirmed these findings using density functional theory (DFT) calculations, which predict a comparable upfield shift (on average −2.6 ppm, Table S1). The upfield shifted T N3 chemical shift is also consistent with previous studies of DNA triplexes containing WC and HG base pairs.10,11 Unlike T N3, G N1 is not H-bonded in HG G•C+ base pairs. As a result, line broadening due to proton exchange with solvent can make it challenging to directly observe G N1 by proton-detected NMR methods. Nevertheless, we expect a sizeable upfield shift for G N1 based on DFT calculations (on average −2.5 ppm, Table S1) and prior NMR observation of a G(anti)-T(anti)•A(anti) base triple in DNA,10 where G N1 is similarly not H-bonded but semi-protected from proton exchange with solvent as it would likely be in HG base pairs.
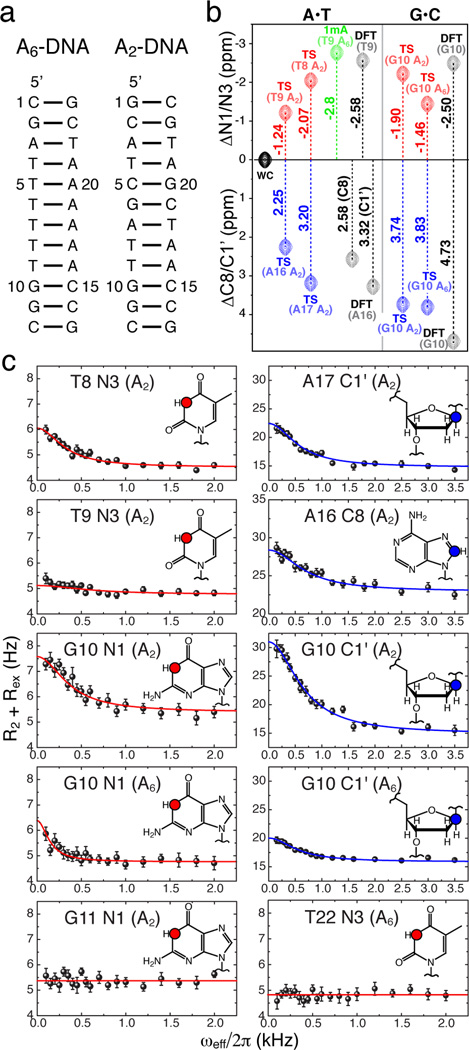
The sizeable differences in the imino nitrogen chemical shifts between WC and HG base pairs should give rise to significant and detectable 15N relaxation dispersion. We tested this hypothesis, and the assignment of HG base pairs as the transient state, by measuring 15N R1ρ relaxation dispersion data for the same base pairs within CA steps that previously showed carbon relaxation dispersion. These represent the first measurements of 15N relaxation dispersion data in nucleic acids. Specifically, we used an offresonance R1ρ relaxation dispersion experiment (Figure S2) that employs selective Hartmann-Hahn cross-polarization transfers to inspect 15N sites one at a time, coupled with low spinlock powers that extend the timescale sensitivity and a 1D acquisition scheme that saves significant time as compared to routinely used 2D experiments.9,12,13 This pulse sequence was adapted from a protein-based experiment designed to target backbone nitrogens9 and is analogous to the 13C R1ρ dispersion experiment that we used to detect transient HG base pairs.1 Using this experiment, we measured 15N chemical exchange at A•T and G•C base pairs in two DNA constructs containing two-(A2-DNA) and six-(A6-DNA) membered adenine tracts (Figure 2a, see Supporting Information (SI)).1
Figure 2.
15N and 13C R1ρ relaxation dispersion sense the same DNA transient states. (a) DNA constructs. (b) Chemical shift comparison between imino 15N (red) or sugar/base 13C (blue) of transient states (TS) with a trapped A•T HG base pair (1mA, green) average values for N1/N3 or as reported before1 for C8/C1’ from DFT calculations (DFT), in A6-DNA (A6) and A2-DNA (A2). (c) On-resonance R1ρ relaxation dispersion profiles showing coupled chemical exchange for 15N and 13C sites (insets) in A•T and G•C base pairs at the CA step and neighboring base pairs and absence of exchange in other residues (G11 and T22), highlighted as in b).
As expected, marked 15N relaxation dispersion was observed in both T and G residues (Figure 2 and Figure S3). Moreover, the 15N relaxation dispersion data exhibited the same trend of enhanced chemical exchange with decreasing pH, expected for a transient protonated G•C+ HG base pair, as observed for 13C data (data not shown).1 Analysis of off-resonance 15N profiles yielded a transient state with lifetimes (1/kB ~0.30 – 1.35 ms) and populations (pB~0.7 – 10%) that, within error, are in very good agreement with those obtained from 13C relaxation dispersion (~0.25 – 1.35 ms and ~0.5 – 1.0% respectively) (Table S3). Indeed, the two data sets can be combined in a single global fit with shared lifetimes and populations, indicating that the carbon and nitrogen nuclei report on the same transient state (Table S3). It should be noted however, that the much smaller observed chemical shift difference between ground and transient state for 15N (ΔωAB <150 Hz) versus 13C (ΔωAB ~350 – 550 Hz) makes it more difficult to discern and accurately determine the mutually dependent pB/ΔωAB parameters from 15N profiles alone and explains the overestimation of transient state populations seen above (see SI and Figure S4). While the relaxation dispersion at C8/C1’ sites is more sensitive to anti-to-syn nucleobase transitions observed here than the relaxation dispersion at N1/N3 sites, this will not necessarily be the case for other structural transitions involving H-bonding dynamics such as 15N protonation equilibria, base pair breaking and complex formation with ligands and proteins that may not incur significant 13C chemical shift changes.
Importantly, the T N3 chemical shifts determined for the transient state, either from individual fitting of 15N data or global fitting of 15N/13C data, was upfield shifted relative to WC by up to −2.1 ppm, in very good agreement with the upfield shift that we expect (−2.5 to −2.8 ppm) for a transient state involving HG type N-H⋯N type H-bonding (Figure 2b, Table S1). The upfield shifted G N1 chemical shift for the transient state is also consistent with the loss of (C)N3-H3⋯N1(G) H-bond and exposure to solvent, as expected for an HG base pair, and as judged based on DFT calculations of HG G•C+ conformations (Figure 2b, Table S1).
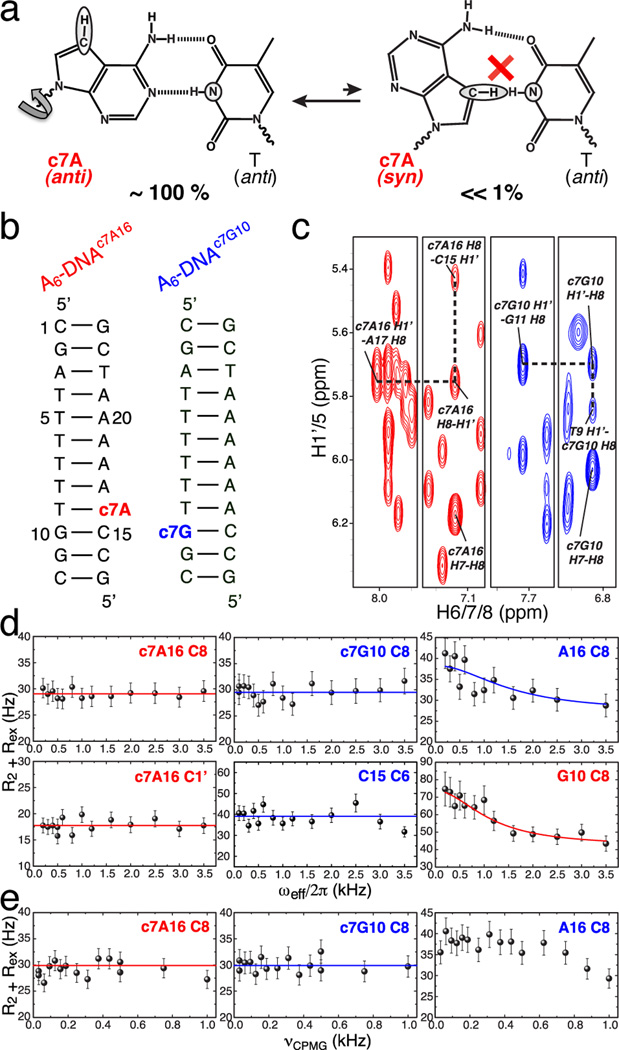
The 15N relaxation dispersion data alone cannot be used to unequivocally establish the existence/absence of a transient H-bond. For example, although larger in magnitude, an upfield shift is also expected for non-H-bonded T N3 sites (> 3 ppm),8 such as those found in DNA loops (> 3.5 ppm).10 We therefore developed a complementary approach to more robustly establish transient changes in N-H⋯N H-bonds. In this approach, we examine the impact of substituting the H-bond acceptor nitrogen (N7) with a carbon (C7H7) on both 13C and 15N relaxation dispersion. Specifically, we swapped A or G with its isosteric base analog, 7-deazaadenine (c7A) or 7-deazaguanine (c7G), that contains an N7 to C7H7 (c7) modification. This single atom substitution knocks out a single hydrogen bond in HG A•T and G•C+ base pairs without affecting WC H-bonds (Figure 3a). Thus, it is expected to strongly disfavor transitions towards HG base pairs. Indeed, 7-deazapurines were originally used to prevent DNA triplex hybridization via HG base pairing14 and have been successfully used to demonstrate that human DNA Polymerase Iota (Polι) replicates DNA via HG base-pairing.15 Although the strength of a single WC or HG N3-H3⋯N7 H-bond has not been established experimentally, individual N-H⋯N H-bonds have been predicted by computational methods to contribute at least 7 kcal/mol towards WC base pair stability.16 Such loss in stability should make transient HG states in c7-modified DNA undetectable by relaxation dispersion, even if the predicted values overestimate the actual ones by twofold.
Figure 3.
C7-purine substitution preserves WC geometry but suppresses 13C chemical exchange. (a) Substitution of N7 with C7H7 in 7-deazaadenine (c7A) eliminates the potential for N-H⋯N H-bond and inhibits transient HG base pairs (T N3 and c7A C7H7 are highlighted). (b) c7-modified constructs A6-DNAc7A16 and A6-DNAc7G10, modified residues are highlighted in red and blue respectively. (c) NOE cross-peaks between H8/H6/H1’ of c7A16 or c7G10 with adjacent nucleotides supporting WC geometry, highlighted as in (b). (d) R1ρ and (e) CPMG 13C relaxation dispersion profiles (effective R1ρ or R2,CPMG designated as R2 + Rex) showing chemical exchange is suppressed at c7A16•T9 in A6-DNAc7A16 (pH 6.8) and c7G10•C15 in A6-DNAc7G10 (pH 5.2) but retained at the unmodified neighboring base pairs G10•C15 and A16•T9 (pH 5.2). Solid lines represent average R2+Rex in the case of no detectable exchange or best fits to Eq. S2 (A16 C8 CPMG profile not fitted due to too fast exchange).
The use of c7 analogs of adenine and guanine precluded preparation of NMR samples, in which the modified residue is 13C/15N-enriched. Fortunately, we could take advantage of the much greater sensitivity and timesaving afforded by our 1D 13C experiment to collect data at natural abundance, as described previously for damaged DNA.17 To this end, we prepared two unlabeled samples containing the A6-DNA target sequence,1 A6-DNAc7A16 and A6-DNAc7G10, where A16 and G10 were replaced with their c7-analogs, c7A16 and c7G10 (Figure 3b). We confirmed using NOE connectivity and chemical shifts that the modified base pairs adopt the expected WC conformation in agreement with recent high-resolution structural studies of a c7G-containing duplex showing minimal perturbations to the WC helical framework (Figure 3c, Figure S5).18 However, the modification did give rise to chemical shift perturbations (< 0.5 ppm) in surrounding nucleotides as well as resulted in greater imino proton exchange with solvent at and near the modified site (Figure S5). These data suggest that the single-atom substitution retains WC base pairing, but may lead to structural and dynamic perturbations likely due to altered hydration and stacking, which can transmit to distant residues. These findings are in agreement with recent biophysical studies showing that c7-purines, while not impairing WC geometry, enhance local base pair dynamics and noticeably destabilize DNA duplexes (~2 kcal/mol per c7G) primarily due to unfavorable enthalpy.18,19
We examined the impact of the c7 substitution on the 13C R1ρ relaxation dispersion profiles measured at natural abundance (Figure 3d). Strikingly, the single-atom substitution completely quenched the 13C chemical exchange at the modified base pair. This includes the sugar C1’ and base C8 of the modified purine base, and in the case of c7G10, the modification also quenched the 13C chemical exchange at the complementary cytosine base (C15) (Figure 3d).1 We confirmed the latter observation in a DNA sample, where C15 was 13C/15N-labeled, to improve sensitivity (Figure S6). The lack of chemical exchange at C C6 suggests suppression of C N3 protonation, normally required for optimal HG base pairing. As an internal control, we verified that the introduction of the c7 modification at one base pair in the CA step did not impact relaxation dispersion observed at the neighboring base pair using unlabeled and 13C/15N-enriched samples (Figure 3d, Figure S5). Moreover, the neighboring G•C base pair retained pH dependent line broadening in samples containing a c7A•T base pair, as expected for transient G•C+ HG base pairing, while no pH dependent line broadening was observed for the c7G•C base pair (Figure S6). These data establish that chemical exchange can indeed be accurately detected at natural abundance in these samples and that the effect of the c7 substitution is mainly localized and not distributed over the entire duplex. Unfortunately, we were unable to measure 15N relaxation dispersion data at N1 of the unlabeled c7G due to prohibitively low sensitivity of 15N natural abundance experiments, or for N3 in a 13C/15N-enriched T across an unlabeled c7A owing to rapid proton exchange with solvent and excessive line broadening (Figure S5).
Although highly unlikely, the flat R1ρ carbon relaxation dispersion profiles could reflect a slower exchange process (due to stabilization of both the ground and transient states and/or destabilization of the transition state) that falls outside the detection limits of our method rather than a reduction in the fractional population of the transient state due to its energetic destabilization. However, flat R1ρ relaxation dispersion profiles were observed over a range of temperatures (8 to 26°C). Moreover, flat profiles were also observed when measuring Carr-Purcell-Meiboom-Gill (CPMG) 13C relaxation dispersion data that extends the upper timescale sensitivity deeper into the millisecond (ms) regime potentially to tens of ms compared to < 5 ms for on-resonance R1ρ dispersion alone (Figure 3e, Figure S6). The flat profiles are unlikely due to selective stabilization of the modified WC base pairs, which are observed to have impaired rather than improved stability, or to reduction in the chemical shift difference between exchanging states (Table S1). Rather, the results are consistent with at least a fivefold reduction in the fraction of the transient HG state (pB), which is in agreement with the larger expected decline in HG versus WC base pair stability as a result of the c7 substitution.16,19 These results also help further rule out alternative flipped-out conformations, whose absolute stability will likely remain unaffected by the c7 substitution.
In conclusion, we have developed two independent approaches for dissecting transient changes in N-H⋯N hydrogen bonds in nucleic acids. Our results provide direct evidence for transient Hoogsteen base pairs that are stabilized by N-H⋯N type H-bonds. The approach can be extended to target other H-bond donors and acceptors and directly bonded sites, including amino nitrogens and carbonyl carbons, which can be applied in concert with other single-atom substitutions. These approaches provide a new basis for exploring transient changes in Hbonds, which are a defining feature of DNA and RNA structure.
Supplementary Material
ACKNOWLEDGMENT
We thank Alexandar Hansen for valuable discussions and help with pulse sequence setup, Jameson Bothe for critical review of the manuscript, and members of the Al-Hashimi lab for insightful comments. We gratefully acknowledge the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for support in the purchase of a 600 MHz spectrometer and thank Alex Kurochkin for maintenance of the NMR instrument. This work was supported by a NIH grant (RO1 GM089846) awarded to H.M.A.
ABBREVIATIONS
- NMR
Nuclear magnetic resonance
- WC
Watson-Crick
- HG
Hoogsteen
- c7
7-deaza
- CPMG
Carr-Purcell-Meiboom-Gill
- ppm
parts per million
- NOE
nuclear Overhauser effect
Footnotes
ASSOCIATED CONTENT
Supporting Information. Details of sample preparation, resonance assignments, relaxation dispersion experiments and profiles, DFT calculations and simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM. Nature. 2011;470:498. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu L, Yi C, Jian X, Zheng G, He C. Nucleic Acids Res. 2010;38:4415. doi: 10.1093/nar/gkq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. Genes Dev. 1999;13:3217. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C. Nucleic Acids Res. 2002;30:5244. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Nat Struct Mol Biol. 2010;17:423. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ughetto G, Wang AH, Quigley GJ, van der Marel GA, van Boom JH, Rich A. Nucleic Acids Res. 1985;13:2305. doi: 10.1093/nar/13.7.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert DE, van der Marel GA, van Boom JH, Feigon J. Proc Natl Acad Sci U S A. 1989;86:3006. doi: 10.1073/pnas.86.9.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czernek J, Fiala R, Sklenar V. J Magn Reson. 2000;145:142. doi: 10.1006/jmre.2000.2091. [DOI] [PubMed] [Google Scholar]
- 9.Korzhnev DM, Orekhov VY, Kay LE. J Am Chem Soc. 2005;127:713. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 10.Radhakrishnan I, Gao X, de los Santos C, Live D, Patel DJ. Biochemistry. 1991;30:9022. doi: 10.1021/bi00101a016. [DOI] [PubMed] [Google Scholar]
- 11.Dingley AJ, Masse JE, Peterson RD, Barfield M, Feigon J, Grzesiek S. J Am Chem Soc. 1999;121:6019. [Google Scholar]
- 12.Massi F, Johnson E, Wang C, Rance M, Palmer AG., 3rd J Am Chem Soc. 2004;126:2247. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 13.Palmer AG, 3rd, Massi F. Chem Rev. 2006;106:1700. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 14.Latimer LJ, Lee JS. J Biol Chem. 1991;266:13849. [PubMed] [Google Scholar]
- 15.Johnson RE, Prakash L, Prakash S. Proc Natl Acad Sci U S A. 2005;102:10466. doi: 10.1073/pnas.0503859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szatylowicz H, Sadlej-Sosnowska N. J Chem Inf Model. 50:2151. doi: 10.1021/ci100288h. [DOI] [PubMed] [Google Scholar]
- 17.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. J Am Chem Soc. 2009;131:3818. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Li F, Ganguly M, Marky LA, Gold B, Egli M, Stone MP. Biochemistry. 2008;47:7147. doi: 10.1021/bi800375m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganguly M, Wang F, Kaushik M, Stone MP, Marky LA, Gold B. Nucleic Acids Res. 2007;35:6181. doi: 10.1093/nar/gkm670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.