Abstract
Marker positions on nine genetic linkage, radiation hybrid, and integrated maps of human chromosome 22 were compared with their corresponding positions in the completed DNA sequence. The proportion of markers whose map position is <250 kb from their respective sequence positions ranges from 100% to 35%. Several discordant markers were identified, as well as four regions that show common inconsistencies across multiple maps. These shared discordant regions surround duplicated DNA segments and may indicate mapping or assembly errors due to sequence homology. Recombination-rate distributions along the chromosome were also evaluated, with male and female meioses showing significantly different patterns of recombination, including an 8-Mb male recombination desert. The distributions of radiation-induced chromosome breakage for the GB4 and the G3 radiation hybrid panels were also evaluated. Both panels show fluctuations in breakage intensity, with different regions of significantly elevated rates of breakage. These results provide support for the common assumption that radiation-induced breaks are generally randomly distributed. The present studies detail the limitations of these important map resources and should prove useful for clarifying potential problems in the human maps and sequence assemblies, as well as for mapping and sequencing projects in and across other species.
Introduction
Genetic linkage (GL) (meiotic) maps and radiation hybrid (RH) maps are tremendously valuable resources for many types of genetic and genomic studies. These maps provide relative positional information for tens of thousands of DNA markers, as well as map distances on a centimorgan or centiray (cR) scale. GL and RH maps greatly facilitate the localization and cloning of disease genes, the prediction of risk of inherited diseases, and the construction of cross-species comparative maps. Also of importance is the use of some of these maps in the human genomewide sequencing projects, in which large-insert clones were selected for DNA sequencing on the basis of their map positions, and maps were one of many tools used in the assembly and validation of the sequenced contigs (Lander et al. 2001; Venter et al. 2001). For example, the National Center for Biotechnology Information’s frequent updates of the public assembly of the human genome uses maps for prevention of incorrect sequence joins during contig construction and for ordering and orienting the contigs on each chromosome (G. Schuler, personal communication). Maps are similarly being used in the mouse-sequencing project and will likely facilitate sequencing projects in other species as well. The information provided on GL and RH maps must be as accurate as possible to optimize these endeavors.
As with any tool based on experimental methods, there are many ways that errors can be introduced. Despite commonly invoked procedures to minimize error, including the removal of genotypes that violate Mendelian rules of inheritance and the duplicate scoring of all RH markers, it is well established that significant levels of genotyping and RH scoring errors (estimated at 1%–8%) exist in public data sets (Brzustowicz et al. 1993; Hudson et al. 1995; Schuler 1997; Stewart et al. 1997; Broman et al. 1998). Therefore, a complete understanding of the accuracy and limitations of maps constructed by these methods and from these types of data is critical.
Historically, manual annotation of chromosomal maps by expert users has produced maps of excellent quality. Long stretches of contiguous DNA sequence are now available, and it is possible to directly compare the marker order that is determined by mapping methods with the marker order that is observed in the sequence. In particular, with the publication of the complete DNA sequence of human chromosomes 21 and 22 (Dunham et al. 1999; Hattori et al. 2000), one can begin to evaluate the accuracy of GL and RH maps on a chromosomewide level. Specific markers or chromosomal regions that have been mismapped or mispositioned in the sequence assembly may now be identified, and a greater understanding of what causes such discrepancies may lead to improvements in both mapping and sequence-assembly methods. The availability of nearly complete DNA sequences for chromosomes 21 and 22 also allows for a chromosomewide comparison of linkage- and RH-based estimated map distances with true physical distances, thereby facilitating the identification of chromosome-breakage jungles and deserts and providing insight into how recombination- and radiation-induced breaks are distributed along the chromosome. In addition, the ability to study breakage intensities for both linkage and RH maps provides a unique opportunity to look for shared regions of either increased or decreased breakage frequency.
The three main objectives of this study were to evaluate how well marker positions on GL, RH, and integrated maps compare to their positions as identified in the sequence, to identify specific markers or chromosomal regions that may have been incorrectly mapped or assembled, and to compare intermarker map-based distances with actual physical distances. Under the assumption that the sequence-based marker order is largely correct, an indirect result of these studies is an objective comparison of the relative accuracy of several different GL and RH maps. These studies will provide a greater understanding of the value and limitations of GL and RH maps and may ultimately lead to improvements in the techniques used in map construction and/or sequence assembly. The results should prove useful, not only for clarifying potential problems with the previously constructed maps and assemblies in humans but also for mapping and sequencing projects now under way in many other species. Finally, these results will be of particular interest to researchers studying specific regions of human chromosome 22 and homologous nonhuman chromosomal regions.
Material and Methods
Selection of Maps and Marker Sets
Nine comprehensive, genomewide, and frequently cited GL, RH, and integrated maps of human chromosome 22 were used for the present study (table 1). The three GL maps were produced at the Cooperative Human Linkage Center (CHLC) (Murray et al. 1994), Généthon (Dib et al. 1996), and the Marshfield Center for Medical Genetics (Broman et al. 1998). The four RH maps consisted of the individual Genebridge 4 (GB4) and Stanford G3 maps that combine to form the GeneMap’99 human transcript map (GM99-GB4 and GM99-G3) (Deloukas et al. 1998), the GB4 map produced at the Whitehead Institute Center for Genetics Research (WI) (Hudson et al. 1995), and the G3 map produced at the Stanford Human Genome Center (SHGC) (Stewart et al. 1997). The two integrated maps—the Genetic Location Database (LDB) (Collins et al. 1996) and the Unified Database for Human Genome Mapping (UDB) (Chalifa-Caspi et al. 1998)—derive map positions on the basis of a combination of information from cytogenetic, GL, RH, cross-species comparative, and physical maps. All nine of these maps consist primarily of PCR-based STS and EST markers. For the GL and RH maps, only markers that were assigned a specific map position were included, because markers assigned to map bins or intervals cannot be fairly evaluated. In addition, whenever possible for the GL and RH maps, only markers whose map positions were statistically well supported (LOD >3) were included in the present analysis. All markers on the integrated maps for which primers and amplimer sizes were available were included in the analyses. A total of 2,160 markers, representing 1,062 unique primer pairs, were evaluated over all nine maps. Marker information (aliases, map positions, and primer sequences) was obtained from the Genome Database or, for the integrated maps, directly from the LDB and UDB Web sites.
Table 1.
Description of the Maps Used for Analysis
| Map Type and Source | Data | No. of Markersa | Map Resolution (kb)b | Reference |
| GL: | ||||
| CHLC | CEPH | 16 | 1,732 | Murray et al. 1994 |
| Généthon | CEPH | 56 | 563 | Dib et al. 1996 |
| Marshfield | CEPH | 85 | 384 | Broman et al. 1998 |
| RH: | ||||
| GM99-G3 | G3 | 88 | 340 | Deloukas et al. 1998 |
| SHGC | G3 | 105 | 301 | Stewart et al. 1997 |
| GM99-GB4 | GB4 | 158 | 227 | Deloukas et al. 1998 |
| WI | GB4 | 166 | 211 | Hudson et al. 1995 |
| Integrated: | ||||
| LDB | Mixed | 613 | 58 | Collins et al. 1996 |
| UDB | Mixed | 605 | 58 | Chalifa-Caspi et al. 1998 |
No. of markers for which sequence positions were identified using e-PCR.
Physical length of the map divided by no. of map intervals (no. of markers − 1).
Identification of Markers in DNA Sequence
The May 19, 2000, chromosome 22 DNA sequence available at the Sanger Centre was used for the present studies. This current release consisted of 12 disjoint contigs spanning 34.6 Mb and is an updated version of the originally reported completed sequence (Dunham et al. 1999). Electronic PCR (e-PCR) (Schuler 1997) was used to identify the sequence positions of markers in each of the evaluated maps. The number of allowed primer-base mismatches (N) and the allowable variation from the reported amplicon length (M) were tested against a training set of markers. Values of N >2 substantially increased the ratio of false-positive to true-positive results, whereas values of M >1,000 had no consequential effect upon match totals. Accordingly, the complete set of primer sequences was queried against the chromosome 22 sequence, using six sets of parameters (N=0, 1, or 2; M=50 or 1,000). The six different sets of N and M parameters were applied in a gradient from most stringent (N=0; M=50) to least stringent (N=2; M=1,000) (table 2), using only the e-PCR match found at the highest stringency for each marker. Only a small fraction of markers that were not identified by e-PCR could be unambiguously identified in the sequence by use of the BLAST sequence–alignment algorithm; therefore, analyses were restricted to the set of e-PCR–based matches.
Table 2.
e-PCR Method and Cumulative Results
| N | M | No. (%) of Identified Markers |
| 0 | 50 | 765 (85.3) |
| 1 | 50 | 839 (93.5) |
| 2 | 50 | 872 (97.2) |
| 0 | 1,000 | 893 (99.6) |
| 1 | 1,000 | 895 (99.8) |
| 2 | 1,000 | 897 (100) |
Comparison of Map Positions
In the present study, one map in each comparison was either a GL, an RH, or an integrated map, and the other (sequence) map consisted of the DNA sequence positions of the markers on the first map. Two maps of the same region can be compared using a number of methods, each providing a different picture of how the marker positions on each map compare. For the present study, two different types of comparisons were used (one purely visual, i.e., qualitative, and the other quantitative), but neither approach alone provided enough detail to appropriately summarize the comparisons. Therefore, the results from both analyses, as well as detailed descriptive text, are provided.
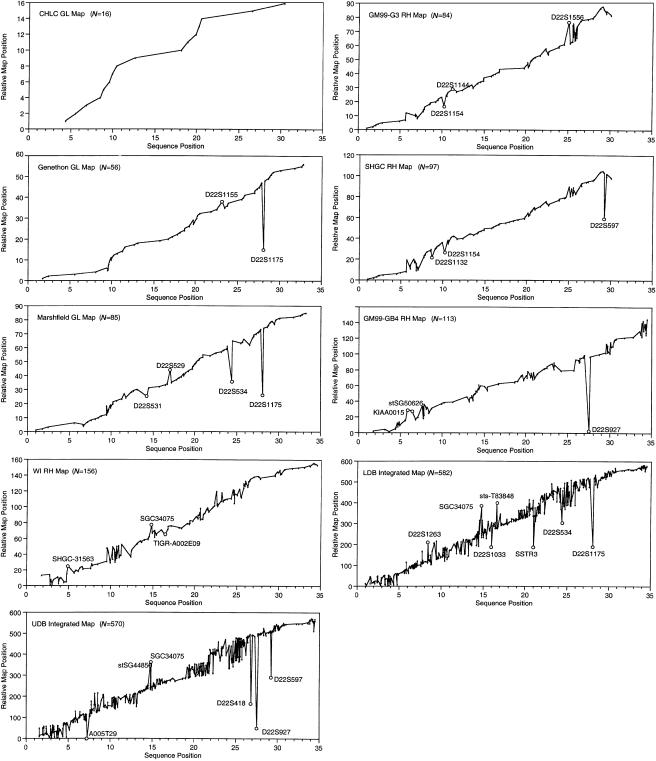
The visual comparisons were obtained by plotting the linear order of markers on each map against the position of the same markers on the sequence (fig. 1). For each map-versus-sequence comparison, the sequence positions were assumed to be correct and the markers were sorted according to their sequence position. The corresponding relative position of each marker on the maps was identified, and these positions (absolute sequence position, relative map position) make up the X and Y values of each point that is plotted on the graph. Therefore, the scale of the X-axis is the same across all comparisons, facilitating evaluation across multiple maps. The position of markers along the X-axis reflects the actual marker density, as identified in the sequence, whereas the markers are evenly spaced along the Y-axis. Markers that follow an increasing slope have map orders that are consistent with sequence order. Single markers that are out of position, inverted sets of markers, and other inconsistent sets of markers are clearly discerned by deviations from an increasing slope. Map comparisons plotted in this manner are particularly useful for comparative evaluation across multiple maps.
Figure 1.
Plots of marker positions on nine maps versus sequence-based positions. For each map, values on the X-axis represent the observed sequence position of each marker, and values on the Y-axis represent the relative position of each marker on the map. The names of isolated markers whose map positions are displaced by >1 Mb from their sequence positions are indicated on the graphs. The DNA sequence starts near the centromere (X-axis position 0) and proceeds to the telomere.
A number of recently applied methods were considered for quantitative comparisons of each map with the sequence map (Agarwala et al. 2000; Olivier et al. 2001; Tapper et al. 2001). However, none of these methods fully quantifies the complex nature of map-versus-sequence comparisons. Many types of discrepancies are observed: single markers that are out of position, pairs or triplets of markers whose relative order is reversed, larger sets of markers whose orders are scrambled, and various combinations of these types of inconsistencies. We used a method similar to the one outlined by Tapper et al. (2001) to identify approximate sequence positions that correspond to each map position. Specifically, markers were sorted according to both the sequence data and the map results. Implied sequence positions for discordant markers were calculated by interpolation. For example, suppose markers a and b have the same order in the sequence and on a given map and further suppose that marker z maps between a and b in a position that is discordant with its identified sequence position. If the map locations for a, b, and z are M_a, M_b, and M_z and if the sequence locations are S_a, S_b, and S_z, respectively, then the implied sequence position for z is S_z = S_a + (S_b − S_a) (M_a − M_b)/(M_z − M_a). The algorithm proceeds iteratively, with positions being interpolated for the most-discordant markers before less-discordant markers (Morton et al. 1992). The algorithm terminates when all implied marker positions agree with observed sequence-based positions. These interpolations assume that the observed sequence positions for the concordant markers are correct and that the observed map distances are correct.
Once implied sequence positions have been identified for all markers in a given map, these can be compared with observed sequence positions. We identified the percentages of markers whose implied map positions are within 250 kb, 500 kb, or 1 Mb of their observed sequence position. To objectively evaluate each map, the sequence was used as an index against which each map was compared. In some cases, the number of markers used for this comparison is smaller than the number for which sequence positions were identified. This is because markers for which more than one sequence position was identified must be excluded from the analysis. The length of the longest contiguous or adjacent set of markers for which the relative map and sequence orders agree exactly was also determined, as an additional objective measure of map quality.
Map Distances versus Physical Distances
For a comparison of GL map distance versus physical distance, the set of Généthon markers from chromosome 22 was analyzed. Of the 67 Généthon markers on chromosome 22 with single map positions, sequence positions for 56 were identified using e-PCR. To help ensure accurate estimates of map distance, under the assumption that the sequence-based order of these markers is correct, five markers whose map positions were discordant with their corresponding sequence positions were removed (see “Comparison of Map Positions” section). Linkage analyses, recombination frequency, and Kosambi map distances were computed for this map, using the MultiMap program (Matise et al. 1994), and the sex-specific map distances were then used for comparison with sequence-based intermarker distances. Genotype data were obtained from the public database at CEPH.
Two sets of markers were used for comparisons of the RH map distance versus physical distance: a set of markers scored in the low-resolution GB4 panel (Gyapay et al. 1996) and a set of markers scored in the medium-resolution G3 panel (Stewart et al. 1997). For the GB4 comparison, a subset of markers was chosen at ∼1-Mb resolution, reflecting the approximate mean resolving power of this panel. Similarly, for the G3 panel, a subset of markers was chosen at ∼500-kb resolution. In both cases, the chosen subset was restricted to markers whose order on the RH map was consistent with their order in the sequence. Linkage analyses, frequencies of breaks, and map distances were computed using the MultiMap program (Matise et al. 1994; Matise and Chakravarti 1995). Radiation hybrid scores were obtained from the Radiation Hybrid Database (Rodriguez-Tome and Lijnzaad 2001).
The rates of recombination and chromosome breakage per unit of physical distance (breakage intensities) were initially determined as the map size of each interval divided by the base pair length of each map interval. Breakage intensities are expressed as centimorgans per megabase for the recombination-based analyses and as centirays per megabase for the radiation-based analyses. Owing both to errors present in the data and to the well-known tendency of derivative estimators to amplify noise, plots of the raw breakage intensity showed a highly fluctuating pattern (data not shown). To more precisely determine the rate of change of genetic distance relative to sequence distance, the software LOCFIT (Loader 1999) was used to fit a local quadratic regression to the map distance as a function of the sequence distance. The local quadratic functions were based on weighted sliding windows. The linear component, or the first derivative, was extracted from the local quadratic fit at each marker position. This local slope can be thought of as a good approximation of the first derivative of the underlying function relating map distance to sequence distance. We used generalized cross-validation to select the best values of the tuning parameter (Craven and Wahba 1979). Critical values for a significance level of α=.05 were determined by use of a Monte Carlo technique. For each data set, the total number of observed chromosomal breaks was randomly generated from the uniform distribution. The original marker locations were used to create a new genetic distance function and to identify the local slopes. The minimum and maximum values were recorded and the entire process was then repeated 10,000 times. The 5th percentile of the minimums and the 95th percentile of the maximums were used for the critical values. In addition, the Kolmogorov-Smirnov goodness-of-fit test was used to compare the observed patterns of recombination and radiation-induced breakage with each other and with a random uniform distribution.
Results
Identification of Markers in Sequence
Nine comprehensive and frequently cited chromosome 22 GL, RH, and integrated maps (together representing 1,062 unique markers) were used for the present study (table 1). By use of our system of gradient e-PCR, positions for 897 (84%) of these markers were identified in the finished chromosome 22 DNA sequence, a success rate comparable to other similar studies (Olivier et al. 2001). There are several possible reasons why we were unable to detect the remaining 16% of markers, including the possibility that some may lie in genomic regions not yet sequenced (which could be as much as 8 Mb [Tapper 2001]) and that errors may be present in reported primer sequences, amplimer lengths, and local sequence assembly. The vast majority (86%) of markers identified by e-PCR were identified using the most stringent parameters (table 2), with the remainder being identified only with less stringent parameters. A summary of the numbers of markers that were identified by each of the six e-PCR parameter iterations is shown in table 2. The markers and associated e-PCR results for all 897 markers are provided in a table at the authors' Web site.
Comparison of Map Positions
The linear X,Y-plots of maps versus sequence are shown in figure 1. These provide a quick visual comparison of each map versus its corresponding sequence map. An increasing slope indicates map positions that are concordant with sequence positions, whereas deviations indicate markers or groups of markers whose map and sequence positions are discrepant. These graphs clearly show that the CHLC GL map fits its corresponding sequence map perfectly and that marker positions on the other maps differ from corresponding sequence positions to varying degrees. The number of inconsistent markers generally rises with increased marker density.
The proportion of markers whose map positions are within 250 kb, 500 kb, and 1 Mb of their assumed correct position provides an objective, quantitative measure of concordance that is used here to compare marker positions between maps and sequence. The proportions are 100%–35% (table 3) and are significantly correlated with the number of markers on each map (250 kb, P=.0164; 500 kb, P=.0013; 1 Mb, P=.0029), with the least-dense maps showing the highest percentage of markers near their estimated sequence positions. The longest adjacent set of markers for which the relative map and sequence orders agree exactly is also shown in table 3 and was relatively short for most maps.
Table 3.
Results of Map Evaluations
|
Percent with Displacement of |
||||||||
| Map Type and Source | Size(Mb) | StartPosition | EndPosition | No.Compared | <250 kb | <500 kb | <1 Mb | No. in LCSa |
| GL: | ||||||||
| CHLC | 26.0 | 4350294 | 30325144 | 16 | 100 | 100 | 100 | 16 |
| Généthon | 31.0 | 1803371 | 32798620 | 56 | 70 | 86 | 95 | 11 |
| Marshfield | 32.2 | 1011913 | 33235613 | 85 | 53 | 73 | 87 | 9 |
| RH: | ||||||||
| GM99-G3 | 28.9 | 1113027 | 30013100 | 84 | 78 | 85 | 89 | 12 |
| SHGC | 28.9 | 1113027 | 30013100 | 97 | 69 | 81 | 90 | 15 |
| GM99-GB4 | 32.7 | 1803371 | 34486007 | 113 | 62 | 79 | 88 | 5 |
| WI | 32.7 | 1803398 | 34455098 | 156 | 44 | 67 | 87 | 4 |
| Integrated: | ||||||||
| LDB | 33.5 | 1011913 | 34486007 | 582 | 38 | 56 | 74 | 4 |
| UDB | 32.9 | 1537694 | 34486007 | 570 | 35 | 48 | 67 | 4 |
LCS = longest contiguous set of markers for which the relative map and sequence orders agree exactly.
Detailed Analysis of Nine Maps
Because neither of the comparative methods described above can fully capture the complex nature of the types of discrepancies observed between marker map and sequence positions, a detailed manual comparison was also done. As the density of markers increases, it becomes much more difficult to classify observed inconsistencies. Detection of specific markers with inconsistent positions was straightforward for GL maps. On the more dense RH and integrated maps, it is possible to identify some markers that are clearly misplaced in either the map or the sequence, but the bulk of the discrepancies are larger groups of inconsistently ordered markers, separated by short stretches of consistent markers. Characterization of the observed discrepancies is given in table 4, with additional comments provided in the paragraphs immediately below. Specific markers that are obviously misplaced are described in table 4. The sizes of the displacements were calculated under the assumption that the sequence order is correct and that the discrepancies represent errors in the maps.
Table 4.
Detailed Description of Observed Inconsistencies between Maps and DNA Sequence
|
No. of |
|||||
| Map | Inversionsa | InconsistentGroups (Markers)a | Misplaced Markers | Misplaced Marker | EstimatedDisplacement(Mb)b |
| Généthon | 2 | 0 (0) | 3 | D22S1175 | −15.7 |
| D22S1155 | +1.9 | ||||
| D22S277 | −.25 | ||||
| Marshfield | 3 | 0 (0) | 7 | D22S1175 | −15.2 |
| D22S534 | −7.6 | ||||
| D22S529 | +2.2 | ||||
| D22S531 | −1.3 | ||||
| D22S444 | −.346 | ||||
| D22S1163 | −.329 | ||||
| D22S533 | −.466 | ||||
| GM99-G3 | 1 | 4 (27) | 4 | D22S1154 | −2.1 |
| D22S1144 | +1.6 | ||||
| D22S1556 | +1.3 | ||||
| SHGC-30811 | +.472 | ||||
| SHGC | 1 | 5 (27) | 8 | D22S597 | −9.5 |
| D22S1154 | −2.2 | ||||
| D22S1132 | −2.1 | ||||
| SGC34055 | −.98 | ||||
| D22S1556 | −.929 | ||||
| D22S678 | −.925 | ||||
| D22S1674 | +.828 | ||||
| SHGC-7765 | +.242 | ||||
| GM99-GB4 | 3 | 8 (72) | 10 | D22S927 | −24 |
| KIAA0015 | +2.3 | ||||
| stSG50626 | +2.0 | ||||
| Ib1320 | +.736 | ||||
| EMBL-T95789 | −.590 | ||||
| stSG30356 | −.391 | ||||
| TIGR-A004X26 | −.383 | ||||
| D22S1257 | −.216 | ||||
| sts-N72133 | +.211 | ||||
| stSG4190 | −.203 | ||||
| WI | 2 | 9 (120) | 4 | SGC34075 | +3.8 |
| SHGC-31563 | +3.0 | ||||
| TIGR-A002E09 | −1.2 | ||||
| WI-15873 | −.195 | ||||
| LDB | NA | NA | 7 | D22S1175 | −15.4 |
| SSTR3 | −9.6 | ||||
| D22S1263 | +5.1 | ||||
| D22S534 | −6.4 | ||||
| sts-T83848 | +5.2 | ||||
| SGC34075 | +6.8 | ||||
| D22S1033 | −4.6 | ||||
| UDB | NA | NA | 6 | D22S927 | −23.7 |
| D22S418 | −16 | ||||
| D22S597 | −9.8 | ||||
| SGC34075 | +8.6 | ||||
| stSG44859 | +8.5 | ||||
| A005T29 | −7.3 | ||||
NA = inconsistencies too complex to characterize.
+ = positive displacements (map position closer to telomere than sequence position); − = negative displacements.
CHLC GL map
All 16 of the framework markers on this map are in positions consistent with their sequence-based positions.
Généthon GL map
The marker D22S1175 has been previously identified as having a map position that is inconsistent with its sequence location (Dunham et al. 1999).
Marshfield GL map
Interestingly, four of the seven misplaced markers were genotyped in only four CEPH pedigrees, but the majority (83%) of markers were scored in a larger sample of eight CEPH pedigrees.
LDB integrated map
It should be noted that the LDB does provide a “rank” value for each marker, which gives an indication of the degree of support for each marker’s position on the map. Because only a small minority of markers receives the higher ranking, all markers were included in our analysis, regardless of their rank. Recently, the LDB has computed a new, sequence-based map of chromosome 22 that should provide a more useful general map resource for this chromosome (Tapper et al. 2001).
Comparisons across maps
The seven GL and RH maps were scanned for shared regions of marker-order discrepancies, which might indicate regions with common mapping or sequence assembly problems. The CHLC and Généthon GL maps did not contribute to the analysis, because their marker density was too low. The integrated maps could not be used for the present analysis, because they contain a high degree of order inconsistency. There are four chromosomal regions that appear to show order discrepancies involving different markers across multiple maps. Because each map contains different markers and different marker densities, the endpoints of each shared inconsistent region are approximate. Regions were noted only if they were observed across both GL and RH maps.
The first such region shows varying types of marker-order discrepancies on five maps (Marshfield, GM99-G3, SHGC, GM99-GB4, and WI) and spans 5.6–7.0 Mb (Sanger Centre Chromosome 22 Gene Annotation Group, unpublished data). On the Marshfield GL map, there are three markers in this region, and their order on the map is simply inverted compared with their order as observed in the sequence. The GM99-G3 RH map has five markers in this region that also generally show an inverted order. The SHGC (G3) RH map has 12 markers in this region whose map order is completely scrambled compared with sequence order. The GM99-GB4 RH map has five markers here; this includes an insertion of two markers, each displaced by ∼2 Mb. The WI RH map has eight markers in this area, six of which have a scrambled map order in comparison with sequence order. This region includes the locally duplicated immunoglobulin l gene family, which contains >36 potentially functional gene segments and >90 pseudogenes or other related segments spanning positions 5913524–6717195 (Dunham et al. 1999). This region is also located within the 22q11 low-copy-repeat family (positions 2.5–8.7 Mb) that has been associated with chromosomal rearrangements leading to the 22q11.2 deletion syndrome causing the DiGeorge and velocardiofacial syndromes.
A second shared segment of inconsistency occurs at ∼20.0–20.8 Mb (Sanger Centre Chromosome 22 Gene Annotation Group, unpublished data). The two G3-RH maps have the same four markers in this region with the same rearrangement, whereas the two GB4-RH maps have more markers in this region with completely scrambled map orders in comparison with sequence order. There are two known sets of repeated genes that map to this region: the APOL2 (20073029–20085188) and APOL (20102771–20111826) set and the CSF2RB (20739089–20757347) and CSF2RB2 (20767438–20770631) set, which is a partial inverted duplication of CSF2RB.
A third shared region of inconsistency lies near two CYP2D genes: CYP2D7P (25944997–25948183) and CYP2D8P (25954605–25959721) (Sanger Centre Chromosome 22 Gene Annotation Group, unpublished data). Each of the four RH maps shows a substantial number of markers whose map order disagrees with the sequence order surrounding these loci.
A fourth segment spans positions at ∼13.3–13.7 Mb and shows various order discrepancies on all four RH maps. No repeated segments have been identified in this region in the Sanger annotation. However, a recent study of segmental duplications does show a duplicated segment in this region (Bailey et al. 2001), with a significant homology between DNA at equivalent positions 13.4–13.5 Mb (13 Mb of DNA that was added to the public consortium sequence to represent the p arm and pericentromeric region has been removed to align with the Sanger sequence of chromosome 22). This region contains one member of the RFPL cluster of three genes (Lander et al. 2001).
Map Distances versus Physical Distances
Recombination-induced chromosome breakage
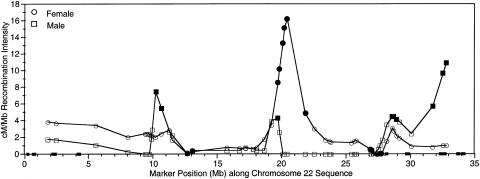
A set of Généthon markers was used to compare recombination-based map distances with physical distance. This subset of 51 markers consisted of all the Généthon markers for which sequence positions were identified by e-PCR, excluding five markers whose map positions were inconsistent with sequence position. Deletion of these markers decreased the estimated sex-averaged map length by 20%, from 70 cM to 56 cM, and the likelihood of the map order improved by several orders of magnitude. The average map interval was 1.1 cM or 620 kb. To plot the rate of recombination versus corresponding sequence distances, or the breakage intensity, we fit local quadratic regressions to the map as functions of the sequence distance. The linear component at each marker position is plotted in Figure 2, with male and female map distances studied separately. A Monte Carlo technique was used to identify those markers whose local breakage intensity was greater than the 95th percentile of the maximums (regions of increased recombination) or was lower than the 5th percentile of the minimums (regions of decreased recombination).
Figure 2.
Recombination intensity plot. Squares represent the rate of recombination in males, circles represent female recombination. Blackened symbols indicate values above the 95th percentile critical values. Small blackened boxes on the X-axis indicate the location and size of the 11 remaining sequence gaps. The DNA sequence starts near the centromere (X-axis position 0) and proceeds to the long arm telomere.
In males the regression curves (fig. 2) showed a range of 0–10.9 cM/Mb, a mean of 1.8 cM/Mb (or 556 kb/cM), and 5th and 95th percentile critical values of 0 and 4.0 cM/Mb. Although the lower critical value was 0, the critical value at the 26.5th percentile was also 0, so that a breakage intensity value of 0 does not represent a significant result. In females the rate of recombination was 0–16.7 cM/Mb, with a mean of 2.8 cM/Mb (or 357 kb/cM), and 5th and 95th percentile critical values of 0.35 and 4.68 cM/Mb.
Although the overall patterns of recombination in male and female meioses appear similar, the markers with significantly increased recombination in males usually did not overlap with those in females (fig. 2), and the distributions of recombination are significantly different (P=.0066). Males have four regions of significantly increased recombination, and the observed pattern of recombination intensity differs significantly from a uniform random distribution (P=.002). The most significant recombination levels in males are seen at the three most telomeric markers (31724302–32798620), where the relative recombination rate is 3–6 times greater than the male average rate and is 7–11 times greater than the local rate in females. Males also show significant breakage intensities spanning two marker positions at 10188627–10602716, where the regression curve shows a relative rate of recombination that is 2.1–2.5 times greater in males than in females, at position 1972362, which falls within a region of significantly increased recombination in females, and also at positions 28592697–28711779. In addition, there is a very notable difference between the male and female rates of recombination in the large 7.9-Mb region at 19723620–27647660, where there is no recombination observed in males. Although a single marker showing a local rate of recombination equal to zero is not statistically significant in males, our simulations showed that it is extremely unlikely that four or more consecutive zero recombination intensities would be observed by chance (P<.0001). This recombination desert partially overlaps the region of greatest breakage intensity in females. Females show six consecutive significant intensities spanning positions 19723620–21802638, with breakage intensities that are two to six times greater than the female average rate and as much as 16 times greater than in males. The observed pattern of recombination intensity in females does not differ significantly from a uniform random distribution (P=.1). The largest sequence gap is estimated to be 200 kb (1/150 the size of the entire chromosome). The small remaining sequence gaps are beyond the resolution of this study.
Radiation-induced chromosome breakage
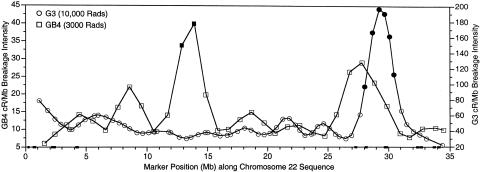
Of the markers scored in the GB4 panel whose sequence positions were identified and whose order matched the corresponding sequence order, a subset of 34 markers, at ∼1-Mb resolution, were chosen for breakage-intensity analysis. A similar procedure was used to select a subset of 65 markers at ∼500-kb resolution scored in the G3 panel. The fitted regression curves of breakage intensities across the chromosome for the G3 and GB4 panel are shown in figure 3. In the GB4 panel, breakage intensities were 6–40 cR3000/Mb, with a mean of 15 (corresponding to 67 kb/cR3000). The 5th and 95th percentile critical values were 1.8 and 30.3 cR3000/Mb, respectively. In the G3 panel, breakage intensities were 22–196 cR10000/Mb, with a mean of 54 (corresponding to 18.5 kb/cR10000), and 5th and 95th percentile critical values of 21.6 and 86.5 cR10000/Mb, respectively. No regions of significantly reduced breakage were observed in either panel.
Figure 3.
Radiation hybrid breakage intensity plot. Squares represent the rate of breakage in the GB4 RH panel; circles represent breakage in the G3 RH panel. Blackened symbols indicate values above the 95th percentile critical values. Small blackened boxes on the X-axis indicate the location and size of the 11 remaining sequence gaps. The DNA sequence starts near the centromere (X-axis position 0) and proceeds to the long arm telomere.
The regression curves for breakage intensity on the GB4 and G3 panels are very different from each other. Each panel shows significantly increased breakage in different regions of the chromosome, and, in other areas, the less significant peaks and valleys do not match. The GB4 panel has two consecutive markers that show locally increased rates of breakage at positions 12890651–13915270, whereas the G3 panel has six consecutive markers that show increased breakage at positions 28063555–30448976. For both the GB4 and the G3 panels, the observed patterns of recombination intensity do not differ significantly from a uniform random distribution (P=.995 and P=.135, respectively). The small remaining sequence gaps are beyond the resolution of this study.
Discussion
The analyses presented here provide the first in-depth look at the level of accuracy of nine commonly used GL, RH, and integrated maps of human chromosome 22. In addition, they provide a detailed examination of the distribution of recombination and radiation-mediated chromosome breakage along this chromosome. Because maps play significant, and sometimes critical, roles in sequence assembly—as well as in validation, gene localization, genetic disease risk prediction, and comparative mapping—it is important to appreciate their limitations and to understand how breakage intensities may fluctuate along the chromosome. Although there are differences in sequence composition and complexity between chromosome 22 and other chromosomes, many of the results presented here should generally extend to the majority of the human and other eukaryotic genomes.
It is important to point out some limitations of our analyses. Although chromosome 22 is considered a “finished” chromosome, its sequence is not yet 100% complete, and there likely remain some localized errors in the sequence assembly. The analyses presented here focused on the maps and sometimes required the assumption that the sequence assembly is correct. Violation of this assumption might somewhat change the observed percentages of markers mapping to within 250 and 500 kb of their assumed correct positions (table 2), but the same relative effects would be observed across all of the maps and would likely have little effect on our overall results. It is quite unlikely that the existence of 11 small remaining gaps in the sequence analyzed here has any major effect on any of our results (the largest gap being only 200 kb). It is safe to assume that, on average, the sequence assembly is probably more correct than are many of the maps. Therefore, many of the misplacements we observe are likely to be due to errors in mapping rather than errors in assembly.
On average, the GL maps show approximately the same level of concordancy with sequence as do the RH maps (excluding the CHLC GL map that matched the sequence order perfectly). Naturally, the proportion of markers whose map position is within 1 Mb of their assumed correct positions is higher than the proportion that map to within 500 or 250 kb. Most of the GL and RH maps give reasonably high concordance at the 1-Mb level (∼90%), fair concordance at the 500-kb level (78%–80%), and poor performance at the level of 250 kb (∼63%). The integrated maps show fair-to-poor concordance at all three levels. Any localized misassembly errors would have greatest impact on the comparisons with the higher-density integrated maps. For all of the GL and RH maps, if the markers that were excluded because of low statistical support for order were instead included in the present analysis, the concordance with sequence positions would decrease substantially. These results confirm what we and others have suspected for some time: that very dense maps, which appear to depict “best” marker orders but include marker positions that are not statistically well supported, are potentially misleading to investigators unfamiliar with the limitations of, or level of statistical support for, a given map. There is clearly a trade-off in precision when trying to maximize the number of markers placed on a map; all maps serve as good localization tools, but not all provide high precision.
These comparisons identify several isolated markers whose map positions are most likely incorrect. Most of these are specific to each map and do not represent any systematic or common error. One exception involves the marker D22S1175 (AFM331WC9), which appears to be mislocalized on all maps that include it (Généthon and Marshfield GL maps and the LDB integrated map). This marker was noted as being incorrectly mapped in the publication of the complete sequence of chromosome 22 (Dunham et al. 1999). As noted by Dunham and colleagues, the actual sequence position of this marker lies within a segment of DNA that has a homologous counterpart 12 Mb proximal. However, the map position of this marker gives an estimated sequence position that is 16–17 Mb proximal and does not include the location of the duplicated segment. Therefore, the presence of this large long-range duplication may not be the sole possible explanation for the misplacement of this marker on these maps. Another marker that is misplaced on multiple maps is D22S927 (AFM320YG5). It is displaced by 24 Mb on the GeneMap’99-GB4 RH map and on the UDB integrated map. This marker is not present on the other RH or integrated maps but is correctly mapped on the Généthon and Marshfield GL maps. Investigation of this marker suggested a marker misnaming error that resulted in the primers for D22S427 being incorrectly assigned to marker D22S927 either during construction of, or after reporting of, GeneMap’99. Other probable data errors include the two markers D22S1154 and D22S1156, which are misplaced on both the GeneMap’99-G3 and the SHGC RH maps, and SGC34075, which is misplaced by WI, LDB, and UDB.
In addition, these analyses reveal four regions with marker discrepancies present on multiple maps. These regions all occur near the positions of locally duplicated genes and/or DNA segments. The presence of duplicated DNA could contribute to mapping and/or assembly errors. Further in-depth evaluation of the sequence and the markers localized in these regions could lead to the identification and correction of these errors.
The present study also provides an in-depth view of the distribution of both recombination- and radiation-induced chromosome breakage along chromosome 22. It delineates clear regions of increased and decreased recombination and rates of recombination that differ between male meioses and female meioses. These patterns of sex-specific breakage match previous sex-averaged observations (Dunham et al. 1999; Majewski and Ott 2000), but follow a pattern quite different from that observed on chromosome 21 (Lynn et al. 2000). The mean rates of recombination across human chromosome 22 are very similar to those observed on chromosome 21: 2.8 cM/Mb in females and 1.8 cM/Mb in males on chromosome 22, compared with 2.4 cM/Mb and 1.6 cM/Mb on chromosome 21, as reported elsewhere by Lynn et al. 2000. This revious study of the distribution of recombination along chromosome 21 showed increasing male recombination from the centromere to the telomere but relatively constant female recombination. However, the distribution of recombination along chromosome 22 is quite different, with each sex showing regions of significantly elevated recombination separated by areas of near zero or zero recombination; there is even an 8-Mb region that shows zero recombination in males but spans 25 cM in females. The same large region of extremely low recombination in males is also seen on the sequence-based integrated map in the LDB (Tapper et al. 2001). The LDB map includes a much greater density of polymorphic markers, several of which have been scored in considerably more families than the standard set of eight used to genotype most Généthon markers. Here, the low-recombination region spans 7.5 Mb, and 1 cM of meiotic distance is observed in males, compared with 22 cM in females.
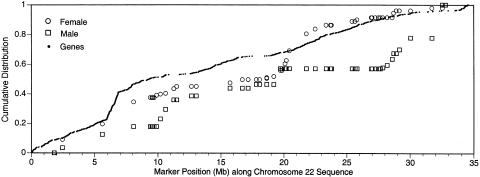
Recombination is greatly elevated at the telomere in males, in comparison with females, as previously noted (Brennan et al. 2000), and this male-to-female disparity at the telomere is among the most extreme of all human chromosomes (A. Lynn and A. Chakravarti, personal communication). A recent chromosome 21 study (Lynn et al. 2000) identified a positive correlation between gene density and recombination intensity in males; but for chromosome 22, no correlation is observed between gene density and either female (P=.001) or male (P<.0001) recombination intensity (fig. 4). There are many region-specific and sex-specific factors postulated to contribute to recombination, including chromatin accessibility, sex-specific gene expression, and gene density (A. Lynn and A. Chakravarti, personal communication). A good understanding of the differences in the distribution of recombination between chromosome 21 and chromosome 22 may facilitate the identification and characterization of some of these factors.
Figure 4.
Plot of cumulative gene density, female, and male recombination along chromosome 22
The comparisons of RH map versus sequence presented here provide the first opportunity to directly assess whether radiation-induced breaks are generally randomly distributed along chromosomes. Initially, it was assumed that radiation-induced chromosome breakage would be generally randomly distributed. However, work by Teague and colleagues indicated that the observed patterns of breakage along human chromosome 21 are not random (Teague et al. 1996). The existence of marked, and occasionally significant, peaks and valleys on the plots of breakage intensity for the G3 and GB4 map (fig. 3) implies that the observed distribution of breaks is not perfectly random. However, the observation that the locations of the peaks and valleys differ between the G3 and the GB4 panel supports the premise that any apparent nonrandom breakage is due to noise in the data rather than the existence of chromosomal regions that are significantly more or less prone to breakage. Furthermore, a test of goodness-of-fit between the observed distributions of breakage and a uniform random distribution supports the hypothesis of randomly distributed breaks at the α=.05 level.
We also compared the patterns of radiation-induced breakage with the male and female patterns of recombination-induced breakage, to address whether any regions of chromosome 22 might be exceptionally susceptible or resistant to both types of breakage. In general, the locations of increased and decreased breakage on the male and female GL maps and on the G3 and GB4 RH maps do not overlap. One possible exception to this observation is the region at 28–29 Mb. Although the breakage intensity in this region is significantly elevated only for the male GL map and the G3 RH map, all four maps do show some degree of elevated breakage in this region.
The mean estimates of breakage per unit of physical distance (cR/Mb) determined from this study are somewhat different from previous observations for the GB4 and G3 panels (Hudson et al. 1995; Stewart et al. 1997), but those earlier studies used an estimated chromosome length of 41 Mb instead of 33.5–34 Mb and may have had different coverage of the chromosome. The mean breakage intensities presented here (GB4:15 cR3000/Mb; G3:54 cR10000/Mb) reflect a more accurate measure of the relationship between RH map distances and physical distances for chromosome 22 for these RH panels.
In summary, a close comparison of genomewide maps has revealed wide variation in the agreement of marker placements between maps and “finished” sequence. This variation appears to be affected to differing extents by marker density, experimental technique, and experimental design. Because maps remain very useful tools for sequence assembly, positional cloning, risk prediction, and comparative genomics, a clear understanding of the relative accuracy of various maps is critical. Accurate maps will also be essential for understanding the regulation of recombination rates and chromosomal breakage between and among chromosomes. The raw results of our sequence analyses are available on the authors' Web site for this project. As sequencing of additional chromosomes nears completion, the reporting of similar comparative analyses would be a first step toward an open forum for dissemination and discussion of mapping and sequence discrepancies.
Acknowledgments
We thank Shahriar Sabuktagin, for valuable help in programming, Greg Schuler, James Ostell, Linda Brzustowicz, Jeff Bailey, Audrey Lynn, and Aravinda Chakravarti, for helpful discussions and additional data, and Victoria Appleton and Ingrid Burgetz, for technical assistance. We are especially appreciative of the helpful comments from two anonymous reviewers. This work was supported in part by National Institutes of Health grants HG01691 (to T.C.M.) and MH60240 (to P.S.W.)
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CEPH, http://www.cephb.fr/cephdb/
- Chromosome 22 Map Comparisons, http://compgen.rutgers.edu/c22maps/e-PCR_results.html
- Genome Database, http://www.gdb.org
- Genetic Location Database, http://cedar.genetics.soton.ac.uk/public_html/ldb.html
- LOCFIT, http://cm.bell-labs.com/cm/ms/departments/sia/project/locfit/ (for software to determine rate of change of genetic distance)
- Radiation Hybrid Database, http://www.ebi.ac.uk/RHdb/ (for radiation hybrid scores)
- Sanger Centre Chromosome 22 Gene Annotation Group, http://www.sanger.ac.uk/HGP/Chr22
- Unified Database for Human Genome Mapping, http://bioinformatics.weizmann.ac.il/udb
References
- Agarwala R, Applegate DL, Maglott D, Schuler GD, Schaffer AA (2000) A fast and scalable radiation hybrid map construction and integration strategy. Genome Res 10:350–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M, Neibergs H, Phillips K, Moseley S (2000) Polymorphic markers for the arylsulfatase A gene reveal a greatly expanded meiotic map for the human 22q telomeric region. Genomics 63:430–432 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Merette C, Xie X, Townsend L, Gilliam TC, Ott J (1993) Molecular and statistical approaches to the detection and correction of errors in genotype databases. Am J Hum Genet 53:1137–1145 [PMC free article] [PubMed] [Google Scholar]
- Chalifa-Caspi V, Prilusky J, Lancet D (1998) The unified database. Rehovot, Israel, Weizmann Institute of Science, Bioinformatics Unit and Genome Center [Google Scholar]
- Collins A, Frezal J, Teague J, Morton N (1996) A metric map of humans, 23,500 loci in 850 bands. Proc Natl Acad Sci USA 93:14771–14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven P, Wahba G (1979) Smoothing noisy data with spline functions. Numerische Mathematik 31:377–403 [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, et al (1998) A physical map of 30,000 human genes. Science 282:744–746 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillet D, Muselet D, Prud'homme J, Dib C, Auffray C, Morissette J, Weissenbach J, Goodfellow PN (1996) A radiation hybrid map of the human genome. Hum Mol Genet 5:339–358 [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, et al (2000) The DNA sequence of human chromosome 21: the chromosome 21 mapping and sequencing consortium. Nature 405:311–319 [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety S, Ma J, Castle A, Silva J, Slonim D, Baptista R, Kruglyak L, Xu S, Hu X (1995) An STS-based map of the human genome. Science 270:1945–1954 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Loader C (1999) Local regression and likelihood. Springer Verlag, New York, pp 101–105 [Google Scholar]
- Lynn A, Kashuk C, Petersen MB, Bailey JA, Cox DR, Antonarakis SE, Chakravarti A (2000) Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res 10:1319–1332 [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J (2000) GT repeats are associated with recombination on human chromosome 22. Genome Res 10:1108–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Chakravarti A (1995) Automated construction of radiation hybrid maps using MultiMap. Am J Hum Genet Suppl 57:A15 [Google Scholar]
- Matise TC, Perlin M, Chakravarti A (1994) Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet 6:384–390 [DOI] [PubMed] [Google Scholar]
- Morton NE, Collins A, Lawrence S, Shields DC (1992) Algorithms for a location database. Ann Hum Genet 56:223–232 [DOI] [PubMed] [Google Scholar]
- Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, Manion F, Quillen J, et al (1994) A comprehensive human linkage map with centimorgan density. Science 265:2049–2054 [DOI] [PubMed] [Google Scholar]
- Olivier M, Aggarwal A, Allen J, Almendras AA, Bajorek ES, Beasley EM, Brady SD, et al (2001) A high-resolution radiation hybrid map of the human genome draft sequence. Science 291:1298–1302 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Tome P, Lijnzaad P (2001) RHdb: the Radiation Hybrid database. Nucleic Acids Res 29:165–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD (1997) Sequence mapping by electronic PCR. Genome Res 7:541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, McKusick K, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, et al (1997) An STS-based radiation hybrid map of the human genome. Genome Res 7:422–433 [DOI] [PubMed] [Google Scholar]
- Tapper WJ, Morton NE, Dunham I, Ke X, Collins A (2001) A sequence-based integrated map of chromosome 22. Genome Res 11:1290–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague J, Collins A, Morton N (1996) Studies on locus content mapping. Proc Natl Acad Sci USA 93:11814–11818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]