Abstract
The effects of interleukin-33 (IL-33) on the immune system have been clearly demonstrated; however, in cardiovascular diseases, especially in coronary artery disease (CAD), these effects have not yet been clarified. In this study, we investigate the genetic role of the IL-33-ST2L pathway in CAD. We performed three-stage case-control association analyses on a total of 4,521 individuals with CAD and 4,809 controls via tag SNPs in the genes encoding IL-33 and ST2L-IL-1RL1. One tag SNP in each gene was significantly associated with CAD (rs7025417T in IL33, padj = 1.19 × 10−28, OR = 1.39, 95% CI: 1.31–1.47; rs11685424G in IL1RL1, padj = 6.93 × 10−30, OR = 1.40, 95% CI: 1.32–1.48). Combining significant variants in two genes, the risk for CAD increased nearly 5-fold (padj = 8.90 × 10−21, OR = 4.98, 95% CI: 3.56–6.97). Traditional risk factors for CAD were adjusted for the association studies by SPSS with logistic regression analysis. With the two variants above, both located within the gene promoter regions, reporter gene analysis indicated that the rs7025417 C>T and rs11685424 A>G changes resulted in altered regulation of IL33 and IL1RL1 gene expression, respectively (p < 0.005). Further studies revealed that the rs7025417 genotype was significantly associated with plasma IL-33 levels in the detectable subjects (n = 227, R2 = 0.276, p = 1.77 × 10−17): the level of IL-33 protein increased with the number of rs7025417 risk (T) alleles. Based on genetic evidence in humans, the IL-33-ST2L pathway appears to have a causal role in the development of CAD, highlighting this pathway as a valuable target for the prevention and treatment of CAD.
Introduction
Coronary artery disease (CAD [MIM 608320, 610947, 611139, 608901, 607339, 608316]), a complex trait that results from multiple genetic and environmental factors and their interactions, is the leading cause of morbidity and mortality worldwide.1,2 In 2002 and 2007, Zdravkovic et al. estimated that the total heritability of CAD was 0.57 in men and 0.38 in women for death3 and 0.39 in men and 0.43 in women for angina, based on a longitudinal study of more than 20,000 Swedish twins.4 Recently, large-scale genome-wide association studies (GWASs) have identified approximately 50 risk loci for CAD.5–15 However, these risk loci primarily have modest effect sizes (odds ratios = 1.04–1.28) and collectively explain only approximately 10.6% of the total CAD heritability.15–17 Therefore, a large proportion of the heritability of CAD remains unexplained and is called “missing heritability.”17
Inflammation has been demonstrated to play a key role in the pathogenesis of atherosclerotic CAD.18 Considerable evidence from animal studies has indicated that many cytokines are involved in the development of atherosclerosis.19 Most population-based studies have observed that variants in the genes encoding interleukin-1 receptor antagonist, IL-6, IL-10, IL-16, IL-17A, and IL-18 are also genetically associated with atherosclerosis and CAD.20–24
Recently, it was confirmed that the novel cytokine IL-33, a member of the IL-1 family, and its receptor ST2L-IL-1RL1 play important roles in inflammatory diseases. Furthermore, Miller et al. demonstrated that the IL-33-ST2L pathway might inhibit the development of atherosclerosis.25 Therefore, we speculated that the IL-33-ST2L pathway might be genetically associated with CAD. To confirm this possibility, we performed the following steps: (1) a case-control association study for CAD based on the GeneID Chinese Han population, by using tag SNPs covering IL33 (MIM 608678) and IL1RL1 (MIM 601203) with a large discovery sample size; (2) an interaction analysis between the most significant variants of IL33 and IL1RL1 in the association with CAD; (3) a reporter gene assay to investigate the function of the reference variants; and (4) a circulation level study of IL-33 in the individuals with CAD.
Material and Methods
Study Population
The GeneID Chinese Han population is an ongoing database that was established for studying the genetic basis of various cardiovascular diseases.13,26–28 The study subjects were 4,521 CAD cases and 4,809 controls, who were acquired from this database in three stages: first, the discovery population of 860 cases and 707 controls was assembled from eastern China; second, the validation population of 575 cases and 471 controls was assembled from northern China; and third, two replication populations with much larger sample sizes (2,016 cases and 1,647 controls in replication 1 and 1,070 cases and 1,984 controls in replication 2) were assembled from central China.
The criterion for the enrolment as a CAD case was >70% luminal stenosis in at least one main vessel, as identified by coronary angiography, coronary artery bypass graft, percutaneous coronary intervention, and/or myocardial infarction. Individuals were excluded from the study if they had experienced myocardial spasms or had a myocardial bridge, as identified by angiography, or had congenital heart disease, childhood hypertension, or type 1 diabetes mellitus.
The control subjects in the discovery and validation studies were defined as individuals without atherosclerotic lesions, as confirmed by angiography, and without any history of CAD. In the large replication studies, the control subjects were selected from the general population among individuals undergoing physical examinations. Study subjects who had notes of potential CAD or myocardial infarction in their medical records or from electrocardiographic tests were excluded.
The disease status of hypertension and diabetes mellitus were evaluated, and the lipid profile (total cholesterol [Tch], low-density lipoprotein cholesterol [LDL-c], high-density lipoprotein cholesterol [HDL-c], and triglyceride [TG]) was measured according to guidelines and standard methods.29–31 Other clinical data, such as age, gender, body mass index (BMI), and smoking history, were obtained via direct interviews or a medical records review.
The study complied with the ethical principles set forth by the Declaration of Helsinki, and approval was obtained from the local institution review boards on human subject research. Informed consents were obtained from all of the participants.
Genetic Analysis
The DNA samples were collected from peripheral blood. Tag SNPs were selected according to the following principles: (1) linkage disequilibrium (LD) between SNPs according to Haploview (v.4.2) based on HapMap CHB and JPT data sets (v.3, release 2) with the thresholds of r2 > 0.4 and D′ > 0.7 to reduce the redundancy;32,33 (2) potential functional sites predicted by bioinformatics (Promoter and Genevar); and (3) a minor allele frequency (MAF) threshold of greater than 0.05. Data were excluded if the allele call rate was less than 95% or the Hardy-Weinberg equilibrium (HWE) χ2 p value was less than 0.001. The selected tag SNPs were rs7025417, rs10975514, and rs10975519 in IL33 and rs11685424 and rs3771180 in IL1RL1 (Figure 1). rs7025417 and rs11685424 were located in the predicted promoter regions, which might regulate the expression of the genes.
Figure 1.
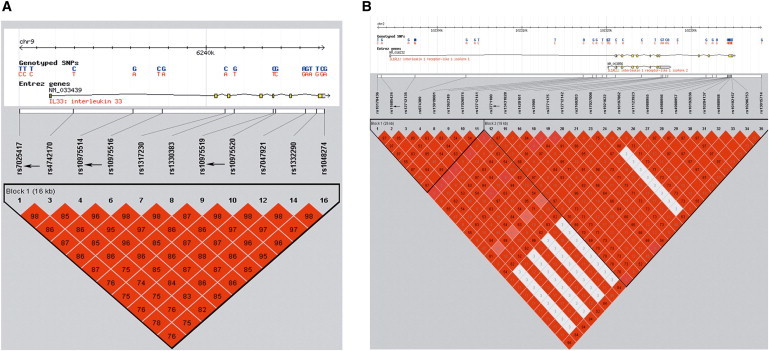
LD Blocks of IL33 and IL1RL1 Based on the HapMap CHB and JPT Data Sets
LD blocks of (A) IL33 and (B) IL1RL1 are shown. The arrows indicate the selected tag SNPs. Each diamond represents the LD degree between the SNPs. The color indicates the D′ (a redder color represents a higher D′) and the numbers within the diamonds are the r2 values (r2 > 0.55 and D′ > 0.83 in the blocks of both genes).
Genotyping was performed with a Roto-gene 6000 High-Resolution Melt (HRM) system (Corbett Life Science),26–28 with a total of 25 μl PCR volume containing 1 μl of LC Green dye, 5 pmol of each primer, 25 ng of genomic DNA, 2.5 μl of 10× PCR buffer with 1.5 mmol/l MgCl2, 5 mmol deoxynucleotide triphosphates, and 1 unit of Taq polymerase. Two positive controls for each genotype were included in each run, and the data with call rates of less than 95% were excluded. For each SNP, a total of 48 cases and controls were randomly selected for the verification of genotyping results by direct DNA sequencing analysis. DNA sequence analysis was performed with forward and/or reverse primers with the BigDye Terminator v3.1 Cycle Sequencing Kits on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Reporter Gene Assay
Because rs7025417 and rs11685424 were both located in the promoter region, which could influence gene expression, we conducted a reporter gene assay.34 We first subcloned a 1,556 bp 5′ UTR sequence of IL33 containing the rs7025417 C allele into a pGL3 basic vector (Promega Corporation). After confirmation by restriction mapping and direct sequencing, the pGL3 vector containing the “C” allele of rs7025417 was used as a template to generate the insert containing the T allele by using a site-directed mutagenesis kit (Promega). Direct sequencing was used to verify the two plasmids (IL33-C and IL33-T). The human lung adenocarcinoma cell line A549 and the human embryonic kidney cell line 293 (HEK293) (American Type Culture Collection [ATCC]) were transfected with 500 ng of each plasmid simultaneously with 50 ng of a pRL-TK vector (Promega) using Lipofectamine 2000 (Invitrogen). After 48 hr, firefly luciferase activity was measured with a Dual Luciferase Reporter Assay Kit (Promega). Three independent experiments were conducted, and each was performed in triplicate. The empty pGL3-basic vector was used as the negative control.
For IL1RL1, the constructed plasmids contained the rs11685424 A or G allele and the distal promoter sequence, totaling 1,218 bp continuous with exon 1a of IL1RL1. The two plasmids carrying IL1RL1 (IL1RL1-A and IL1RL1-G) were transfected into the human mast cell line CHMAS (ATCC) by nucleofection (Amaxa Cell Line Nucleofector Kit R).
Circulation Level Test for IL-33
Blood samples for the determination of IL-33 were collected in tubes containing potassium EDTA at the time that the subjects enrolled in the GeneID Chinese Han Population database. After centrifugation, plasma samples were frozen at −80°C for less than 2 months. The concentrations of IL-33 were measured in 440 individuals with CAD by enzyme-linked immunosorbent assay (ELISA) with commercially available LEGEND MAX kits (BioLegend). The detection sensitivity was 4.14 pg/ml.
Statistical Analyses
Hardy-Weinberg disequilibrium tests were performed among controls for each population by PLINK. The allelic and genotypic associations for each SNP were analyzed by Pearson’s 2 × 2 and 2 × 3 contingency table chi-square tests, and the odds ratio (OR) and 95% confidence interval (CI) were also calculated. Traditional risk factors for CAD, including age, gender, BMI, hypertension, diabetes mellitus, smoking history, Tch, HDL-c, LDL-c, and TG, were analyzed as covariates by multiple logistic regression models. Testing for interactions used logistic regression analysis under the genotypic model as suggested.35 Statistical power analysis and sample size estimation for validation and replication populations were performed with a free power and sample size calculation program (PS v.3.0.12) for single-variant association analysis and a free software program, QANTO, for interaction analysis (QANTO v.1.2.4). The luciferase activity difference was tested with Student’s t test (independent samples test, four degrees of freedom), and statistical significance was defined as a p value of less than 0.05. Analysis of association between the rs7025417 genotypes and IL-33 plasma concentrations was performed with a linear regression model, assuming an additive effect of the allele. The Mann-Whitney U test was used to test for differences in the IL-33 levels in the subjects with different genotypes of rs7025417.
Results
Population Characteristics
Table 1 illustrates the detailed clinical features of the three cohorts. The age, gender, and BMI did not differ between the case subjects and control subjects; the LDL-c, smoking, hypertension, and diabetes mellitus were significantly different. The case individuals had higher levels of LDL-c and a higher incidence of smoking, hypertension, and diabetes mellitus than those of the controls in all four populations.
Table 1.
Clinical Characteristics of the Studied GeneID Chinese Han Population
| Characteristics |
GeneID Discovery |
p |
GeneID Validation |
p |
GeneID Replication 1 |
p |
GeneID Replication 2 |
p |
GeneID Combined |
p | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAD | Control | CAD | Control | CAD | Control | CAD | Control | CAD | Control | ||||||
| Number of subjects | 860 | 707 | – | 575 | 471 | – | 2,016 | 1,647 | – | 1,070 | 1,984 | – | 4,521 | 4,809 | – |
| Age (years)a | 59.4 ± 11.2 | 58.2 ± 13.7 | 0.12 | 60.8 ± 11.0 | 59.9 ± 11.5 | 0.24 | 60.5 ± 9.08 | 59.9 ± 8.49 | 0.14 | 60.2 ± 10.2 | 59.6 ± 9.12 | 0.10 | 60.2 ± 9.85 | 59.7 ± 9.25 | 0.10 |
| Percent of subjects male | 56.8 | 59.1 | 0.36 | 69.7 | 65.7 | 0.17 | 61.7 | 63.9 | 0.16 | 61.5 | 59.6 | 0.21 | 61.9 | 61.6 | 0.80 |
| Percent of subjects smoking | 44 | 19 | 0.00 | 21.7 | 6.4 | 0.00 | 24.1 | 0.4 | 0.00 | 23.8 | 7.1 | 0.00 | 27.5 | 5.8 | 0.00 |
| BMI (kg/m2) | 24.7 ± 3.49 | 23.8 ± 2.92 | 0.34 | 24.5 ± 2.80 | 23.7 ± 9.41 | 0.39 | 24.8 ± 2.93 | 23.5 ± 3.09 | 0.46 | 26.3 ± 2.10 | 25.7 ± 8.24 | 0.11 | 25.1 ± 3.31 | 24.5 ± 5.16 | 0.10 |
| Hypertension (%) | 61 | 29.6 | 0.00 | 48.6 | 16 | 0.00 | 69.6 | 1.2 | 0.00 | 49.3 | 11 | 0.00 | 60.5 | 10.7 | 0.00 |
| DM | 16.8 | 9.9 | 0.00 | 17.5 | 6 | 0.00 | 23.1 | 0.4 | 0.00 | 16.8 | 3 | 0.00 | 19.7 | 3.4 | 0.00 |
| Tch (mmol/l) | 4.49 ± 1.07 | 4.35 ± 0.89 | 0.00 | 4.59 ± 1.11 | 4.39 ± 0.93 | 0.00 | 4.57 ± 1.17 | 4.53 ± 0.85 | 0.13 | 4.69 ± 1.21 | 4.55 ± 0.88 | 0.00 | 4.58 ± 1.10 | 4.50 ± 0.73 | 0.00 |
| TG (mmol/l) | 1.84 ± 1.36 | 1.52 ± 0.88 | 0.00 | 1.92 ± 1.48 | 1.65 ± 0.90 | 0.00 | 1.86 ± 1.39 | 1.57 ± 0.89 | 0.00 | 1.74 ± 1.10 | 1.46 ± 0.70 | 0.00 | 1.84 ± 1.16 | 1.53 ± 0.65 | 0.00 |
| HDL-c (mmol/l) | 1.08 ± 0.24 | 1.11 ± 0.24 | 0.08 | 1.10 ± 0.21 | 1.16 ± 0.22 | 0.00 | 1.11 ± 0.23 | 1.14 ± 0.25 | 0.00 | 1.14 ± 0.27 | 1.23 ± 0.20 | 0.00 | 1.12 ± 0.32 | 1.18 ± 0.21 | 0.00 |
| LDL-c (mmol/l) | 2.66 ± 0.77 | 2.41 ± 0.83 | 0.01 | 2.57 ± 0.98 | 2.40 ± 0.82 | 0.00 | 2.60 ± 0.84 | 2.37 ± 0.84 | 0.00 | 2.67 ± 0.91 | 2.52 ± 0.65 | 0.00 | 2.60 ± 0.84 | 2.44 ± 0.60 | 0.00 |
The data are presented as the means ± standard deviation or percent.
Age for the case subject is at diagnosis; age for the control subject is at enrollment.
Under the population parameter settings of the effect size (OR = 1.2 for CAD),28 the allele frequency of 0.46 (HapMap CHB and JPT data sets, the minimum minor allele frequency for rs11685424 between the two significant variants in the discovery study), our sample size provides a statistical power of 60% in the validation population, 97.3% in the replication 1 population, and 92.4% in the replication 2 population.
The prior statistical power used to detect the interaction between rs7025417 in IL33 and rs11685424 in IL1RL1 under the genotypic model for their association with CAD was greater than 80% in the combined population.
Association Analysis of Single Variants with CAD in the GeneID Chinese Han Population
There was no deviation from the Hardy-Weinberg equilibrium for all of the SNPs in the control subjects. In the GeneID-discovery population, both IL33 and IL1RL1 possessed a promoter variant that was significantly associated with CAD (rs7025417T in IL33, padj = 1.38 × 10−4, OR = 1.32, 95% CI: 1.14–1.52; rs11685424G in IL1RL1, padj = 2.12 × 10−3, OR = 1.25, 95% CI: 1.08–1.44). In the GeneID-validation population, the SNPs rs7025417 and rs11685424 were also significantly associated with CAD (rs7025417T in IL33, padj = 7.69 × 10−6, OR = 1.49, 95% CI: 1.25–1.77; rs11685424G in IL1RL1, padj = 1.25 × 10−4, OR = 1.40, 95% CI: 1.18–1.67). In the large GeneID-replication 1 population, the associations between the two variants and CAD were still significant (rs7025417T in IL33, padj = 2.32 × 10−7, OR = 1.28, 95% CI: 1.16–1.40; rs11685424G in IL1RL1, padj = 1.15 × 10−12, OR = 1.40, 95% CI: 1.28–1.53). In the GeneID-replication 2 population, the associations between the two variants and CAD was confirmed (rs7025417T in IL33, padj = 1.06 × 10−12, OR = 1.47, 95% CI: 1.33–1.66; rs11685424G in IL1RL1, padj = 2.17 × 10−13, OR = 1.49, 95% CI: 1.34–1.65) (Table 2). In addition, genotypic association analysis also demonstrated that rs7025417 in IL33 and rs11685424 in IL1RL1 were significantly associated with CAD in all three population stages, as well as in the combined population (Table S1 available online).
Table 2.
Allelic Association Analysis of rs7025417 in IL33 and rs11685424 in IL1RL1 with CAD in the GeneID Chinese Han Population
| Population |
n |
Gene, SNP (Reference Allele) |
Frequency |
phwe | pobs | padj | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Case | Control | ||||||
| GeneID Discovery | 860 | 707 | IL33, rs7025417T | 0.585 | 0.516 | 0.259 | 1.02 × 10−4 | 1.38 × 10−4 | 1.32 (1.14–1.52) |
| IL1RL1, rs11685424G | 0.567 | 0.515 | 0.293 | 3.63 × 10−3 | 2.12 × 10−3 | 1.25 (1.08–1.44) | |||
| GeneID Validation | 575 | 471 | IL33, rs7025417T | 0.584 | 0.486 | 0.4 | 7.42 × 10−6 | 7.69 × 10−6 | 1.49 (1.25–1.77) |
| IL1RL1, rs11685424G | 0.57 | 0.487 | 0.453 | 1.48 × 10−4 | 1.25 × 10−4 | 1.40 (1.18–1.67) | |||
| GeneID Replication 1 | 2,016 | 1,647 | IL33, rs7025417T | 0.589 | 0.528 | 0.008 | 2.28 × 10−7 | 2.32 × 10−7 | 1.28 (1.16–1.40) |
| IL1RL1, rs11685424G | 0.566 | 0.483 | 0.776 | 1.08 × 10−12 | 1.15 × 10−12 | 1.40 (1.28–1.53) | |||
| GeneID Replication 2 | 1,070 | 1,984 | IL33, rs7025417T | 0.594 | 0.497 | 0.367 | 4.39 × 10−13 | 1.06 × 10−12 | 1.47 (1.33–1.66) |
| IL1RL1, rs11685424G | 0.579 | 0.481 | 0.118 | 1.65 × 10−13 | 2.17 × 10−13 | 1.49 (1.34–1.65) | |||
| GeneID Combined | 4,521 | 4,809 | IL33, rs7025417T | 0.591 | 0.509 | 0.001 | 1.41 × 10−27 | 1.19 × 10−28 | 1.39 (1.31–1.47) |
| IL1RL1, rs11685424G | 0.573 | 0.487 | 0.027 | 6.25 × 10−30 | 6.93 × 10−30 | 1.40 (1.32–1.48) | |||
| GeneID Anatomical CAD | 1,598 | 4,809 | IL33, rs7025417T | 0.579 | 0.509 | 0.001 | 6.06 × 10−12 | 3.79 × 10−11 | 1.32 (1.22–1.44) |
| IL1RL1, rs11685424G | 0.565 | 0.487 | 0.027 | 2.37 × 10−14 | 2.73 × 10−13 | 1.36 (1.25–1.48) | |||
| GeneID Clinical CAD | 2,923 | 4,809 | IL33, rs7025417T | 0.594 | 0.509 | 0.001 | 2.19 × 10−24 | 8.57 × 10−22 | 1.39 (1.30–1.49) |
| IL1RL1, rs11685424G | 0.573 | 0.487 | 0.027 | 4.73 × 10−25 | 9.48 × 10−22 | 1.39 (1.30–1.49) | |||
Abbreviations are as follows: phwe, p value from Hardy-Weinberg equilibrium tests; pobs, observed p value; padj, p value adjusted by covariates; OR, odds ratio after adjustment.
rs10975514 and rs10975519 in IL33 and rs3771180 in IL1RL1, which failed to show significant associations with CAD in the GeneID-discovery population (the lowest adjusted p value of the three SNPs is 0.20 for rs10975514) (Table S2), were excluded from the next two stages of the study.
Association Analysis of rs7025417 in IL33 and rs11685424 in IL1RL1 with Subgroups of CAD in the GeneID-Combined Population
We separated the individuals with CAD into two subgroups: GeneID-anatomical-CAD, n = 1,598, individuals with anatomical disease (severe coronary stenosis); and GeneID-clinical-CAD, n = 2,923, individuals with clinical disease (myocardial infarction or revascularization). In GeneID-anatomical-CAD, the associations between the two SNPs and CAD were significant, with OR values of more than 1.3 for both (rs7025417T in IL33, padj = 3.79 × 10−11, OR = 1.32, 95% CI: 1.22–1.44; rs11685424G in IL1RL1, padj = 2.73 × 10−13, OR = 1.36, 95% CI: 1.25–1.48). In GeneID-clinical-CAD, the associations of the two SNPs with myocardial infarction or revascularization were also significant, with OR values of approximately 1.4 for both variants (rs7025417T in IL33, padj = 8.57 × 10−22, OR = 1.39, 95% CI: 1.30–1.49; rs11685424G in IL1RL1, padj = 9.48 × 10−22, OR = 1.39, 95% CI: 1.30–1.49) (Table 2). Genotypic association analysis also demonstrated that the two SNPs were significantly associated with CAD in both subgroups (Table S1).
Interaction Analysis between rs7025417 in IL33 and rs11685424 in IL1RL1 under the Genotypic Model in the GeneID-Combined Population
Because the sample size in the GeneID-combined population was sufficiently large to provide valuable statistical power, we performed an interaction analysis between rs7025417 in IL33 and rs11685424 in IL1RL1 under the genotypic model in the combined population. The interaction of the two SNPs (rs7025417 in IL33 and rs11685424 in IL1RL1) in the association with CAD was highly significant, with a p value of 2.81 × 10−32 under the genotypic model. Additionally, the combination with the largest effect under the genotypic model was “TT/GG,” which provided a nearly 5-fold increase in the risk for CAD (padj = 8.90 × 10−21, OR = 4.98, 95% CI: 3.56–6.97) (Table 3, Figure 2A). Figure 2B directly illustrates that the effect of the combination TT/GG was much stronger than that of a single variant in the association with CAD.
Table 3.
Interaction Analysis between rs7025417 in IL33 and rs11685424 in IL1RL1 under the Genotypic Model in the GeneID Combined Population
| Types | Case (%) | Control (%) | padj | OR (95% CI) |
|---|---|---|---|---|
| TT/GG | 675 (14.9) | 416 (8.7) | 8.90 × 10−21 | 4.98 (3.56-6.97) |
| TT/GA | 594 (13.1) | 479 (10.0) | 5.20 × 10−9 | 2.39 (1.78-3.20) |
| TT/AA | 210 (4.6) | 288 (6.0) | 3.53 × 10−2 | 1.43 (1.02-1.98) |
| TC/GG | 577 (12.8) | 490 (10.2) | 4.18 × 10−9 | 2.4 (1.79–3.22) |
| TC/GA | 1,345 (29.8) | 1,518 (31.6) | 5.00 × 10−6 | 1.92 (1.45–2.54) |
| TC/AA | 442 (9.8) | 524 (10.9) | 1.35 × 10−3 | 1.64 (1.21–2.21) |
| CC/GG | 190 (4.2) | 188 (3.9) | 2.23 × 10−3 | 1.71 (1.21–2.40) |
| CC/GA | 332 (7.3) | 499 (10.4) | 6.50 × 10−2 | 1.34 (0.98–1.83) |
| CC/AA | 156 (3.5) | 407 (8.5) | – | 1 |
| ADD/ADD | – | – | 2.81 × 10−32 | – |
The population CAD combined consisted of 4,521 cases and 4,809 controls. Abbreviations are as follows: padj, p value after adjustment for covariates, such as age, gender, smoking, BMI, hypertension, DM, Tch, TG, HDL-c, and LDL-c; ADD, additive model, rs7025417_TT/TC/CC, rs11685424_GG/GA/AA.
Figure 2.
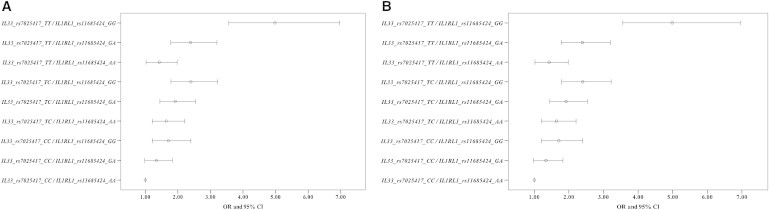
Comparison between OR Values
(A) OR values with a 95% confidence interval (95% CI) of the combination types under the genotypic model.
(B) The comparison of the OR values with a 95% confidence interval (95% CI) between the combination type “TT/GG” and the genotypic models of single variants.
ADD, additive model, rs7025417_TT/TC/CC, rs11685424_GG/GA/AA; DOM, dominant model, rs7025417_TT+TC/CC, rs11685424_GG+GA/AA; REC, recessive model, rs7025417_TT/TC+CC, rs11685424_GG/GA+AA.
Reporter Gene Analysis
Given that rs7025417 and rs11685424 were both located within the gene promoter region and could therefore interfere with the regulation of gene expression, we constructed plasmids carrying IL33-C/T and IL1RL1-A/G. The assay was performed in triplicate, and a representative result of the three independent experiments is illustrated as the relative luciferase activity in Figure 3. The luciferase activity in the cells transfected with IL33-T plasmid was twice that in the cells transfected with IL33-C plasmid, and the luciferase activity in the cells transfected with the IL1RL1-G plasmid was four times that in the cells transfected with the IL1RL1-A plasmid (p < 0.005, Figure 3). These results indicate that the rs7025417 C>T change may increase the expression of IL33 and that the rs11685424 A>G change may increase the expression of IL1RL1. The results exhibited for IL33 were tested in A549, and those for IL1RL1 were tested in CHMAS. The results found in HEK293 cells for IL33 were similar to those found in A549 cells (data not shown).
Figure 3.
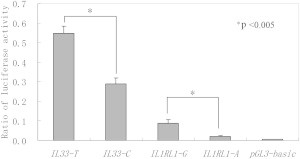
The Reporter Gene Analysis
The luciferase activity was tested by means of cellular extracts. p values less than 0.005 indicate that the difference was statistically significant between the plasmids. Mean ± SD of the relative luciferase activity is shown.
Association Analysis between the IL-33 Circulation Level and rs7025417 Genotype
The detection rate of the plasma IL-33 concentration was 51.6% in the 440 individuals with CAD (median, 233.67 pg/ml; range, 12–3,807 pg/ml). As shown in Figure 4A, rs7025417 genotype was significantly associated with plasma IL-33 levels in subjects with a detectable IL-33 level (n = 227, R2 = 0.276, p = 1.77 × 10−17). The IL-33 protein levels increased with the number of risk (T) alleles. The comparison between each pair of the three genotypes by the Mann-Whitney U test confirmed this trend, with p values that were less than 0.001 (Figure 4B). The results suggested that rs7025417 could affect the circulation level of IL-33.
Figure 4.
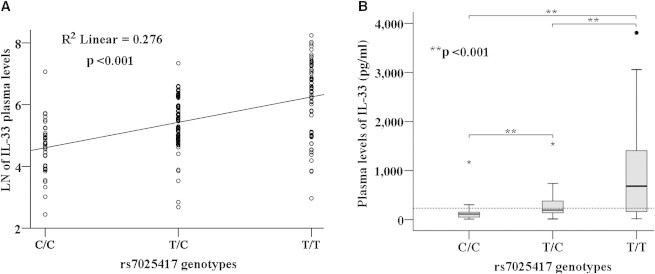
Circulation Level Analysis of IL-33
(A) Association analysis between the circulation levels of IL-33 and rs7025417 genotype in a linear regression model.
(B) The comparison between every pair of the three genotypes, via the Mann-Whitney U test. Special symbols (asterisk or circle) mark the outliers of the values in different genotypes. A broken line indicates the median value of 233.67 pg/ml for IL-33 in the detectable CAD individuals.
Discussion
In the present study, we demonstrated that rs7025417 in IL33 and rs11685424 in IL1RL1 are significantly associated with CAD in three stages of a case-control association study based on the GeneID Chinese Han population. The interaction between the two variants exhibited a much greater effect than that of a single variant in the association with CAD, which indicated that these variants could interact with each other in a biological manner. Given that the two variants were located within the gene promoter regions, we assessed whether these variants could influence gene expression by using reporter gene analysis and found that they did. In addition, rs7025417 genotype affected the level of circulating IL-33, which indicates that rs7025417 might influence the development of CAD by regulating the expression of IL33.
IL-33, a cytokine that belongs to the IL-1 family, has been shown to be expressed in the nucleus of human vascular endothelial cells of most healthy tissues and tumor tissues, as well as epithelial cells exposed to the external environment and fibroblasts of lymphoid tissues;36 however, of all the blood cell types, only activated dendritic cells and activated macrophages express IL-33 at low levels.37 IL-1RL1, previously known as the only receptor for the cytokine of IL-33, has two main protein forms: membrane-bound ST2L and soluble ST2 (sST2). It has been demonstrated in vitro that IL-33 can activate many immune cells by binding to ST2L-IL-1RL1 on the surface of many cells, including Th2 cells, mast cells, basophils, eosinophils, and natural killer cells.36,38–42 In vivo, the administration of IL-33 can influence multiple inflammatory-immune diseases, such as arthritis, asthma, and inflammatory bowel disease.43–45 This important evidence, demonstrating the involvement of the IL-33-ST2L pathway in the immune-inflammatory system, indicated that this pathway could also have important effects in the development of atherosclerosis and CAD.
Studies that have addressed the role of the IL-33-ST2L pathway in atherosclerosis and CAD have yielded conflicting results. Animal studies have indicated that the IL-33-ST2L pathway reduces macrophage foam cell formation46 and inhibits the development of atherosclerosis in apolipoprotein E-deficient (APOE−/−) mice.25 However, other studies have reported that the IL-33-ST2L pathway promotes angiogenesis and vascular leakage and induces adhesion molecule and proinflammatory cytokine expression in human endothelial cells,47–49 suggesting that the IL-33-ST2L pathway enhances the development of CAD by inducing the activation and injury of the coronary artery endothelium. To our knowledge, there have been few genetic or epidemiological studies on the association between the IL-33-ST2L pathway and CAD. Though Tsapaki et al. investigated the association between the distal promoter variants in IL1RL1 and CAD with small sample size, their significant results indicated that the IL-33-ST2L pathway might genetically associate with CAD.50 In the present study, with a large discovery sample size with sufficient statistical power, we detected that two tag SNPs in the two genes analyzed (rs7025417 in IL33 and rs11685424 in IL1RL1) were significantly associated with CAD not only in the subgroup of anatomical disease (severe coronary stenosis) but also in the subgroup of clinical disease (myocardial infarction or revascularization). Furthermore, additional functional studies using reporter gene and circulation level analysis suggested that the rs7025417 genotype affects the level of circulating IL-33, which indicates that rs7025417 might influence the development of CAD by regulating the expression of IL33. These results are in agreement with the above in vitro studies with human endothelial cells, suggesting that the IL-33-ST2L pathway enhances the development of CAD.
The conflicting results about the role of the IL-33-ST2L pathway in the development of atherosclerosis and CAD can be explained as follows. First, evidence provided by Demyanets et al.,47 Choi et al.,48 and Aoki et al.49 suggested that the IL-33-ST2L pathway could contribute to the early events of atherosclerosis development via activation and injury of the coronary artery endothelium. Miller et al.,25 in contrast, obtained results based on a model in which atherosclerosis had already developed. Second, given that the contradictory findings were obtained from experiments that used different species, IL-33 could function differently in human and in mouse tissues. The present study was performed with human cells, producing results that were consistent with those of the human studies described above. The influence of genetic variants on human diseases commences before the occurrence of the diseases and can begin as early as birth.
In 2006, Kabesch et al. reported that individual SNPs might interact with each other in a biological way if their interaction resulted in a more than multiplicative effect.51 In 2011, Demyanets et al. observed that endothelial cells express both IL-33 and ST2L in the nucleus and that the mRNA expression of these molecules is significantly correlated in the carotid atherosclerotic tissue,47 suggesting that the crosstalk of IL-33 and ST2L-IL-1RL1 significantly enhances the risk of atherosclerosis. In the present study, we determined that the OR values for the combination types of the two SNPs were much higher than the values for a single one in the associations with CAD (Table 3 and Figure 2). Therefore, we speculated that the two SNPs might interact each other in a biological way, which might influence the development of CAD by regulating the expression of their genes.
There are some limitations to our study. First, although we performed a three-stage population study, a larger sample size in different populations is required to replicate the association results. Second, although the reporter gene analysis and the circulation study suggested that the two promoter variants might regulate the gene expression, a thorough analysis of the loci represented by the two variants is necessary to detect the causal variants that have much larger effect sizes.
In conclusion, this study demonstrated that the two promoter SNPs rs7025417 in IL33 and rs11685424 in IL1RL1 are significantly associated with CAD. This finding implies that the IL-33-ST2L pathway may strongly influence the development of CAD, highlighting the IL-33-ST2L pathway as a valuable target for the prevention and treatment of CAD.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (973 Program: 2013CB531103 and 2012CB517800), the National Natural Science Foundation of China (No. 81222002, 81170303, 81070106, and 81270163), the Program for New Century Excellent Talents at the University of China (NCET-09-0380 and NCET-11-0181), and a Hubei Province Natural Science Key Program (2008CDA047).
Contributor Information
Qing K. Wang, Email: qkwang@mail.hust.edu.cn.
Xiang Cheng, Email: nathancx@mail.hust.edu.cn.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Genevar (Gene Expression Variation), http://www.sanger.ac.uk/resources/software/genevar/
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
Promoter 2.0 Prediction Server, http://www.cbs.dtu.dk/services/Promoter/
PS program, http://biostat.mc.vanderbilt.edu/wiki
QANTO, http://hydra.usc.edu/gxe
References
- 1.He J., Gu D., Wu X., Reynolds K., Duan X., Yao C., Wang J., Chen C.S., Chen J., Wildman R.P. Major causes of death among men and women in China. N. Engl. J. Med. 2005;353:1124–1134. doi: 10.1056/NEJMsa050467. [DOI] [PubMed] [Google Scholar]
- 2.Roger V.L., Go A.S., Lloyd-Jones D.M., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics—2012 update: a report from the American Heart Association. Circulation. 2012;125:188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 3.Zdravkovic S., Wienke A., Pedersen N.L., Marenberg M.E., Yashin A.I., De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J. Intern. Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 4.Zdravkovic S., Wienke A., Pedersen N.L., de Faire U. Genetic influences on angina pectoris and its impact on coronary heart disease. Eur. J. Hum. Genet. 2007;15:872–877. doi: 10.1038/sj.ejhg.5201846. [DOI] [PubMed] [Google Scholar]
- 5.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke R., Peden J.F., Hopewell J.C., Kyriakou T., Goel A., Heath S.C., Parish S., Barlera S., Franzosi M.G., Rust S., PROCARDIS Consortium Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 7.Erdmann J., Grosshennig A., Braund P.S., König I.R., Hengstenberg C., Hall A.S., Linsel-Nitschke P., Kathiresan S., Wright B., Trégouët D.A., Italian Atherosclerosis, Thrombosis, and Vascular Biology Working Group. Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium. Cardiogenics Consortium New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., Schunkert H., Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soranzo N., Spector T.D., Mangino M., Kühnel B., Rendon A., Teumer A., Willenborg C., Wright B., Chen L., Li M. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdmann J., Willenborg C., Nahrstaedt J., Preuss M., König I.R., Baumert J., Linsel-Nitschke P., Gieger C., Tennstedt S., Belcredi P. Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur. Heart J. 2011;32:158–168. doi: 10.1093/eurheartj/ehq405. [DOI] [PubMed] [Google Scholar]
- 11.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disease C.A., Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 13.Wang F., Xu C.Q., He Q., Cai J.P., Li X.C., Wang D., Xiong X., Liao Y.H., Zeng Q.T., Yang Y.Z. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat. Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 14.Lu X., Wang L., Chen S., He L., Yang X., Shi Y., Cheng J., Zhang L., Gu C.C., Huang J., Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B.A., CARDIoGRAMplusC4D Consortium. DIAGRAM Consortium. CARDIOGENICS Consortium. MuTHER Consortium. Wellcome Trust Case Control Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peden J.F., Farrall M. Thirty-five common variants for coronary artery disease: the fruits of much collaborative labour. Hum. Mol. Genet. 2011;20(R2):R198–R205. doi: 10.1093/hmg/ddr384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prins B.P., Lagou V., Asselbergs F.W., Snieder H., Fu J. Genetics of coronary artery disease: genome-wide association studies and beyond. Atherosclerosis. 2012;225:1–10. doi: 10.1016/j.atherosclerosis.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Hansson G.K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 19.Kleemann R., Zadelaar S., Kooistra T. Cytokines and atherosclerosis: a comprehensive review of studies in mice. Cardiovasc. Res. 2008;79:360–376. doi: 10.1093/cvr/cvn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Minkelen R., Wettinger S.B., de Visser M.C., Vos H.L., Reitsma P.H., Rosendaal F.R., Bertina R.M., Doggen C.J. Haplotypes of the interleukin-1 receptor antagonist gene, interleukin-1 receptor antagonist mRNA levels and the risk of myocardial infarction. Atherosclerosis. 2009;203:201–205. doi: 10.1016/j.atherosclerosis.2008.06.029. [DOI] [PubMed] [Google Scholar]
- 21.Blagodatskikh K.A., Evdokimova M.A., Agapkina IuV., Nikitin A.G., Brovkin A.N., Pushkov A.A., Blagodatskikh E.G., Kudriasheva O.Iu., Osmolovskaia V.S., Minushkina L.O. [Gene IL6 G(-174)C and gene IL10 G(-1082)A polymorphisms are associated with unfavourable outcomes in patients with acute coronary syndrome] Mol. Biol. (Mosk.) 2010;44:839–846. [PubMed] [Google Scholar]
- 22.Chen Y., Huang H., Liu S., Pan L.A., Zhou B., Zhang L., Zeng Z. IL-16 rs11556218 gene polymorphism is associated with coronary artery disease in the Chinese Han population. Clin. Biochem. 2011;44:1041–1044. doi: 10.1016/j.clinbiochem.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., Pei F., Zhang M., Yan C., Huang M., Wang T., Han Y. Interleukin-17A gene variants and risk of coronary artery disease: a large angiography-based study. Clin. Chim. Acta. 2011;412:327–331. doi: 10.1016/j.cca.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Tiret L., Godefroy T., Lubos E., Nicaud V., Tregouet D.A., Barbaux S., Schnabel R., Bickel C., Espinola-Klein C., Poirier O., AtheroGene Investigators Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation. 2005;112:643–650. doi: 10.1161/CIRCULATIONAHA.104.519702. [DOI] [PubMed] [Google Scholar]
- 25.Miller A.M., Xu D., Asquith D.L., Denby L., Li Y., Sattar N., Baker A.H., McInnes I.B., Liew F.Y. IL-33 reduces the development of atherosclerosis. J. Exp. Med. 2008;205:339–346. doi: 10.1084/jem.20071868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L., Li C., Wang C., Xia Y., Wu G., Wang F., Xu C., Wang P., Li X., Wang D. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Hum. Genet. 2009;126:843–849. doi: 10.1007/s00439-009-0737-3. [DOI] [PubMed] [Google Scholar]
- 27.Xu C., Wang F., Wang B., Li X., Li C., Wang D., Xiong X., Wang P., Lu Q., Wang X. Minor allele C of chromosome 1p32 single nucleotide polymorphism rs11206510 confers risk of ischemic stroke in the Chinese Han population. Stroke. 2010;41:1587–1592. doi: 10.1161/STROKEAHA.110.583096. [DOI] [PubMed] [Google Scholar]
- 28.Cheng X., Shi L., Nie S., Wang F., Li X., Xu C., Wang P., Yang B., Li Q., Pan Z. The same chromosome 9p21.3 locus is associated with type 2 diabetes and coronary artery disease in a Chinese Han population. Diabetes. 2011;60:680–684. doi: 10.2337/db10-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapur A., Hall R.J., Malik I.S., Qureshi A.C., Butts J., de Belder M., Baumbach A., Angelini G., de Belder A., Oldroyd K.G. Randomized comparison of percutaneous coronary intervention with coronary artery bypass grafting in diabetic patients. 1-year results of the CARDia (Coronary Artery Revascularization in Diabetes) trial. J. Am. Coll. Cardiol. 2010;55:432–440. doi: 10.1016/j.jacc.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Ye S., Willeit J., Kronenberg F., Xu Q., Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J. Am. Coll. Cardiol. 2008;52:378–384. doi: 10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 31.Mancia G., De Backer G., Dominiczak A., Cifkova R., Fagard R., Germano G., Grassi G., Heagerty A.M., Kjeldsen S.E., Laurent S., Management of Arterial Hypertension of the European Society of Hypertension. European Society of Cardiology 2007 Guidelines for the Management of Arterial Hypertension: The Task Force for the Management of Arterial Hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC) J. Hypertens. 2007;25:1105–1187. doi: 10.1097/HJH.0b013e3281fc975a. [DOI] [PubMed] [Google Scholar]
- 32.John S., Shephard N., Liu G.Y., Zeggini E., Cao M.Q., Chen W.W., Vasavda N., Mills T., Barton A., Hinks A. Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. Am. J. Hum. Genet. 2004;75:54–64. doi: 10.1086/422195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaid D.J., Guenther J.C., Christensen G.B., Hebbring S., Rosenow C., Hilker C.A., McDonnell S.K., Cunningham J.M., Slager S.L., Blute M.L., Thibodeau S.N. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility Loci. Am. J. Hum. Genet. 2004;75:948–965. doi: 10.1086/425870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu M., Matsuda A., Yanagisawa K., Hirota T., Akahoshi M., Inomata N., Ebe K., Tanaka K., Sugiura H., Nakashima K. Functional SNPs in the distal promoter of the ST2 gene are associated with atopic dermatitis. Hum. Mol. Genet. 2005;14:2919–2927. doi: 10.1093/hmg/ddi323. [DOI] [PubMed] [Google Scholar]
- 35.Cordell H.J. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moussion C., Ortega N., Girard J.P. The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: a novel ‘alarmin’? PLoS ONE. 2008;3:e3331. doi: 10.1371/journal.pone.0003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T.K., Zurawski G., Moshrefi M., Qin J., Li X. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Komai-Koma M., Xu D., Li Y., McKenzie A.N., McInnes I.B., Liew F.Y. IL-33 is a chemoattractant for human Th2 cells. Eur. J. Immunol. 2007;37:2779–2786. doi: 10.1002/eji.200737547. [DOI] [PubMed] [Google Scholar]
- 39.Drube S., Heink S., Walter S., Löhn T., Grusser M., Gerbaulet A., Berod L., Schons J., Dudeck A., Freitag J. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood. 2010;115:3899–3906. doi: 10.1182/blood-2009-10-247411. [DOI] [PubMed] [Google Scholar]
- 40.Pecaric-Petkovic T., Didichenko S.A., Kaempfer S., Spiegl N., Dahinden C.A. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood. 2009;113:1526–1534. doi: 10.1182/blood-2008-05-157818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valent P. Interleukin-33: a regulator of basophils. Blood. 2009;113:1396–1397. doi: 10.1182/blood-2008-11-189811. [DOI] [PubMed] [Google Scholar]
- 42.Smithgall M.D., Comeau M.R., Yoon B.R., Kaufman D., Armitage R., Smith D.E. IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int. Immunol. 2008;20:1019–1030. doi: 10.1093/intimm/dxn060. [DOI] [PubMed] [Google Scholar]
- 43.Xu D., Jiang H.R., Kewin P., Li Y., Mu R., Fraser A.R., Pitman N., Kurowska-Stolarska M., McKenzie A.N., McInnes I.B., Liew F.Y. IL-33 exacerbates antigen-induced arthritis by activating mast cells. Proc. Natl. Acad. Sci. USA. 2008;105:10913–10918. doi: 10.1073/pnas.0801898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liew F.Y., Pitman N.I., McInnes I.B. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat. Rev. Immunol. 2010;10:103–110. doi: 10.1038/nri2692. [DOI] [PubMed] [Google Scholar]
- 45.Sedhom M.A., Pichery M., Murdoch J.R., Foligné B., Ortega N., Normand S., Mertz K., Sanmugalingam D., Brault L., Grandjean T. Neutralisation of the interleukin-33/ST2 pathway ameliorates experimental colitis through enhancement of mucosal healing in mice. Gut. 2012 doi: 10.1136/gutjnl-2011-301785. Published online November 21, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaren J.E., Michael D.R., Salter R.C., Ashlin T.G., Calder C.J., Miller A.M., Liew F.Y., Ramji D.P. IL-33 reduces macrophage foam cell formation. J. Immunol. 2010;185:1222–1229. doi: 10.4049/jimmunol.1000520. [DOI] [PubMed] [Google Scholar]
- 47.Demyanets S., Konya V., Kastl S.P., Kaun C., Rauscher S., Niessner A., Pentz R., Pfaffenberger S., Rychli K., Lemberger C.E. Interleukin-33 induces expression of adhesion molecules and inflammatory activation in human endothelial cells and in human atherosclerotic plaques. Arterioscler. Thromb. Vasc. Biol. 2011;31:2080–2089. doi: 10.1161/ATVBAHA.111.231431. [DOI] [PubMed] [Google Scholar]
- 48.Choi Y.S., Choi H.J., Min J.K., Pyun B.J., Maeng Y.S., Park H., Kim J., Kim Y.M., Kwon Y.G. Interleukin-33 induces angiogenesis and vascular permeability through ST2/TRAF6-mediated endothelial nitric oxide production. Blood. 2009;114:3117–3126. doi: 10.1182/blood-2009-02-203372. [DOI] [PubMed] [Google Scholar]
- 49.Aoki S., Hayakawa M., Ozaki H., Takezako N., Obata H., Ibaraki N., Tsuru T., Tominaga S., Yanagisawa K. ST2 gene expression is proliferation-dependent and its ligand, IL-33, induces inflammatory reaction in endothelial cells. Mol. Cell. Biochem. 2010;335:75–81. doi: 10.1007/s11010-009-0244-9. [DOI] [PubMed] [Google Scholar]
- 50.Tsapaki A., Zaravinos A., Apostolakis S., Voudris K., Vogiatzi K., Kochiadakis G.E., Spandidos D.A. Genetic variability of the distal promoter of the ST2 gene is associated with angiographic severity of coronary artery disease. J. Thromb. Thrombolysis. 2010;30:365–371. doi: 10.1007/s11239-010-0496-y. [DOI] [PubMed] [Google Scholar]
- 51.Kabesch M., Schedel M., Carr D., Woitsch B., Fritzsch C., Weiland S.K., von Mutius E. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J. Allergy Clin. Immunol. 2006;117:269–274. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


