Abstract
Non random segregation of sister chromatids has been implicated to help specify daughter cell fate (the Silent Sister Hypothesis [1]) or to protect the genome of long-lived stem cells (the Immortal Strand Hypothesis [2]). The idea that sister chromatids are non-randomly segregated into specific daughter cells is only marginally supported by data in sporadic and often contradictory studies. As a result, the field has moved forward rather slowly. The advent of being able to directly label and differentiate sister chromatids in vivo using fluorescence in situ hybridization [3] was a significant advance for such studies. However, this approach is limited by the need for large tracks of unidirectional repeats on chromosomes and the reliance on quantitative imaging of fluorescent probes and rigorous statistical analysis to discern between the two competing hypotheses. A novel method called Strand-seq which uses next-generation sequencing to assay sister chromatid inheritance patterns independently for each chromosome [4] offers a comprehensive approach to test for non-random segregation. In addition Strand-seq enables studies on the deposition of chromatin marks in relation to DNA replication. This method is expected to help unify the field by testing previous claims of non-random segregation in an unbiased way in many model systems in vitro and in vivo.
Keywords: asymmetric, non-random, template, Strand-seq, Watson, Crick
1. Introduction
The process of cell division relies on faithful DNA replication, such that the resulting daughter cells each receive an exact copy of the parental cell genome. The importance of creating two identical copies of each chromosome prior to cell division and their segregation into daughter cells is reflected in the very complex and precise mechanism that controls semi-conservative replication of complementary template strands [5], as well as the various post-replicative mechanisms that protect the genome from replication errors, genome rearrangements, and mutations. Problems with any of these processes can result in abnormal daughter cells, uncontrolled proliferation, or cell death [6]. Though DNA replication is often thought to create carbon copies of chromosomes, some replication errors do occur and telomere repeat length diversity is generated at every cell division [7]. Nevertheless, the mechanism of DNA replication is typically thought to result in two sister chromatids which are genetically and functionally identical.
Another complex regulatory network ensures that each daughter of a dividing cell gets only one of these “identical” sister chromatids for every chromosome, thereby maintaining the integrity of the genome in daughter cells [8]. Based on the fidelity and importance of DNA replication and spindle checkpoints, the dogma has emerged that “identical” sister chromatids are randomly segregated to daughter cells. This dogma has been accepted without much debate for many decades and was reinforced because there has been no cause to question the functional uniformity of sister chromatids and by a lack of evidence to the contrary. The idea that sister chromatids are not necessarily equivalent was initially proposed based on observations that some cells appear to retain radiolabeled DNA, even after multiple cell divisions (reviewed in [9]). Subsequently, Cairns proposed the Immortal Strand Hypothesis (ISH) as a mechanism whereby some long-lived cells selectively retain template strand DNA in order to prevent the accumulation of replication errors in tissue-regenerating stem cells that are required throughout the life of an organism [2]. Shortly after, Cairns and colleagues reported that intestinal epithelial stem cells, inferred to be dividing during a period of tissue regeneration, selectively retain radiolabeled template strands [10]. Similar studies followed [11], though the caveat that the label-retaining cells could have been quiescent cells rather than repopulating stem cells remains a concern, since both quiescent and active stem cells could occupy the same niche [12].
2. Pulse-chase experiments fuel the immortal strand hypothesis
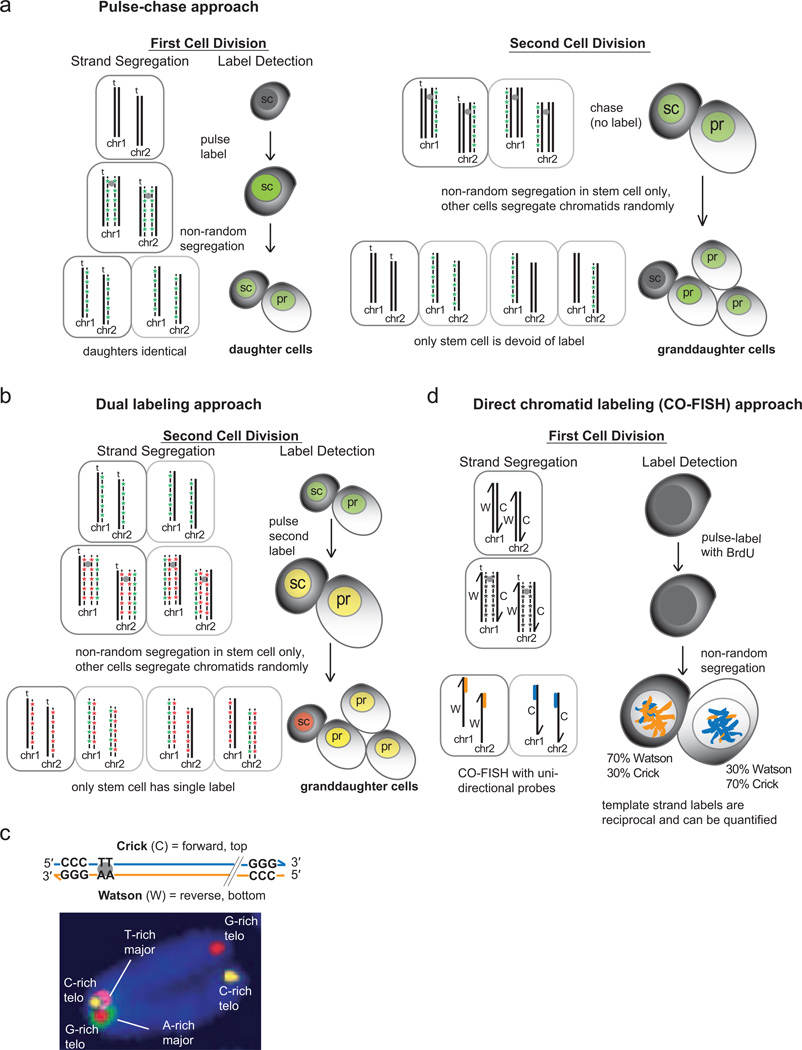
Most studies which followed up on Cairns’ early findings attempted to directly test the ISH with pulse-chase experiments which label nascent DNA with a nucleotide analog during one round of replication in asymmetrically dividing stem cells. The segregation pattern of unlabeled DNA (the template strand) is followed through two sequential cell divisions (Fig. 1a). Of the four resultant granddaughter cells, the absence of detectable label in one cell (the presumed stem cell) and its proximity to labeled neighbouring cells is used as the assay criteria. To circumvent objections over “negative labeling”, a similar approach uses two different nucleotide analogs in the sequential cell divisions to differentially label the self-renewing stem cell from the other three granddaughter cells (Fig. 1b). Favored models in which the pulse-chase approach has been tested have included regeneration of intestinal epithelium by stem cells of the murine colon crypt, neural stem cells, regeneration of injured muscle tissue by muscle satellite stem cells, among others. The application of pulse-chase approaches in these and other model systems has been reviewed in detail [13, 14]. Though these pulse-chase approaches became more frequent, they all essentially rely on DNA labeling followed by imaging, often without quantitative data. An improvement combines staining for stem cell and progenitor markers with the pulse-chase experiments to correlate template strand retention and cell identity [15–17]. Nevertheless, the non-random segregation of original parental template strands in all pulse-chase approaches is always inferred by the absence of the label administered during the first of two cell divisions. Remarkably, many of these studies show a complete absence of this first label in the stem cell, which is necessary in order to support the ISH. It is important to note that some studies using the same approach have shown no evidence of complete template strand retention (immortal strands) in hematopoietic stem cells [18], epidermal stem cells [19] Drosophila male germline stem cells [20] and stem cells and progenitor cells of the colon crypt [21–23].
Figure 1.
Overview of pulse-chase and CO-FISH approaches for detecting non-random chromosome segregation in recently divided cells. a. Single-label pulse-chase approach to test the immortal strand hypothesis requires two cell divisions to discern non-random segregation of chromatids. In an asymmetrically-dividing stem cell (sc), one round of DNA replication is pulsed with a nucleotide analog to label nascent DNA strands (green), leaving original template strands (including the “immortal” template strand, t) unlabeled. The subsequent asymmetric cell division results in two sister cells with hemi-labeled chromosomes that cannot be distinguished based on detection of the label (left panel). The immortal template strands (t) are presumed to end up in the daughter stem cell. A second cell division without the nucleotide analog results in four granddaughter cells, one pair resulting from an asymmetric division of the daughter stem cell (sc) and the other pair from the presumed symmetric division of the other daughter cell (pr, progenitor). Only the granddaughter stem cell is expected to be devoid of detectable label, as the original unlabeled immortal template strands (t) are all co-segregated to this cell, while other non-immortal strands and nascent DNA strands labeled in the first division are randomly distributed to the other three granddaughters. b. A dual-labeling approach pulses a different nucleotide analog during the second cell division (red). In the resultant four granddaughter cells, only the stem cell is expected to exhibit a single label from the second division (red cell) while the other three granddaughters are expected to be labeled with both the first and second labels (yellow cells). c. Designation of “Crick” and “Watson” to denote forward and reverse strands of chromosomes, respectively (top panel). In murine chromosomes, the orientation of tandem major satellite repeats is fixed in relation to telomeric repeats, therefore Crick and Watson template strands of sister chromatids can be identified by hybridization with unidirectional probes using CO-FISH (bottom panel, see ref. [3]). d. CO-FISH can directly assay non-random segregation of sister chromatids by quantifying the distribution of major satellite probes in daughter cells following cell division (Portion of Figure 1c reprinted with permission from ref [3].
The major theoretical objections to the ISH are that immortal strand retention cannot protect more than one stem cell, posing a problem for stem cell self-renewal. In addition, since the complementary DNA strands of all chromosomes in an “immortal” cell differ from each other by at least one generation, only half the genome is truly protected from replication errors [1]. Neither strand is protected from non-replication-based damage induced by genotoxic stresses from the environment, byproducts of cellular metabolism, or spontaneous hydrolysis of nucleotide residues [1, 24]. Consequently, by retaining the old template strands, the immortal stem cell would accumulate these other types of damage over time [25]. This potential accumulation of other mutations in the retained template strand is in direct conflict to the genome protection mechanism proposed by the ISH. Finally, all sister chromatid exchange mechanisms must be completely suppressed in order to prevent mixing of the template and non-template strands. Despite these theoretical objections, the caveats of technical artifacts in labeling experiments and the experimental evidence contradictory to the ISH, it remains a prevalent hypothesis.
3. Non-random segregation as a fate determining mechanism
One of the earliest notable exceptions to the pulse-chase approaches and a complementary hypothesis to the ISH is described in the report that a murine embryonic stem (mES) cell line selectively segregates the template strands of chromosome 7 [26]. This study took advantage of existing inducible recombination cassettes on chromosomes 7 and 11 in the cell line to test for non-random segregation pattern for these chromosomes only. After DNA replication, two sister chromatids from homologous chromosomes are induced to recombine at a specific site, and the recombinant chromatids were shown to segregate to opposite daughter cells in all cases, and never co-segregated to any daughter cell. At the time, this was the first study to distinguish sister chromatids in cells and directly follow their segregation pattern. However, this approach could not become comprehensive because it was limited to testing two chromosomes in one particular engineered cell line and is also not feasible in vivo. Nevertheless, this study provided some evidence that non-random segregation is not obligate for all chromosomes, and negated the objectin that a DNA label itself could be causing or affecting strand segregation patterns. Furthermore, it proposed that co-segregation of expressed or silenced genes to daughter cells could be a mechanism of differentiation, rather than genome protection.
We proposed an alternative to the ISH to help explain not only the phenomenon of non-random segregation, but also the likelihood that not all chromosomes are obliged to segregate non-randomly[1]. The Silent Sister Hypothesis (SSH) proposes that during replication, epigenetic marks at key genes are not replicated to both sister chromatids. The resulting epigenetic differences between sister chromatids could be translated to different gene expression patterns in the daughter cells after cell division. For example, master regulators at the head of transcriptional cascades could initiate differential cell fate programs in daughter cells upon the completion of cytokinesis if their differential expression pattern is specified during DNA replication in the parent cell. This type of mitotic imprinting could add another level of complexity to asymmetric cell divisions, on top of mechanisms that asymmetrically distribute mRNAs and proteins to daughter cells. The notion that epigenetic differences could exist between sister chromatids has previously been proposed [26–29] but remains controversial because it is assumed that all epigenetic marks are faithfully copied to both sister chromatids during DNA replication. Currently there are no methods to rigorously test this assumption. However, it was recently reported that the H2A.Z histone variants differ between sister chromatids replicated from immortal DNA strands versus mortal strands [30]. In addition, regions of strand-specific bias in the distribution of 5-methylcytosine and 5-hydroxymethylcytosine in murine embryonic stem cells has been shown [31] and could be a candidate for initiating epigenetic differences between sister chromatids after replication. Interestingly, it was recently reported that the N-terminal tails of the two histone H3 proteins within a single nucleosome in ES cells can bear different modifications, generating an asymmetry within the nucleosome that correlates with bivalent chromatin domains and pluripotency [32]. Therefore the activating and repressive chromatin marks which coexist in bivalent chromatin domains [33], are now known to originate from the same nucleosome. It has also been proposed previously that in some instances, histone chaperones re-assemble chromatin on newly-replicated DNA by splitting the parental H3H4 tetramer into two H3H4 dimers, allowing for semi-conservative inheritance of histone proteins and therefore epigenetic marks [34]. It is tempting to envision deposition of the activating marks of one H3H4 dimer onto one sister chromatid and the repressive marks of the other H3H4 dimer onto the other sister chromatid at key genes, followed by differential gene expression after cell division. Though the exact mechanisms of chromatin assembly and epigenetic mark inheritance are not yet resolved, such a scenario is predicted by the SSH. The SSH further predicts that only a subset of all chromosomes could be sufficient to help specify different cell fates after cell division. Therefore, not all chromosomes are obliged to segregate non-randomly as in the Immortal Strand Hypothesis.
4. Chromosome-orientation fluorescence in situ hybridization (CO-FISH) directly tests strand segregation patterns of all chromosomes in vivo
To test the SSH, it is necessary to distinguish sister chromatids and follow their segregation pattern after a single cell division. This is not possible by pulse-chase labeling of bulk nascent DNA because it cannot directly distinguish sister chromatids after one round of replication and relies on detecting the absence of a label in the second of two sequential mitoses to infer random or non-random segregation. In addition, non-random segregation of only a subset of chromatids may not be obvious if detection of nascent DNA label is not sensitive enough to detect low amounts of label above background. To overcome this, we adapted chromosome-orientation fluorescence in situ hybridization (CO-FISH)[35] to directly label sister chromatids after one round of cell division. This approach takes advantage of the fact that major satellite repeats show a striking uniform direction relative to telomeric repeats on each chromosome (Fig. 1c). This uniform orientation was used to designate each strand of complementary DNA as “Crick” or “Watson”, corresponding to the forward (top, plus) or reverse (bottom, minus) strands of reference genomes, respectively [3] (Fig. 1c). Since each sister chromatid is synthesized from either a Crick or Watson template strand, the pair differs from each other in template origin. This difference allows the sister chromatids to be distinguished physically. The uniform configuration allows sister chromatids to be distinguished by CO-FISH using only the major satellite probes, both in vitro and in vivo (Fig. 1d) [3, 36]. We used CO-FISH to track the segregation patterns of sister chromatids in murine colon epithelial cells in vivo, and found that a subset of dividing cells segregate a portion of their chromatids non-randomly. This study represented the first direct and statistically significant evidence for nonrandom segregation of select template strands in vivo. We found that the cells exhibiting non-random segregation corresponded to the transit amplifying cells and not the Lgr5+ stem cells in the colon crypts[37], as identified by their position in tissue sections. Importantly, there were no occurrences of 100% non-random segregation, supporting the idea that non-random segregation is not necessarily obligate for all chromosomes, and is not restricted to long-lived stem cells. The results of this study were incompatible with the ISH. However, a recent study that applied CO-FISH to study sister chromatid segregation in muscle satellite stem cells reported complete asymmetrical retention of template strands to the daughter stem cell [38], supporting the ISH. This highlights that even an improved approach such as CO-FISH where template strands can be directly visualized after only one cell division can support both the SSH and ISH in different studies. In addition, because CO-FISH relies on fluorescent probes to detect template strands that cannot distinguish individual chromosomes after one cell division, it cannot detect if only one or a few chromatids segregate non-randomly. Also, the CO-FISH approach appears to be limited to studies of murine cells or where every Watson and every Crick template strand can be labeled with major satellite probes due to the invariant orientation of large tracts of repetitive DNA (up to 10 Mb) in each chromosome.
5. Strand-seq: an unbiased approach to test template strand segregation in all chromosomes independently
Differences in experimental approaches have contributed to conflicting reports testing nonrandom segregation of sister chromatids in cells from same or similar tissues, with some studies reporting polar opposite results and conclusions. For example in the intestinal epithelium, different techniques identified different subsets of cells, or numbers of cells that exhibit non-random segregation [3, 11, 23]. In addition, reports in which there is evidence for non-random segregation, either all chromosomes segregate non-randomly thereby supporting the ISH, or only a subset of chromosomes do, arguing against the ISH and supporting the alternative SSH. Formally, this dichotomy may be explained by different non-random mechanisms being active in different cell types, or that completely non-random segregation is not activated in all consecutive cell divisions. Undoubtedly the many variables in the model systems, approaches to isolate asymmetric cells and assays for non-random segregation will have contributed to the contradicting evidence that cells exhibit non-random segregation, and whether only some or all chromosomes are involved. In addition, strong objections to experimental approaches and authors’ conclusions occasionally arise [39, 40]. As a result it is difficult to broadly assess all of the studies in a review such as this because all the variables can influence the results and interpretation of each study. Label retention approaches, pulse-chase approaches and even the CO-FISH approach are cumbersome. Even if existing protocols were standardized or combined, template strands would still be analyzed en masse, without the possibility to distinguish or identify individual chromosomes. Based on these considerations we suggest that a combination or improvement of currently available methods is probably not the answer, and that much of the debate within the field will not be settled without a new set of tools to unify results and discriminate between hypotheses.
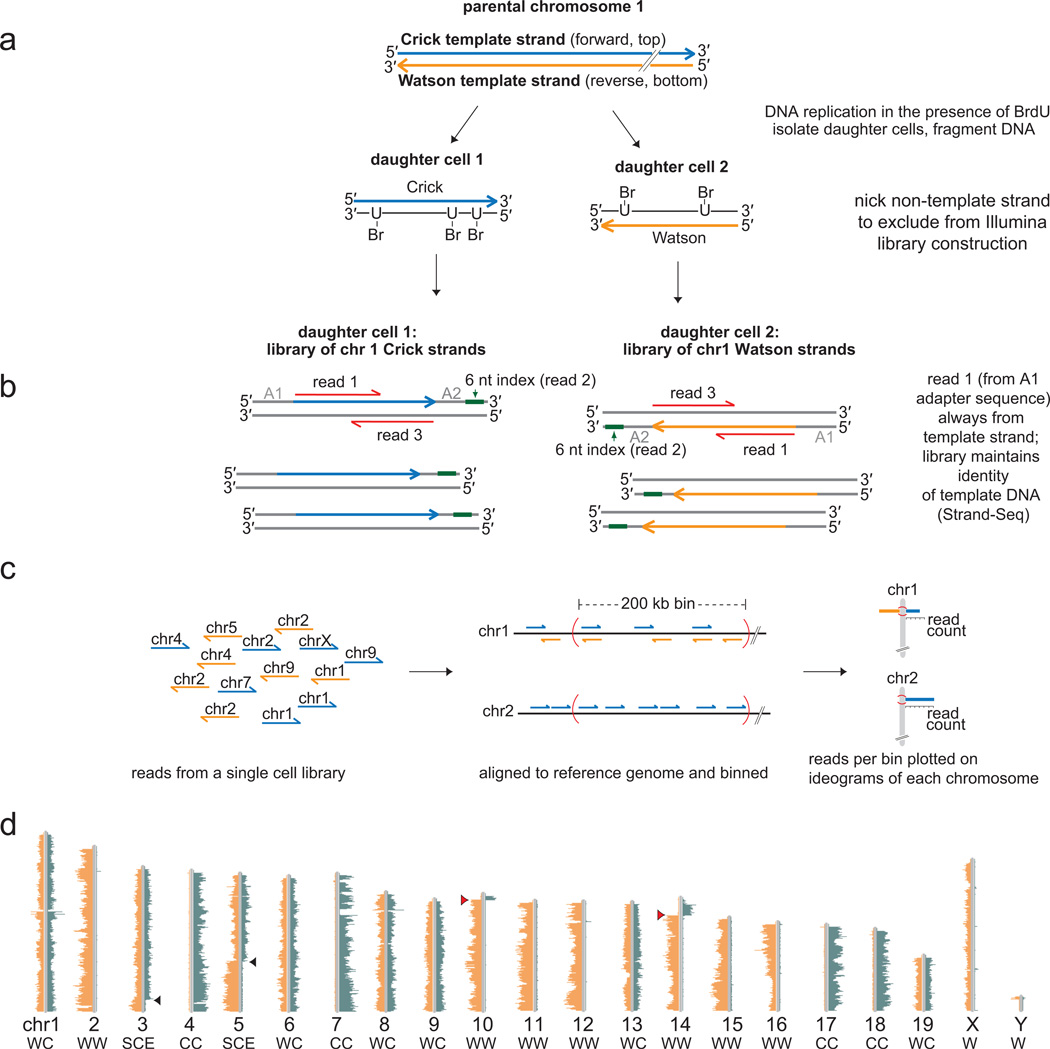
We recently developed a novel sequencing technique called Strand-seq which sequences only parental template strands of all chromosomes from single cells [4]. Identification of sister chromatid inheritance patterns is possible for each individual chromosome independently, since they are identified by short reads from next-generation sequencing platforms and are aligned to a reference genome (Fig. 2). Briefly, the nascent DNA of chromosomes is labeled during S-phase with bromodeoxyuridine (BrdU), leaving the template strands unsubstituted. Single cells are isolated by FACS, the genome is fragmented with micrococcal nuclease, and a standard Illumina paired-end library construction protocol is employed (Fig. 2a). Prior to PCR amplification of the library fragments, the nascent BrdU-substituted DNA is nicked by photolysis with Hoechst 33258 and UV treatment, preventing its amplification in the subsequent PCR reaction. The resulting directional library fragments are sequenced using an Illumina platform (Fig. 2b), and the short sequencing reads that result identify the original template strands inherited from the parental cell (Fig. 2c). A graphical output of these short sequencing reads is generated using a bioinformatic pipeline (BAIT, Bioinformatic Analysis of Inherited Templates) and displays the Watson or Crick parental template strand inheritance pattern (Fig. 2c). A Strand-seq library from a single murine embryonic stem cell clearly shows the template strand inheritance pattern of every chromosome individually (Fig. 2d). In this cell, 7 out of 19 autosomes inherited two Watson template strands, 4 inherited two Crick template strands, and 6 inherited one Watson and one Crick template strand. The two chromosomes which exhibited a sister chromatid exchange (SCE) are excluded from this analysis because it is not possible to determine the “intended” strand. (The impact of SCEs is discussed in more detail in section 8, and in Falconer et al. [36]). In cells from non-inbred genetic backgrounds, the template strand inheritance pattern can be extended to identify parental origin for each template strand using analysis of informative single nucleotide polymorphisms (SNP’s).
Figure 2.
Strand-seq determines template strand inheritance independently for each chromosome in single cells. a. DNA replication in the presence of BrdU results in hemisubstituted sister chromatids which are segregated to daughter cells. Only the newly-formed DNA (black lines) is substituted with BrdU while the original template strands (Watson, orange or Crick, blue) remain unsubstituted. A modified Illumina library construction protocol on isolated daughter cells excludes the BrdU-substituted strands from the final amplification step by creating nicks at the sites of BrdU incorporation. b. The resultant library fragments maintain the genomic directionality of the DNA strands such that read 1 of a paired-end read is always from the template strand (orange or blue) while read 3 is from the complementary strand (grey). Read 2 sequences the 6 nucleotide (nt) index, allowing single-cell libraries to be multiplexed for sequencing. c. A collection of directional library reads from a single cell is processed using BAIT (bioinformatic analysis of inherited templates) software by aligning reads to the reference genome, binning reads, and displaying read counts on ideograms for each chromosome (see ref. [4] for more detailed library construction protocol). d. The BAIT output for a single murine embryonic stem cell library clearly shows the Watson and Crick template strands that were inherited by that cell for each chromosome (listed below each ideogram). Strand-seq can also identify sister chromatid exchanges (SCE) such as those in chromosomes 3 and 5 (black arrowheads) as well as regions where contigs have been mis-oriented in the reference genome (red arrowheads) [4]. Portions of Figure 2 reprinted with permission from ref [4].
6. Testing the immortal strand hypothesis using Strand-seq
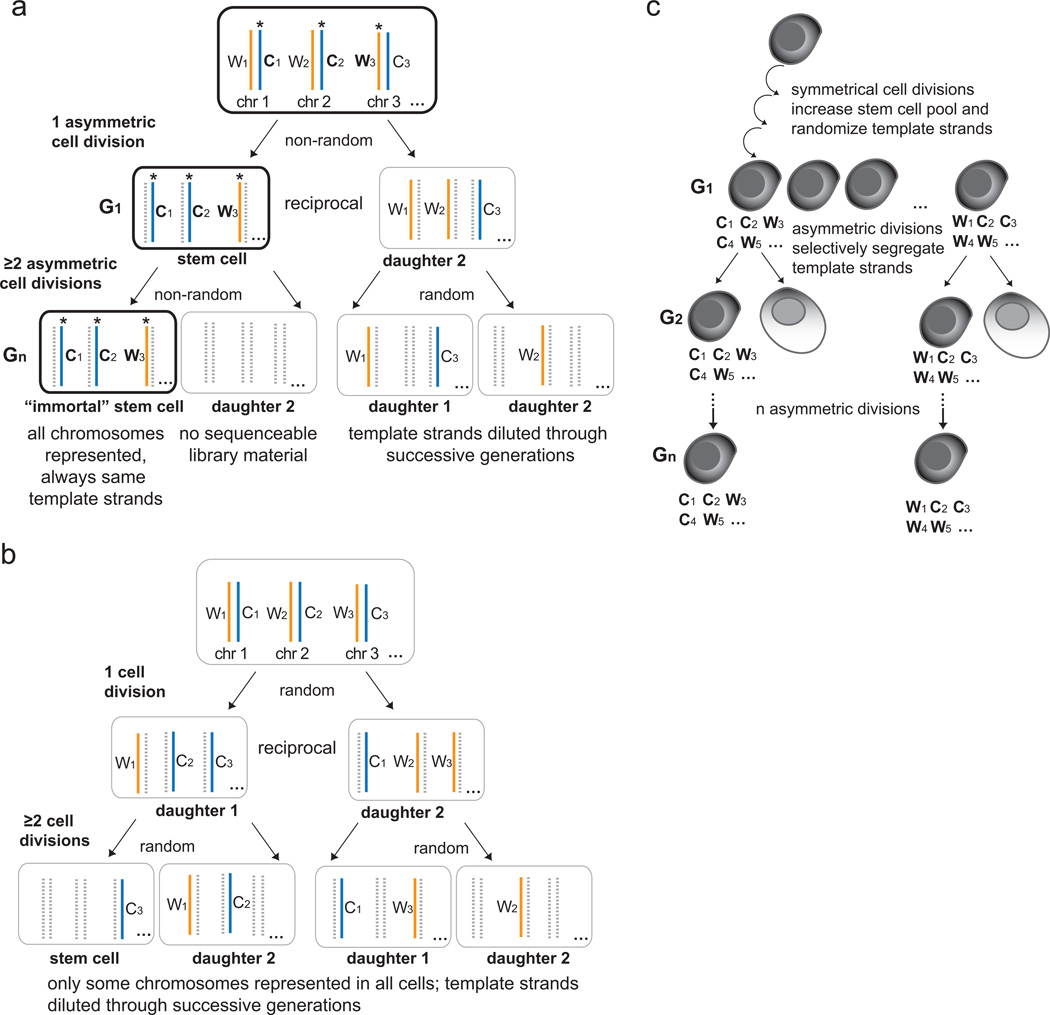
To test the ISH, Strand-seq can identify all template strands inherited by a daughter cell, and could therefore identify patterns of template strand retention through successive cell divisions in vitro (Fig. 3). For example, a self-renewing stem cell expected to retain a set of “immortal” template strands during one asymmetric cell division would also be expected to retain the same set of template strands after more divisions in order to support the ISH (Fig. 3a). This obligate retention of template strands by the self-renewing stem cell lineage means that after two or more generations of culture in BrdU, only the “immortal” cell will have all chromosomes represented in Strand-seq libraries since all other granddaughter cells will have diluted the original template strands by random segregation in successive cell divisions, and all nascent DNA formed during replication in the presence of BrdU will be excluded from the libraries (Fig. 3a, bottom panel). Conversely, if dividing cells segregate template strands randomly, or only a subset of template strands is co-segregated to stem cells, then only a subset of all chromosomes will be represented in a Strand-seq library in all granddaughter cells after two or more generations of culture in BrdU (Fig. 3b). If any cell is shown to exhibit an “immortal” Strand-seq pattern (Fig. 3a), ideally, it should also be identified as a stem cell either phenotypically or with functional assays. However, even with such identification, there is still no available method to compare the accumulation of replication errors in the immortal cell versus the other cells, to formally test the ISH [25].
Figure 3.
Using Strand-seq to test the Immortal Strand Hypothesis. a. In a model of selfrenewing asymmetric cell division, the “immortal” stem cell (black outline) is always expected to inherit the same set of “immortal” Watson and Crick template strands (asterisks) for every chromosome in any number of asymmetric cell divisions (G1-Gn). Since Strand-seq sequences only template strands (solid blue and orange lines) and excludes BrdU-substituted newly-formed strands (dashed grey lines), the first cell division will result in reciprocal Strand-seq libraries (G1). In the second cell division in the presence of BrdU, obligate co-segregation of template strands to the renewed stem cell will result in all chromosomes present only in the Strand-seq library of the granddaughter stem cell. Its sister cell is expected to have no sequenceable library material since it will have inherited a full complement of BrdU-substituted chromosomes. The other two granddaughters will have only a subset of chromosomes detected since the original mortal template strands will have been randomly segregated and diluted through successive generations. b. If there is random segregation, then all cells from the second cell division will result in Strand-seq libraries with only a subset of chromosomes represented in each granddaughter cell. c. Individual stem cells from a population that has expanded through a period of symmetric divisions could have a different set of “immortal” template strands than other stem cells from the same pool (C, Crick; W, Watson). Strand-seq can identify these template strands when applied to multi-generational assays. If the Immortal Strand Hypothesis is true, each individual stem cell lineage will retain the same set of template strands as the previous generation once asymmetric divisions resume (G1-Gn). Note that in this figure only one of the two parental homologs is shown.
It is predicted that symmetrical divisions increase the stem cell pool during development or regeneration of tissues after injury [41] likely via random segregation mechanisms. This would randomize template strands that are to become “immortal” during subsequent asymmetric divisions (Fig. 3c). Therefore, a given stem cell could have a different set of “original” template strands than its sister stem cell or another stem cell from the expanded pool, once it starts to undergo asymmetric divisions. In addition, the complementary Watson and Crick strands of any given chromosome in this expanded stem cell pool will differ in age from each other by at least one replication cycle. For these reasons, Strand-seq would have to be performed on multiple generations of daughter stem cells to test if the pattern of complete template strand co-segregation is maintained in consecutive cell divisions. After one asymmetric division, Strand-seq will show reciprocal template strand patterns in sister cells (Fig. 3a, middle panel) therefore it is not possible to determine which daughter cell contains the “original” template strands unless a subsequent asymmetric division is assayed. It is important to note that in any application of Strand-seq to test the ISH it will be necessary to carefully differentiate symmetric and asymmetric divisions in any model system to be studied.
7. Testing the silent sister hypothesis with Strand-seq
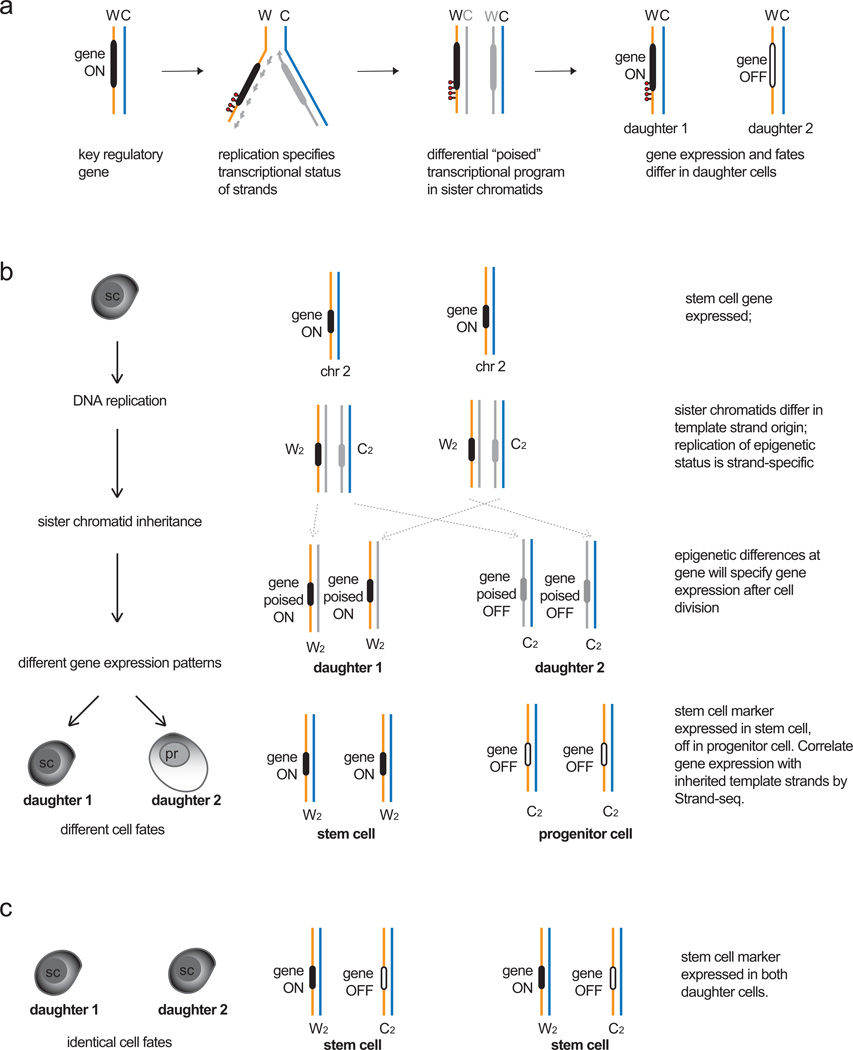
Strand-seq is also ideal to test the silent sister hypothesis (SSH), which predicts that inheritance of certain chromatids with epigenetic signatures distinct from those of its sister will specify different gene expression patterns and thus different cell fates in the two daughter cells upon cell division. For example, the poised transcriptional status of a gene could differ between sister chromatids depending on whether the gene is encoded on a template strand (thereby maintaining its transcriptional status during cell division) or is encoded on the newly formed strand of the other sister chromatid (thereby losing the transcriptional status during cell division, Fig. 4a). Such differences for poised expression states could be encoded by strand-specific epigenetic marks, epigenetic differences established by leading or lagging strand synthesis or unequal distribution or copying of existing epigenetic marks between sister chromatids (such as the examples given in section 3). For asymmetric cell divisions, such mitotic imprinting could be established at master regulatory genes (such as transcription factors at the head of transcriptional hierarchies) to initiate cell fate differentiation upon division. The prediction of the SSH that inheritance of certain template strands with “poised” transcriptional states at key genes can be tested by combining Strand-seq with gene expression assays (such as whole transcriptome libraries or quantitative amplification of preselected transcripts). The combination of Strand-seq and gene expression on daughter cells of asymmetrically dividing cells can determine whether inheritance of a particular template strand correlates with expression of genes encoded on that strand (Fig. 4b–c). As the SSH predicts, if only a subset of template strands (and associated genes) is necessary to specify cell fate, then Strand-seq will be able to identify those template strands independently for each daughter cell in each division. Since only one cell division is required for Strand-seq, this type of application is possible both in vitro and in vivo as long as BrdU can be administered for only one replication cycle and the resultant daughter cells isolated. Since the Strand-seq library of one daughter cell is the mirror image of the Strand-seq library of the other daughter (for example, inheritance of two Crick template strands for chromosome 1 in daughter 1 will mean inheritance of two Watson strands for chromosome 1 in daughter 2), only one of the two daughter cells will need to undergo Strand-seq and the other daughter could undergo functional assays. Therefore it is possible to isolate sister cells after the division of single asymmetrically dividing cells in vitro and test for template strand inheritance, gene expression as well as incorporate phenotypic or functional analysis to identify daughter cell fates. If the isolation of live sister cells is not possible, (as for in vivo experiments) then bulk daughters after cell division can be sorted based on expression of certain phenotypic markers. However, cells sorted based on phenotype must be carefully defined with known cell surface markers or functional assays to ensure a single population is analyzed. Otherwise, a mixed population of cells could affect interpretation of Strand-seq and expression data.
Figure 4.
The Silent Sister Hypothesis. a. Model for establishing different “poised” transcriptional states during DNA replication. For a given gene upon replication (black), strandspecific epigenetic marks (red) or epigenetic differences arising from leading strand (solid grey line) or lagging strand (dashed grey line) synthesis could lead to different poised epigenetic states between sister chromatids after replication is complete. Upon cell division, these differences could be translated as differential gene expression in the daughter cells. b. Differences in daughter cell identity could arise from inheritance of specific template strands and gene expression states in models of asymmetric cell division. If a stem cell expresses a key regulatory gene that specifies stem cell identity, then co-segregation of both Watson template strands encoding the gene and its expression status to the daughter stem cell (daughter 1) would maintain stem cell fate. Daughter 2 (progenitor, pr) no longer expresses the gene, therefore a differentiated fate program can be initiated. A symmetrical cell division could specify the same fate upon expression of the gene in both daughter cells. Strand-seq can identify patterns of template strand inheritance, which could then be correlated with gene expression patterns and cell fate programs in the daughter cells to test the Silent Sister Hypothesis.
Testing the SSH in a multi-generation assay predicts that co-segregation of certain template strands will correlate with expression of genes encoded on that strand for each daughter cell in an asymmetric cell division. For example, all daughter cells fated to be stem cells will co-segregate the same template strands and express the same key stem cell genes, whereas the other daughter fated for downstream differentiation will co-segregate another subset of template strands and express genes specific for its new fate. According to the SSH, this pattern should be maintained for any asymmetric cell division in this self-renewing lineage.
8. The importance of Sister Chromatid Exchange analysis in studies of template strand inheritance
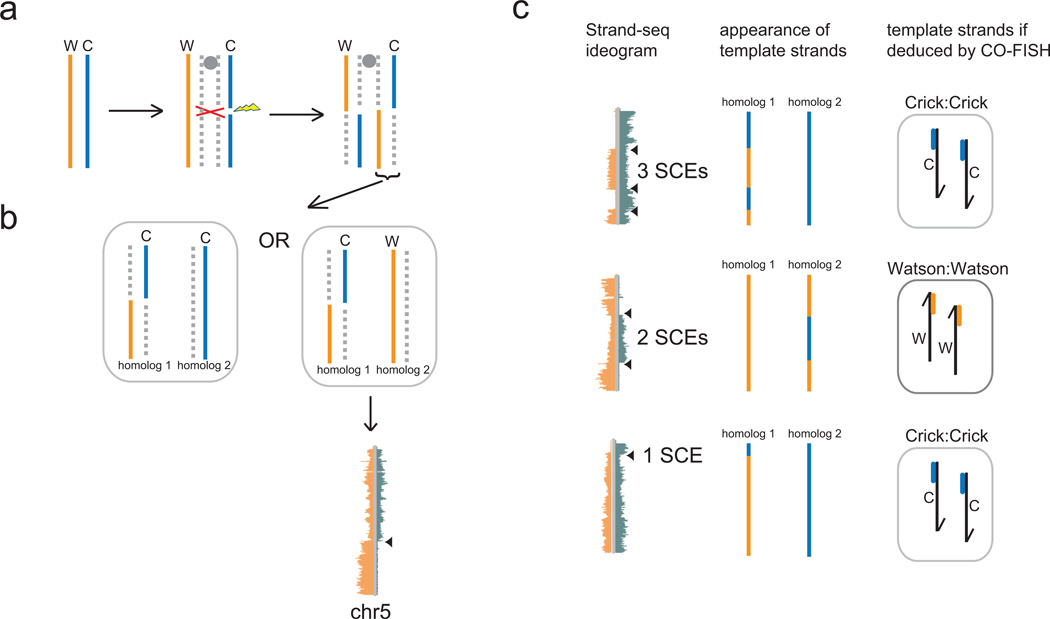
A major assumption of the ISH is that all sister chromatid exchanges are suppressed in asymmetrically dividing stem cells, otherwise a mixing of template and non-template strands would occur prior to chromatid segregation (Fig. 5a) thereby de-protecting the genome. In pulse-chase studies that show obligatory non-random segregation of template strands, it may not be possible to detect SCE events because the small deviations from total asymmetry of the nucleotide analog labels may not be detectable above background. Even with CO-FISH labeling to distinguish sister chromatids [3, 38], it is possible to mis-assign the identity of the inherited template strand because the small probes used to assign Watson or Crick template strands do not represent the entire length of the chromosome (Fig. 5c). Strand-seq of single cells is not only ideal to assay for template strand inheritance, but it simultaneously detects and maps sister chromatid exchanges at high resolution (Fig. 2d and [4]). To use Strand-seq to test the ISH as described in section 6 and Fig. 3, one predicts that SCEs will only be detected in the symmetrically dividing progeny fated for differentiation and not the self-renewing stem cells. One caveat is that the presence of BrdU required for Strand-seq may induce or increase the likelihood of SCEs in the cells being tested [42]. However, if true SCE suppression mechanisms exist in immortal cells, one predicts that no SCEs will be detected by Strand-seq in these model systems. Alternatively, it is predicted that the mechanism of template strand co-segregation involves differentiating the centromeres of sister chromatids and the ensuing kinetochore structures, allowing for selective capture and segregation by the mitotic spindle to the correct pole [13, 43] Therefore, even if SCEs are induced by BrdU and escape the suppression mechanism predicted by the ISH, the “intended” immortal strand (as represented by the centromeric region) will still be co-segregated to the correct daughter stem cell, irrespective of the template strands mixing elsewhere in the chromatid. In that respect, the peri-centromeric sequences of the immortal strand should always be detected by Strand-seq in the daughter stem cell for which it was intended (Fig. 5b). The prevalence of SCEs is more compatible with the SSH because the expression of key genes must be correlated only with the inheritance pattern of the portion of the template strand at that gene. Despite the presence of any number of SCEs on a sister chromatid, as long as the “poised” transcriptional state of key genes are inherited by the intended daughter cell, then the correct transcriptional program will be initiated after cell division.
Figure 5.
Sister chromatid exchange (SCE) analysis is an important consideration when testing both the Immortal Strand and Silent Sister Hypotheses. a. An SCE occurs when a DNA double strand break is repaired post-replication by homologous recombination using the sister chromatid as a template. The result is a mixing of the template and newly-formed strands in both sister chromatids. b. Following mitosis one recombined sister chromatid from a is inherited by one daughter cell along with either an intact Crick or Watson template strand from the other parental homologue. The template strand mixing of the recombined chromatid is apparent in Strand-seq libraries as shown for chromosome 5 (from Figure 2d). c. Sister chromatid exchanges that are visible with Strand-seq (ideograms, left panel) would be undetected if the short unidirectional major satellite probes in the CO-FISH procedure were used to identify the template strands inherited by the daughter cell (right panel) for those particular chromosomes. For all three cases shown, CO-FISH would have identified two Watson (top, bottom) or two Crick (middle) template strands inherited, though a mixture of both were inherited by the cell, due to one or more sister chromatid exchanges. In the bottom case, the majority of the homolog 1 template strand is Watson, though CO-FISH would have detected it as Crick. Portions of Figure 5 adapted from ref [4].
9. Template strand co-segregation and stochasticity
To explain non-random segregation in asymmetrically dividing cells a complex mechanism must exist to distinguish sister chromatids and direct the correct chromatids to the correct poles during cell division. The details of how this mechanism could possibly operate in cells has been reviewed elsewhere [13, 43, 44]. Such models involve a multitude of steps that must be coordinated between the replication machinery at the centromere, the kinetochore complexes, the mitotic spindle and the centrosomes. For the ISH, this mechanism must exist for every chromosome in a stem cell in order to co-segregate all template strands. For the SSH, a less complex mechanism may exist if only a subset of chromatids are selectively segregated. However, an additional possibility exists for the SSH which does not involve directed segregation of chromatids, but is consistent with a stochastic model of segregation. If the transcriptional status of a few key regulatory genes encoded on specific strands must be segregated to a daughter cell, then it is possible that the stochastic inheritance of those transcriptional states is sufficient to direct the correct transcriptional program after cell division and specify cell identity or initiate cell fate programs. The daughter cell that inherits the key “active” transcriptional states of these genes will by default have that programmed fate, irrespective of SCEs. This scenario is only reasonable to imagine in asymmetric divisions if a small number of genes on relatively few chromosomes are involved, or are encoded on opposite strands (Fig. 3b). Unless genes are expressed from only one allele, random segregation of sister chromatids would predict that one in four of the daughter cells will inherit a particular transcriptional state at both alleles (if indeed such a state were linked to a particular template strand during replication). No doubt future studies will shed light on whether gene expression is linked in any way to sister chromatid inheritance.
10. Concluding remarks
Whether epigenetic marks are randomly or non-randomly distributed over sister chromatids remains to be determined. Similarly, the evidence for non-random segregation of sister chromatids remains inconclusive. If the latter were supported by further data it seems certain that the mechanisms of non-random chromosome segregation will be much more complex than the scenarios envisioned thus far. For the time being, a comprehensive method to identify template strand segregation patterns is required, and Strand-seq appears to be the most promising candidate. It cannot be formally excluded that the BrdU required for Strand-seq (and pulse-chase approaches) does not interfere with strand segregation patterns or increase the frequency of sister chromatid exchanges. However, titration of BrdU to minimum levels required for Strand-seq shows a low baseline of SCEs in primary cells (3–4 per cell, data not shown) and are similar to spontaneous SCE levels in studies where BrdU incorporation was not employed[45]. In addition, for Strand-seq, cells only undergo one round of DNA replication in the presence of BrdU, bypassing the requirement for a “chase” cycle or the need for a second DNA label. Therefore, DNA is replicated from original template strands, and not from a nucleotide analog-substituted template, which could itself interfere with strand segregation patterns or induce SCEs.
First, it remains to be seen whether evidence of obligatory non-random segregation previously reported using pulse-chase labeling experiments will be confirmed by Strand-seq when applied to the same model systems. Strand-seq cannot test whether the retained template strands have accumulated less replication errors than the nascent strands distributed in downstream generations. However, since technical shortcomings such as the sensitivity of label detection in other current techniques is eliminated, Strand-seq can determine if non-random segregation in these model systems exists, is obligate for all or only a subset of chromosomes, and whether there is correlation of inheriting certain template strands with gene expression and cell fate. In this respect, Strand-seq promises to be a unifying tool in future studies of non-random chromosome segregation.
Highlights.
non-random segregation proposed as mechanism for genome protection or cell fate specification
chromosome segregation studies are limited by reliance on imaging and inferring template strands
a new method (Strand-seq) can identify all template strands in single cells independently
we model experiments using Strand-seq to test both Immortal Strand and Silent Sister Hypotheses
Acknowledgements
The authors thank Dr. Geraldine Aubert and Ashley Sanders for critical review of this manuscript. Work in the Lansdorp laboratory is supported by grants from the Canadian Institutes of Health Research (RMF-92093 & 105265), the US National Institutes of Health (R01GM094146), and the Terry Fox Foundation (018006) Peter Lansdorp is a recipient of an Advanced Grant from the European Research Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129(7):1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255(5505):197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 3.Falconer E, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 463(7277):93–97. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falconer E, et al. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Methods. 2012 doi: 10.1038/nmeth.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meselson M, Stahl FW. The Replication of DNA in Escherichia Coli. Proc Natl Acad Sci U S A. 1958;44(7):671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443(1):13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- 7.Lansdorp PM, et al. Epigenetic differences between sister chromatids? Ann N Y Acad Sci. 2012;1266(1):1–6. doi: 10.1111/j.1749-6632.2012.06505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decordier I, Cundari E, Kirsch-Volders M. Mitotic checkpoints and the maintenance of the chromosome karyotype. Mutat Res. 2008;651(1–2):3–13. doi: 10.1016/j.mrgentox.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 9.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129(7):1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS, et al. The segregation of DNA in epithelial stem cells. Cell. 1978;15(3):899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 11.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115(Pt 11):2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tajbakhsh S. Stem cell identity and template DNA strand segregation. Curr Opin Cell Biol. 2008;20(6):716–722. doi: 10.1016/j.ceb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Charville GW, Rando TA. Stem cell ageing and non-random chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 2011;366(1561):85–93. doi: 10.1098/rstb.2010.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biology. 2007;5(5):e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinin V, et al. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8(7):677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 17.Karpowicz P, et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170(5):721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007 doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26(11):2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- 20.Yadlapalli S, Cheng J, Yamashita YM. Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J Cell Sci. 2011;124(Pt 6):933–939. doi: 10.1242/jcs.079798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escobar M, et al. Intestinal epithelial stem cells do not protect their genome by asymmetric chromosome segregation. Nat Commun. 2011;2:258. doi: 10.1038/ncomms1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schepers AG, et al. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30(6):1104–1109. doi: 10.1038/emboj.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quyn AJ, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6(2):175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 25.Sherley JL. A new mechanism for aging: chemical "age spots" in immortal DNA strands in distributed stem cells. Breast Dis. 2008;29:37–46. doi: 10.3233/bd-2008-29105. [DOI] [PubMed] [Google Scholar]
- 26.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311(5764):1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 27.Klar AJ. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987;326(6112):466–470. doi: 10.1038/326466a0. [DOI] [PubMed] [Google Scholar]
- 28.Bell CD. Is mitotic chromatid segregation random? Histol Histopathol. 2005;20(4):1313–1320. doi: 10.14670/HH-20.1313. [DOI] [PubMed] [Google Scholar]
- 29.Jablonka E, Lamb MJ. The inheritance of acquired epigenetic variations. J Theor Biol. 1989;139(1):69–83. doi: 10.1016/s0022-5193(89)80058-x. [DOI] [PubMed] [Google Scholar]
- 30.Huh YH, Sherley JL. Molecular cloaking of H2A.Z on mortal DNA chromosomes during nonrandom segregation. Stem Cells. 2011;29(10):1620–1627. doi: 10.1002/stem.707. [DOI] [PubMed] [Google Scholar]
- 31.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 32.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151(1):181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineagecommitted cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140(2):183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey SM, Goodwin EH, Cornforth MN. Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet Genome Res. 2004;107(1–2):14–17. doi: 10.1159/000079565. [DOI] [PubMed] [Google Scholar]
- 36.Falconer E, et al. Chromosome orientation fluorescence in situ hybridization to study sister chromatid segregation in vivo. Nat Protoc. 5(7):1362–1377. doi: 10.1038/nprot.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 38.Rocheteau P, et al. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148(1–2):112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 39.Sherley JL. Overlooked areas need attention for sound evaluation of DNA strand inheritance patterns in Drosophila male germline stem cells. J Cell Sci. 2011;124(Pt 24):4137. doi: 10.1242/jcs.096925. author reply 4138-9. [DOI] [PubMed] [Google Scholar]
- 40.Legraverend C, Escobar M, Jay P. "The immortal DNA strand": difficult to digest? Cell Stem Cell. 2010;6(4):298–299. doi: 10.1016/j.stem.2010.03.005. author reply 299. [DOI] [PubMed] [Google Scholar]
- 41.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441(7097):1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 42.Wilson DM, 3rd, Thompson LH. Molecular mechanisms of sister-chromatid exchange. Mutat Res. 2007;616(1–2):11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Lew DJ, Burke DJ, Dutta A. The immortal strand hypothesis: how could it work? Cell. 2008;133(1):21–23. doi: 10.1016/j.cell.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 44.Tajbakhsh S, Gonzalez C. Biased segregation of DNA and centrosomes: moving together or drifting apart? Nat Rev Mol Cell Biol. 2009;10(11):804–810. doi: 10.1038/nrm2784. [DOI] [PubMed] [Google Scholar]
- 45.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334(6053):194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]