Abstract
We report for the first time, to our knowledge, that the N-terminal loop (N_loop) of amylin (islet amyloid polypeptide (IAPP) residues 1–8) forms extremely long and stable non-β-sheet fibers in solution under the same conditions in which human amylin (hIAPP) forms amyloid fibers. This observation applies to the cyclic, oxidized form of the N_loop but not to the linear, reduced form, which does not form fibers. Our findings indicate a potential role of direct N_loop-N_loop interactions in hIAPP aggregation, which has not been previously explored, with important implications for the mechanism of hIAPP amyloid fiber formation, the inhibitory action of IAPP variants, and the competition between ordered and disordered aggregation in peptides of the calcitonin peptide family.
Introduction
Islet amyloid polypeptide (IAPP), also known as amylin, is a 37-residue intrinsically disordered hormone peptide that is secreted together with insulin by the β cells of the pancreas and is involved in glucose regulation and gastric emptying (1–4). IAPP is implicated in the pathogenesis of diabetes type II due to its deposition in the form of amyloid fibers in the β cells of the pancreas, where insulin is produced (5). It is a member of the calcitonin peptide family (6), which includes calcitonin (Ct), calcitonin-gene-related peptide (CGRP), adrenomedullin (Ad), intermedin, and amylin. These are structurally and genetically related intrinsically disordered hormone peptides, with sequence homology ranging between 20% and 50% (7). All members of the Ct family contain a functional disulfide bond that confers a short ringlike structure (N_loop) to the N-terminus of the peptide (with the exception of Ad, in which the disulfide is located between residues 16 and 21). The sequence of the N_loop is highly conserved among Ct family members.
Although some members of this family are prone to aggregate, others are remarkably soluble. A similarly dramatic difference is also manifest among variants of individual Ct family members. For example, human IAPP (hIAPP) readily forms amyloid fibrils in vitro, whereas the rat variant (rIAPP), differing by six amino acids, does not (Fig. 1)(8). In addition to being highly soluble, rIAPP is an effective inhibitor of hIAPP fibril formation (9). Both these properties of rIAPP have been attributed to the presence of three β-breaking prolines located in the C-terminal half of the peptide (10,11). It has been recently shown that rIAPP interacts with hIAPP during fibril formation through its N-terminal region (12). Although the nature of such an interaction, which does not involve β-sheet formation, has not been identified, association of α-helical intermediates had originally been proposed (9). According to the most recent structural model of hIAPP fibrils, derived from two-dimensional-NMR (2D-NMR) restraints, monomers are aligned at the N-terminus and form in-register parallel β-sheets (13). In this structure, the N_loop is not involved in direct intermolecular interactions. Removal of the N_loop, however, alters both the mass/length distributions of hIAPP fibers (14) and the kinetics of fibril formation, with effects that vary with experimental conditions (15,16).
Figure 1.

Sequences of human IAPP (hIAPP), rat IAPP (rIAPP), and N_loop. The disulfide bond between residues 2 and 7, forming a loop at the N-terminus of the sequences, is indicated by the blue brackets. All sequences have a free N-terminus and amidated C terminus. To see this figure in color, go online.
The mechanism by which the N_loop affects hIAPP aggregation is not yet understood, but it is important for rationalizing the kinetics and potentially developing inhibitors (17,18). The leading hypothesis is that the N_loop alters hIAPP aggregation kinetics indirectly, by affecting the conformational preferences of the monomer in solution (15,16,19). For example, ring opening of the N_loop by chemical reduction decreases the helical content of rIAPP (assumed to be a good structural model of hIAPP) (19,20), and the N_loop has been shown to induce the collapse of rIAPP, hIAPP, and a model hydrophilic peptide (21). Molecular simulations indicate that the N_loop has a rigid structure, suggesting that it may act as a nucleus driving compaction (21). However, a direct role of the N_loop in the aggregation of hIAPP, and in the inhibition of hIAPP by rIAPP, has not yet been explored. The findings presented here indicate that direct association of the N_loop alone is a favorable process at micromolar concentrations.
Methods
Materials
9-Fluorenylmethoxycarbonyl (Fmoc)-protected amino acids were purchased from Novabiochem (Bubendorf, Switzerland). N-hydroxy benzotriazole (HOBt) and O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate (HBTU) were purchased from Genscript. N,N-iisopropylethylamine, or Hünig's base (DIPEA), and N-Methyl-2-pyrrolidone (NMP), used as a base in solid-phase peptide synthesis, were purchased from Sigma Aldrich (St. Louis, MO). Piperidine (Sigma-Aldrich) was used for deprotection. Rink Amide ChemMatrix was purchased from Matrixx Initiatives (Scottsdale, AZ). Dimethyl formamide (DMF), dichloromethane (DCM), and acetonitrile were purchased from Fisher Scientific (Waltham, MA) and were used without further purification.
Peptide synthesis and purification
N1–8 was synthesized on a 0.25 mmol scale using a CEM (Cambridge, MA) Liberty automated microwave peptide synthesizer and PALChem (Angres, France) matrix resin. After synthesis, the peptide was thoroughly washed five times with DMF followed by DCM. After washing, the peptide was stored on the resin at −20°C. For deprotection, the peptide was shaken for 1 h in 20% piperdine, 0.1 M HOBt in DMF. The cleavage cocktail consisted of 95% trifluoroacetic acid (TFA) + 2.5% water + 2.5% triisopropylsilane (TIS) at a ratio of 150 μL/10 mg resin. N1–8 was purified using reverse-phase high-performance liquid chromatography (HPLC) on a Waters 600E system. Crude peptide was purified on a C18 semi-preparative column (Vydac/Grace, Deerfield, IL) at a gradient of 0–14% acetonitrile with 0.1% (v/v) TFA over 45 min.
Disulfide formation and oxidized peptide purification
Lyophilized peptide (1.0 mM) was dissolved in 30% dimethylsulfoxide (DMSO) and 3% acetic acid. The sample was stirred with a magnetic stir bar at 1100 rpm. During this time, the formation of the intramolecular disulfide bonds was monitored via HPLC on a C18 analytical column. The reaction was deemed complete when the reduced peptide’s HPLC peak was no longer apparent: ∼10 h. After this time, the sample was frozen and lyophilized. Oxidized peptide was repurified on a C18 semipreparative column to separate any unreacted peptide.
Calibrated matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) indicated the presence of a −2 Da species (above the margin of error of the instrument), corresponding to the oxidized form of the peptide, N_loop. To further support these findings, a maleimide sulfhydryl reaction was performed.Three-(N-maleimidopropionyl)-biocytin (Cayman Chemical, Ann Arbor, MI) (1 mg) was dissolved in 180 μL 20 mM phosphate-buffered saline (PBS), pH 7.0, and 20 μL acetonitrile. Pure, oxidized N_loop (1 mg) was dissolved in 90 μL 20 mM PBS buffer, pH 7.0, and 10 μL acetonitrile. This solution was combined with the 3-(N-maleimidopropionyl)-biocytin solution and shaken for 4 h. At 1 h increments, 20 μL aliquot was removed and frozen in dry ice. After 4 h, the four aliquots were analyzed via MALDI-TOF MS.
HPLC peaks were analyzed by a Voyager Systems 4320 MALDI-TOF MS (Applied Biosystems, Foster City, CA). The peak corresponding to a molecular weight of 807 Da (corresponding to the amidated and oxidized form of the peptide) was analyzed further for purity by analytical HPLC, using a reverse-phase C18 analytical column 214TP54 (length 250 mm × inner diameter (ID) 4.6 mm, particle size 5 μm) using the same gradient conditions with a 0.9 mL/min flow rate. A single peak eluting at a gradient corresponding to the hydrophobicity of N_loop was collected, immediately frozen in liquid nitrogen, lyophilized, and kept at −20°C.
Fiber preparation
For each measurement, solutions of N_loop were prepared fresh at T = 23°C. The N1–8 samples were prepared in deoxygenated conditions and in the presence of a two-time molar excess of tris(2-carboxyethyl)phosphine hydrochloride (Sigma Aldrich). Buffer was 50 mM sodium acetate at pH 4.9. After adding the buffer to lyophilized peptide in a glass vial, samples were stirred with a glass stirring rod until all peptide was dissolved. The concentration of peptide was measured by absorbance at 205 nm, using an extinction coefficient of ε = 11,040 M−1 cm−1. This was obtained from the linear dependence of absorbance versus concentration measured for solutions of N1–8 in the 60–200 μM range (solutions were prepared by direct dilution from a 200 μM stock solution).
Circular dichroism
Circular dichroism (CD) spectra of the samples were measured in quartz cuvettes (Starna Cells, Atascadero, CA) ranging in pathlength from 10 to 0.1 mm. Stock solutions were diluted with Millipore (Billerica, MA) H2O, resulting in final buffer concentrations ranging from 10 to 25 mM NaAc. A J-710 spectropolarimeter (Jasco, Oklahoma City, OK) was used with a 1 nm bandwidth. For each sample, eight spectra taken with a 0.2 nm pitch at a 50 nm/min scan speed were averaged. Before data analysis, spectra were buffer-subtracted.
Transmission electron microscopy
Carbon Type B (15–25 nm) 200 mesh grids with Formvar coating (Ted Pella, Redding, CA) were glow-discharged using a plasma cleaner immediately before applying the sample. The N_loop sample was diluted to a final concentration of 7–25 mM and allowed to adsorb to the grid for 5 min. The sample was blotted and rinsed with Millipore water using No. 1 Whatman filter paper. Immediately after blotting, 5 μL of freshly filtered 1% uranyl acetate was applied for 2 min, then blotted. Grids were imaged on a Philips CM 12 scanning transmission electron microscope (TEM), operated at 80 kV.
Thioflavin-T assay
Thioflavin-T (ThT) binding assays were used to detect the presence of amyloid fibrils within N_loop samples. Final solutions contained 10 μM ThT, 12.5 mM KCNTATCA in 50 mM sodium acetate, pH 4.9. All ThT experiments were conducted in a QuantaMaster 40 (Photon Technology International, Birmingham, NJ). Emission spectra were monitored at excitation wavelengths of 350 nm and 450 nm, whereas excitation spectra were monitored at emission wavelengths of 450 nm and 480 nm. Excitation and emission slits were kept at 2 nm. ThT was purchased from Pierce, Thermo Fisher Scientific (Rockford, IL).
Fourier transform infrared spectroscopy
N_loop fiber solutions were lyophilized after aging for 1 week at 4°C. Monomeric N_loop samples were lyophilized immediately after HPLC purification. To minimize the possibility of aggregates forming during the lyophilization process, small fractions corresponding to pure N_loop were instantly frozen in liquid nitrogen and maintained in the solid (frozen) phase during the entire lyophilization process. HPLC analysis of samples pre- and postlyophilization show conservation of mass. This observation supports the lack of insoluble aggregate formation during lyophilization. Lyophilized peptide was mixed with oven-dried KBr using a mortar and pestle. The sample was further dried under vacuum for 1 min, and pressed into a pellet. Fourier transform infrared (FTIR) spectra were recorded on a Vertex 70 series instrument (Bruker, Billerica, MA). The optical chamber was flushed with nitrogen for 15 min before each scan. The interferograms were averaged over 512 scans with a resolution of 1 cm−1. Atmospheric compensation (for residual H2O vapor and CO2) and baseline correction (using 10 iterations of concave rubberband correction) were performed using Opus V6.5 software. Control KBr spectra were subtracted from peptide spectra. In the data shown, the fiber absorbance value was rescaled by a constant such that the magnitude of the amide I band matched the absorbance value for the monomer pellet.
Optical microscopy
All imaging was performed on a BH2-UMA optical microscope (Olympus, Center Valley, PA). A quarter waveplate was aligned to 45° for birefringence imaging. All measurements were made at 10× magnification.
NMR spectroscopy
Lyophilized peptide was dissolved in 100% deuterated DMSO (Cambridge Isotopes, Andover, MA) for a final peptide concentration of 1.3 mM. All spectra were acquired at 25°C on a 500 MHz Varian (Palo Alto, CA) spectrometer. One-dimensional NMR spectra of N_loop were recorded before each experiment and did not change, indicating that no significant conformational change or aggregation occurred for the duration of the experiments. Spectra were calibrated to the residual solvent peak for DMSO, 2.49 ppm. Two-dimensional COSY and TOCSY spectra were recorded using a mixing time of 100 ms. 1H chemical shifts determined from the TOCSY spectrum are reported in the Table S1 of the Supporting Material together with the nuclear Overhauser effect (NOE) spectra (Figs. S7 and S8). Secondary chemical shifts, used to identify secondary and residual structure in intrinsically disordered proteins (22), were calculated by subtracting reference random-coil values from De Simone et al. (23) from our measured chemical shifts. Comparison with alternative random-coil libraries (23–25) is reported in Fig. S3. Two-dimensional NOE spectroscopy (NOESY) experiments were measured for both 400 ms and 80 ms mixing times. NOEs were exported from VNMRJ (Varian) and further analyzed in CARA (Keller, Rochus, The Computer Aided Resonance Assignment Tutorial). NOE peak volumes were converted into relative strengths by calibrating them to the crosspeak of the two 1Hβ protons of cysteine 2, assumed to be at a fixed distance of 1.8 Å. NOE strengths were classified into weak (<4.0 Å), medium (<3.3 Å), and strong (<2.6 Å) using cutoffs from Nilges and co-workers (26). Additional NOE data are shown in Fig. S9.
Molecular dynamics simulations
Replica-exchange molecular dynamics (REMD) simulations of the amylin monomer were carried out in explicit TIP3P water, with the Amber ff03∗ force field for the protein and ions (27). Periodic boundary conditions with a 3.5 nm truncated octahedron cell containing the peptide and 1033 water molecules, four sodium, and six chloride ions, were used. Long-range electrostatic interactions were computed using particle mesh Ewald with a real-space cutoff of 0.9 nm and a grid spacing of 0.1 nm. All bond lengths were constrained to their reference values using the LINCS algorithm. This system was equilibrated at constant pressure and a temperature of 295 K, using a Parrinello-Rahman Barostat and Langevin thermostat with a friction coefficient of 1 ps−1 for 200 ps, with an integration time step of 2 fs. After the equilibration, REMD simulations were performed at constant volume, with 32 replicas spanning a temperature range from 278 to 595 K, using a Langevin thermostat to control the temperature of each replica, and with exchange attempts every 2 ps. Structures from the 295 K replica were clustered using the g_cluster utility with the single-linkage method and a cutoff of 0.08 nm. Additional REMD simulations of a pair of amylin monomers were performed using the same simulation parameters, and all structures forming direct contacts (defined as a minimum atom-atom distance between monomers of <0.2 nm) were analyzed.
Results
N_loop fiber formation
We set out to investigate the properties of the isolated N_loop (residues 1–8 of hIAPP) in the oxidized and reduced form. We prepared the linear, reduced form of hIAPP1–8 (N1–8) by solid-phase peptide synthesis. We obtained the oxidized, cyclic form (N_loop) by oxidation in DMSO of N1–8 followed by purification and lyophilization (see Methods). Remarkably, we found that the cyclic peptide immediately forms fibers visible by eye when dissolved in aqueous buffer at peptide concentrations ranging from 0.65 mM to 50 mM (Fig. 2 a) By contrast, the linear peptide N1–8 did not form fibers under the same experimental conditions. Light microscopy imaging of the fibers reveals strong birefringence ( Fig. 2 b), indicating a large degree of supramolecular ordering in the fibers. Consistently, TEM images show large bundles of aligned microfibers ( Fig. 2 c). Fibers of the same morphology where observed in TEM within 1 h of sample preparation, at concentrations as low as 57 μM (Fig. S1). Fibers did not appear to dissolve under highly acidic conditions (pH <2) or after heating to 80°C. Based on a ThT binding assay, the fibers do not appear to be amyloid in nature (28) (Fig. S2). This is further confirmed by the absence of β-sheet features in both CD and FTIR spectra presented below.
Figure 2.
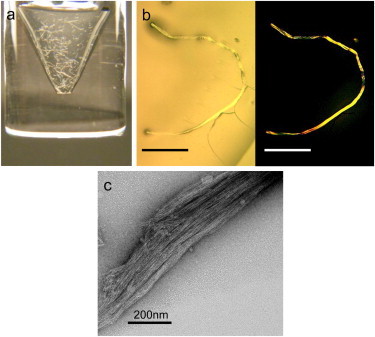
(a) Oxidized N_loop fibers, visible immediately upon dissolving peptide in buffer (50 mM peptide in 50 mM sodium acetate, pH 4.9). Immediate formation of fibers was also observed at concentrations of 57 μM by TEM (Fig. S1). (b) Optical microscopy of N_loop fibers in the absence (left) and presence (right) of crossed polarizers, revealing fiber birefringence (scale bar, 1 mm). (c) TEM image of fibers prepared at 25 mM (scale bar, 200 nm). Fibers of same morphology were observed at concentrations as low as 57 μM (Fig. S1). To see this figure in color, go online.
The sequence of N_loop (Fig. 1) does not contain particularly hydrophobic or insoluble side chains that would suggest such aggregation. We therefore hypothesize that structural features associated with peptide cyclization are conducive to fibril formation. To investigate such features, we analyzed the linear and cyclic monomer as well as the fibers, using CD and FTIR spectroscopy (Fig. 3).
Figure 3.
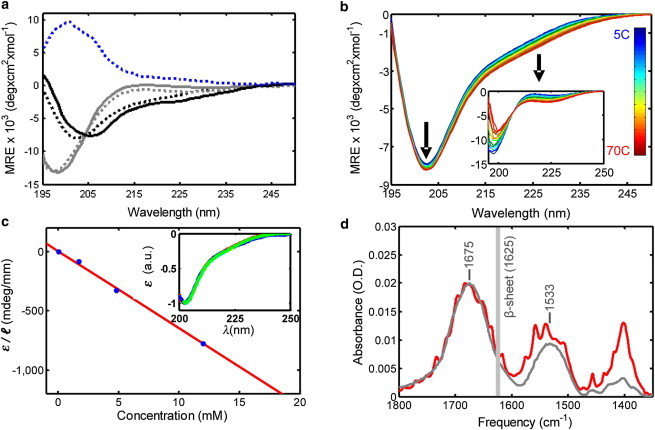
(a) CD spectra of linear N1–8 (gray line) and cyclic N_loop (black line) peptides at 5°C compared to pure component spectra for disordered coil (dotted gray line), type I β-turn (dotted black line), and type II β-turn (dotted blue line) (redrawn from Perczel and Fassman (30)); MRE, mean residue ellipticity. (b) Temperature dependence of CD spectra from 5°C to 70°C of N_loop and N1–8 (inset) (660 μM peptide in 50 mM sodium acetate, pH 4.9). (c) Concentration dependence of N_loop CD signal amplitude at minimum, ε, rescaled by pathlength l (solid circles) and linear fit (line); renormalized CD spectra do not vary with concentration (inset) (20°C, pH 4.9 in 10–25 mM sodium acetate buffer). (d) FTIR spectra of KBr pellets of HPLC-purified N_loop peptide (gray) versus fibers (red) in the amide region. N_loop samples were lyophilized immediately after HPLC purification. N_loop fiber samples were lyophilized after 1 week aging at 4°C (7 mM peptide in 50 mM NaAc, pH 4.9). Peak positions for the amide I (1675 cm−1) and II (1533 cm−1) bands are given for the N_loop spectrum. The vertical gray line indicates the characteristic amide I peak of IAPP amyloid fibrils. All spectra are atmospheric compensated, baseline corrected, and KBr subtracted. To see this figure in color, go online.
The CD spectrum of the linear peptide ( Fig. 3 a) is typical of linear disordered coils (29). In contrast, the spectrum of the cyclic peptide deviates significantly from disordered coils and contains distinct features. We compared it to the pure-component spectra obtained by spectral deconvolution of model cyclic peptides of similar length, in which the structure was determined by x-ray crystallography and solution NMR (30), (31). Fig. 3 a shows the overlay of the type II (blue line) and type I β-turn spectrum (black dotted line) (30) with the N_loop spectrum (black line). The resemblance with the type I β-turn spectrum is striking, considering that in short cyclic peptides CD is sensitive to very small backbone conformational distortions (31). We therefore attribute the CD spectral features of cyclic N_loop to a type I β-turn structure.
We derived further information on the structural rigidity of the peptides by monitoring the change in CD spectra as a function of temperature (Fig. 3 b). Although the spectrum of N1–8 displays significant changes upon increasing temperature, consistent with a noncooperative loss of polyproline II structure (as observed in other disordered proteins (32,33)), the N_loop remains almost unchanged. We conclude that cyclization imparts rigidity to the N_loop, rendering the type I β-turn stable to thermal denaturation.
To gain insight into the structural changes occurring upon fiber formation, we measured the CD spectra as a function of peptide concentration in the 10 μM to 12 mM range, which includes concentrations at which fibers were clearly visible. To our surprise, the signal amplitude followed a simple linear dependence with concentration (Fig. 3 c) and we observed no spectral changes (Fig. 3 c, inset). By contrast, significant spectral changes are typically observed during amyloid fibril formation, as peptides convert from disordered structures to β-sheet structures. Two explanations are possible: either the N-loop monomer does not change conformation upon aggregation or the fibers do not contribute to the CD signal. In the latter case, we would expect to observe a saturation of the signal amplitude at increasing peptide concentrations, as the monomers approach the solubility limit. The linear dependence of the signal at concentrations beyond those needed for fiber formation suggests instead that the monomer structure does not change upon fiber formation, and that the fiber structure does not introduce appreciable supramolecular chirality. There remains a third possibility, that even at 10 μM, the N_loop is present as a very low-molecular-weight oligomer (dimer/trimer/...) rather than as a monomer. In this event, our results would mean that the N_loop monomer has the same conformation in the low-molecular-weight oligomers (rather than monomers) and in the fibers. The question is then whether the structure of the monomer would be much affected by the formation of oligomers. Further evidence from REMD simulations, provided below, supports the contention that the monomer structure is rigid and is not significantly affected by aggregation.
To further test these conclusions, we compared the FTIR spectra of the HPLC purified peptide with those of the fibers in the amide region (Fig. 3 d). The appearance of fine structure in the fiber spectrum (red) indicates quasicrystalline order in the fibers. These data confirm the high degree of microscopic order revealed by birefringence (Fig. 2 b). Further, the observed increase in intensity of the near-amide-III bands (1300–1500 cm−1), which reflects contributions from methylene (CH2) and methyl (CH3) bending vibrations of aliphatic side chains (34), is consistent with packing of these side chains within the fibers. In contrast, the positions of the amide I and II peaks, which reflect backbone secondary structure (35), do not change. Thus, there is no significant backbone structural rearrangement when the N_loop is incorporated into the fibrils. The FTIR spectrum also confirms the absence of β-sheet structures in the fibers, which would appear as a distinct amide I peak at 1620–1630 cm−1 (36–38). The position of the amide I peak at 1675 cm−1 is consistent with a β-turn structure (observed in model linear and cyclic β-turn peptides at 1672–1674 cm−1) (39–42), in agreement with our interpretation of the CD spectrum of the N_loop (Fig. 3a).Taken together, the data presented here indicate that the disulfide bond constrains the N_loop into a fairly rigid, well defined structure containing a type I β-turn. This structure highly favors the formation of stable interpeptide interactions that result in the observed formation of fibers.
Solution NMR
To obtain more detailed structural information on the N_loop, we measured proton chemical shifts and NOEs by 2D-NMR solution spectroscopy. Given the low solubility of N_loop in aqueous solvent, we performed these measurements in 100% deuterated DMSO. We calculated proton secondary chemical shifts by subtracting residue-specific random-coil values (23) from our measured chemical shifts. These values are typically used to identify propensities to populate α-helix versus β-sheet dihedral angles in intrinsically disordered proteins (22). Fig. 4 shows a comparison between values obtained here for N_loop and values previously reported by Yonemoto et al. (16) and Williamson et al. (43) for the corresponding residues of full-length hIAPP and rIAPP, respectively. These data do not indicate significant structural differences between the N_loop and the corresponding residues of full-length IAPP, considering the difference in solvent conditions and the variations originating from the use of different random-coil libraries (a comparison of random-coil libraries (23–25) is shown in Fig. S3). In the Supporting Material, we also consider the effect of aqueous dilutions of DMSO (Figs. S4 and S5).
Figure 4.
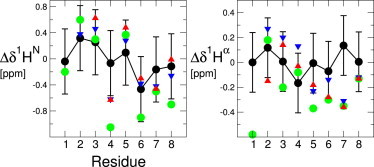
Comparison of secondary chemical shifts measured for N_loop (green), using random coil values of reference De Simone et al. (23), and values reported in Yonemoto et al. (16) and Williamson and Miranker (43), for residues 1–8 of full-length hIAPP (red) and rIAPP (blue). Values computed from REMD simulations using SPARTA+ (black) are also compared with experimental data sets. Error bars on the calculated shifts represent the root mean-squared error in the shift prediction algorithm. To see this figure in color, go online.
In Table 1, we report the strength of amide-amide sequential NOEs obtained from 2D-NOESY experiments, which we have classified as weak, medium, and strong, according to cutoffs reported byNilges et al. (26) (additional NOE data are shown in Fig. S9). As an internal calibration standard, we used the known distance (1.8 Å) between the two β-protons of C2. Analysis of the 80 ms and 400 ms mixing-time spectra yielded similar results (Fig. S9). We note that direct comparison between these values and those reported by Yonemoto et al. would require the use of robust and self-consistent internal calibration standards. Generally speaking, though, our NOE peaks are consistent with those observed by Yonemoto et al. (16), with the exception of the presence of a T4-A5 crosspeak (absent in the Yonemoto study (16)), which appears instead of the T4-A8 crosspeak observed by Yonemoto and co-workers (16). The proximity of T4 and A8 observed in the Yonemoto work (16) was attributed to a stabilizing effect that the N_loop would have on the helical conformational ensemble sampled by residues 8–18 of full-length hIAPP. The absence of the T4-A8 crosspeak in the N_loop, where residues 9–37 are missing, is consistent with this interpretation. In summary, these data do not indicate significant backbone structural changes between the isolated N_loop and the corresponding residues of full-length hIAPP. Although this is not generally expected for linear fragments of full-length proteins, it is not surprising in the case of short, relatively rigid cyclic peptides such as the N_loop.
Table 1.
Comparison between measured NOEs and distances from simulations
| Residues | NOE | Simulation distance (Å) |
|---|---|---|
| 1 and 2 | Medium | 4.05 |
| 2 and 3 | Medium | 4.44 |
| 3 and 4 | Strong | 3.03 |
| 4 and 5 | Medium | 3.13 |
| 5 and 6 | Strong | 2.73 |
| 6 and 7 | Strong | 2.40 |
| 7 and 8 | Strong | 2.88 |
Amide-amide sequential NOE strengths and averaged distances computed as <r3>−1/3 from the REMD simulation replica at 295 K. Additional NOEs and comparison to simulations are shown in Fig. S9.
MD simulations
To provide a molecular description of the possible location of the type I β-turn in the N_loop monomer, we ran REMD simulations of the N_loop sequence studied here (residues 1–8). The N_loop conformations we obtained are similar to those found in our earlier work (21). Because of the constraint introduced by the linker, the N_loop only adopts a limited number of backbone conformations. This rigidity is evident in the Cα root mean-square fluctuations of the backbone, which are only 1.0 Å at 278 K, and increase to 1.15 Å at the highest REMD temperature of 595 K. Conformational clustering with a 0.8 Å cut-off yields two major conformations with populations of 64.0% and 33.3%, with the remaining conformations having very low populations. Secondary-structure assignment using STRIDE indicates that the most populated cluster (Fig. 5 a) has a type I turn between residues 3 and 6 and a type IV turn between residues 4 and 7, whereas the second-most-populated cluster (Fig. 5 b) has a type I turn from residues 4 to 7. Thus, there is expected to be a large population of type I turns, consistent with the experimental findings. Type I β-turns are geometrically defined as a stretch of four residues (i,i + 3), with α carbons of residues i and i + 3 within 7 Å and internal residues i + 1 and i + 2 having (ψ,ϕ) angles of (−30,−60) and (0,−90), respectively (44). We found, based on these geometric criteria (Fig. S6), that a type I β-turn is present at residues 3–6 most of the time, with a significant fraction of type I turn also present at residues 4–7, in agreement with the analysis of the most populated clusters. We have also validated the results of the simulations by comparison with the available NMR data. Calculation of average 1Hα and 1HN chemical shifts using the SPARTA+ shift prediction algorithm (45) results in reasonable agreement with experimental data from several groups, including our own, considering the uncertainty in the shift prediction (Fig. 4). We find that 75% of the predictions lie within the (one-standard-deviation) error bars for the HN shifts and 63% for the Hα shifts. The linear correlation coefficients are 0.84 and 0.1 for the HN and Hα shifts, respectively. Furthermore, the distances between amide protons measured in the simulation correlate well with the intensities of the NOEs measured for the N_loop (Table 1 and Fig. S9).
Figure 5.
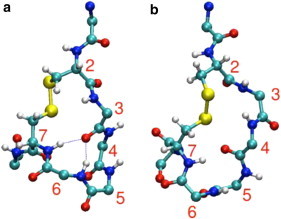
Structure of N_loop from molecular simulations in CPK representation. (a) Most populated conformation (64.0%), with type I turn between residues 3 and 6. (b) Next-most populated conformation (33.3%), with type I turn between residues 4 and 7. To see this figure in color, go online.
To investigate how much the structure of the monomer might be affected by self-association, we performed REMD simulations of a pair of monomers at ∼100 mM concentration and analyzed the configurations where the two molecules are in direct contact. The structures of the monomers in the associated configurations were generally very similar to those of the isolated peptides, with 70% of the monomers having a Cα root mean-square deviation from the central structure of the largest cluster of the monomer simulations of <1.0 Å. Representative clusters of the associated pairs of monomers are given in Fig. S10. These results also shed some light on the type of interactions that may drive aggregation—the peptides associate through a variety of polar interactions but clearly not through a classical amyloid structure. Although there are certainly some intermolecular interactions involving the backbone, because the amide groups are not aligned in the same direction, conventional amyloid structure is not formed.
Discussion
Our experimental observations and REMD simulations concur in showing that the N-terminal fragment (residues 1–8) of hIAPP is constrained into a well-defined, rigid structure containing a type I β-turn. This structure highly favors the formation of stable interpeptide interactions resulting in the observed formation of fibers. Cyclization has the effect of significantly decreasing the conformational entropy cost required for fiber formation, making it much more favorable for the N_loop to aggregate compared to the linear peptide, N1–8. This mechanism is similar to that exploited in the design of cyclized, stable β-hairpin peptides that readily self-assemble into highly ordered nanofibers, rodlike crystals, and other β-sheet aggregates (46–52). To our knowledge, though, this is the first time this mechanism has been observed for a non-β-sheet fiber, formed by type I β-turn structures. Molecular simulations suggest that the amide groups of the N_loop are not correctly aligned for formation of conventional amyloid structure, and simulations of a pair of N_loops did not reveal any amyloidlike association. The rigidity of the N_loop, coupled with the relatively small number of observed modes of interaction, may explain the high propensity to form fibrils.
Our observation of stable N_loop fibers at concentrations as low as 57 μM suggests that the N_loop plays a direct role in IAPP aggregation. According to structural models of hIAPP amyloid fibers, these fibers are formed by intermolecular β-sheets involving residues ∼25–37 (C-terminal sheet) and 8 to ∼17 (N-terminal sheet), and the N_loop (residues 1–7) is not involved in direct intermolecular interactions (13). However, removal of the N_loop alters both the mass/length distributions of hIAPP fibers (14) and the kinetics of fibril formation (15,16). Based on the observed strong tendency of the N_loop to form interpeptide interactions, we propose that N_loop-N_loop interactions occurring at the N-terminus of full-length IAPP may drive the initial association of IAPP, before β-sheet formation. This would be consistent with experimental data on both hIAPP amyloid fiber formation and the disruption of β-sheet interactions in hIAPP fibers in the presence of rIAPP, as explained below.
Initial association of IAPP by N_loop-N_loop interactions could occur at a relatively low entropy cost, unlike the direct association of β-sheets. In sequences that allow extensive β-sheet formation (e.g., in hIAPP, and not in rIAPP), such interactions would favor aggregation into ordered fibrils by aligning the peptides at the N-terminus, consistent with accepted models of hIAPP fibrils (13). This mechanism would be consistent with recent 2D-IR studies indicating that β-sheet structures form in the later stages of hIAPP aggregation (53), and with p-cyanophenilalanine fluorescence studies indicating that side chains of residues 15, 23, and 37 remain partially exposed to solvent during the lag phase, until significant β-sheet structure has formed (54). In addition, it would explain the observed changes in nucleation processes in the absence of the intact N_loop (15,16). We note that N_loop fibers (i.e., fibers formed by N_loop-N_loop interactions) have not been observed so far at equilibrium in full-length hIAPP. Two factors would interfere with the formation of N_loop-based fibers in full-length hIAPP: first, intramolecular interactions between the N_loop and the linear portion of the chain (residues 8–37) will contribute to stabilization of the monomer and compete with N_loop-N_loop interactions. Such intramolecular interactions have been proposed by Vaiana et al. to explain the large degree of compaction observed in full-length hIAPP and rIAPP, and in a model hydrophilic sequence that contained the intact N_loop (21). This model is supported by molecular simulations (21). Second, the distance between the N_loops in the NMR structures of the fiber suggest that formation of N_loop-based fibers may be in competition with the extensive intermolecular hydrogen bonding of the amyloid fibrils.
Our proposed model would also be consistent with the ability of rIAPP to disrupt intermolecular β-sheets in hIAPP amyloid fibers. According to our model, in fact, direct N_loop-N_loop interactions would also occur in solutions containing mixtures of IAPP variants. Variants of hIAPP containing proline substitutions in the C-terminal region, but with identical N-terminal sequences and intact N_loop (e.g., rIAPP, I26P, and G24P), have been shown to inhibit hIAPP amyloid fiber formation, implying a direct interaction between hIAPP and the inhibitor (55,56). Recent results show that interactions between rIAPP and hIAPP initiate in the N-terminal region of the peptides and disrupt the native β-sheet structure of hIAPP amyloid fibers (12). Our present findings suggest that such interactions could be mediated by N_loop-N_loop association, which could act in addition to, or as an alternative to, the originally proposed mechanism of aggregation via α-helical or β-sheet intermediates (9,19,20,57).
Our proposed role of the N_loop in mediating intermolecular interactions of IAPP may in fact be related to the role of this highly conserved sequence in the biological activity of Ct family peptides. Although receptor binding affinity is mainly determined by residues in the 8–37 region, the N_loop is required for Ct peptides to activate cell response when binding to their receptors (58,59), possibly via interactions between the N_loop and the extramembrane region of the CGRP receptor (60). We propose that the rigid type I β-turn structure reported here mediates such interactions. This would be consistent with early work showing that modifications of the disulfide bridge topography (by substitution of cysteinyl residues with penicillamine) can greatly affect CGRP receptor activation upon ligand binding (61).
Summary and Conclusions
In conclusion, the data reported here demonstrate for the first time, to our knowledge, that the N_loop of IAPP forms stable fibers containing a type I β-turn structure and not β-sheets. We propose that the rigid β-turn structure plays a role in both the biological activity of Ct family peptides and in the pathological aggregation of IAPP, a possibility that to our knowledge has not yet been explored. We note that N_loop-N_loop interactions could be exploited to develop novel inhibitors of hIAPP amyloid formation.
Acknowledgments
We thank W. A. Eaton and R. Pappu for helpful discussions, Anindya Roy for help with peptide purification, David Lowry for help with the TEM imaging, Arnab Dutta for help with the FTIR instrumentation, Brian Cherry for help with NMR instrumentation and student training, and Marina Faiella for help with NMR software. We gratefully acknowledge the use of facilities in the LeRoy Eyring Center for Solid State Science, the Proteomics Facility and the Life Sciences EM Facility, and the Magnetic Resonance Research Center at Arizona State University (ASU). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland (http://biowulf.nih.gov).
This work was supported by ASU start-up funds to S.M.V. G.G. and S.S. were supported in part by National Science Foundation CAREER award 0449842. R.B.B. was supported by a Royal Society University Research Fellowship and by the Intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References
- 1.Cooper G.J.S., Leighton B., Reid K.B.M. Amylin found in amyloid deposits in human type 2 diabetes mellitus may be a hormone that regulates glycogen metabolism in skeletal muscle. Proc. Natl. Acad. Sci. USA. 1988;85:7763–7766. doi: 10.1073/pnas.85.20.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reidelberger R.D., Arnelo U., Permert J. Comparative effects of amylin and cholecystokinin on food intake and gastric emptying in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;280:R605–R611. doi: 10.1152/ajpregu.2001.280.3.R605. [DOI] [PubMed] [Google Scholar]
- 3.Kahn S.E., D’Alessio D.A., Porte D., Jr. Evidence of cosecretion of islet amyloid polypeptide and insulin by β-cells. Diabetes. 1990;39:634–638. doi: 10.2337/diab.39.5.634. [DOI] [PubMed] [Google Scholar]
- 4.Westermark P., Andersson A., Westermark G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011;91:795–826. doi: 10.1152/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- 5.Cooper G.J.S., Willis A.C., Reid K.B.M. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. USA. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westermark P., Wernstedt C., Sletten K. A novel peptide in the calcitonin gene related peptide family as an amyloid fibril protein in the endocrine pancreas. Biochem. Biophys. Res. Commun. 1986;140:827–831. doi: 10.1016/0006-291x(86)90708-4. [DOI] [PubMed] [Google Scholar]
- 7.Wimalawansa S.J. Amylin, calcitonin gene-related peptide, calcitonin, and adrenomedullin: a peptide superfamily. Crit. Rev. Neurobiol. 1997;11:167–239. doi: 10.1615/critrevneurobiol.v11.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 8.Westermark P., Engström U., Betsholtz C. Islet amyloid polypeptide: pinpointing amino acid residues linked to amyloid fibril formation. Proc. Natl. Acad. Sci. USA. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao P., Meng F., Raleigh D.P. The ability of rodent islet amyloid polypeptide to inhibit amyloid formation by human islet amyloid polypeptide has important implications for the mechanism of amyloid formation and the design of inhibitors. Biochemistry. 2010;49:872–881. doi: 10.1021/bi901751b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abedini A., Raleigh D.P. Destabilization of human IAPP amyloid fibrils by proline mutations outside of the putative amyloidogenic domain: is there a critical amyloidogenic domain in human IAPP? J. Mol. Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 11.Betsholtz C., Christmansson L., Westermark P. Sequence divergence in a specific region of islet amyloid polypeptide (IAPP) explains differences in islet amyloid formation between species. FEBS Lett. 1989;251:261–264. doi: 10.1016/0014-5793(89)81467-x. [DOI] [PubMed] [Google Scholar]
- 12.Middleton C.T., Marek P., Zanni M.T. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat. Chem. 2012;4:355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luca S., Yau W.M., Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsbury C., Goldie K., Aebi U. Amyloid fibril formation from full-length and fragments of amylin. J. Struct. Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 15.Koo B.W., Miranker A.D. Contribution of the intrinsic disulfide to the assembly mechanism of islet amyloid. Protein Sci. 2005;14:231–239. doi: 10.1110/ps.041051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonemoto I.T., Kroon G.J.A., Kelly J.W. Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrone F.A. Nucleation: the connections between equilibrium and kinetic behavior. Methods Enzymol. 2006;412:285–299. doi: 10.1016/S0076-6879(06)12017-0. [DOI] [PubMed] [Google Scholar]
- 18.Eichner T., Radford S.E. A diversity of assembly mechanisms of a generic amyloid fold. Mol. Cell. 2011;43:8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Williamson J.A., Loria J.P., Miranker A.D. Helix stabilization precedes aqueous and bilayer-catalyzed fiber formation in islet amyloid polypeptide. J. Mol. Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abedini A., Raleigh D.P. A critical assessment of the role of helical intermediates in amyloid formation by natively unfolded proteins and polypeptides. Protein Eng. Des. Sel. 2009;22:453–459. doi: 10.1093/protein/gzp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaiana S.M., Best R.B., Hofrichter J. Evidence for a partially structured state of the amylin monomer. Biophys. J. 2009;97:2948–2957. doi: 10.1016/j.bpj.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjaergaard M., Poulsen F.M. Disordered proteins studied by chemical shifts. Prog. Nucl. Magn. Reson. Spectrosc. 2012;60:42–51. doi: 10.1016/j.pnmrs.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 23.De Simone A., Cavalli A., Vendruscolo M. Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J. Am. Chem. Soc. 2009;131:16332–16333. doi: 10.1021/ja904937a. [DOI] [PubMed] [Google Scholar]
- 24.Tamiola K., Acar B., Mulder F.A.A. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J. Am. Chem. Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- 25.Kjaergaard M., Poulsen F.M. Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR. 2011;50:157–165. doi: 10.1007/s10858-011-9508-2. [DOI] [PubMed] [Google Scholar]
- 26.Nilges M., Clore G.M., Gronenborn A.M. 1H-NMR stereospecific assignments by conformational data-base searches. Biopolymers. 1990;29:813–822. doi: 10.1002/bip.360290415. [DOI] [PubMed] [Google Scholar]
- 27.Best R.B., Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudva Y.C., Mueske C., Eberhardt N.L. A novel assay in vitro of human islet amyloid polypeptide amyloidogenesis and effects of insulin secretory vesicle peptides on amyloid formation. Biochem. J. 1998;331:809–813. doi: 10.1042/bj3310809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenfield N.J. Analysis of circular dichroism data. Methods Enzymol. 2004;383(Pt D):282–317. doi: 10.1016/S0076-6879(04)83012-X. [DOI] [PubMed] [Google Scholar]
- 30.Perczel A., Fasman G.D. Quantitative analysis of cyclic β-turn models. Protein Sci. 1992;1:378–395. doi: 10.1002/pro.5560010310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manning M.C., Illangasekare M., Woody R.W. Circular dichroism studies of distorted α-helices, twisted β-sheets, and β turns. Biophys. Chem. 1988;31:77–86. doi: 10.1016/0301-4622(88)80011-5. [DOI] [PubMed] [Google Scholar]
- 32.Kjaergaard M., Nørholm A.B., Kragelund B.B. Temperature-dependent structural changes in intrinsically disordered proteins: formation of α-helices or loss of polyproline II? Protein Sci. 2010;19:1555–1564. doi: 10.1002/pro.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uversky V.N. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 34.Barth A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 35.Susi H., Timasheff S.N., Stevens L. Infrared spectra and protein conformations in aqueous solutions. I. The amide I band in H2O and D2O solutions. J. Biol. Chem. 1967;242:5460–5466. [PubMed] [Google Scholar]
- 36.Zandomeneghi G., Krebs M.R.H., Fändrich M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson M.R., Raleigh D.P. Analysis of amylin cleavage products provides new insights into the amyloidogenic region of human amylin. J. Mol. Biol. 1999;294:1375–1385. doi: 10.1006/jmbi.1999.3286. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee S., Chowdhury P., Gai F. Effect of dehydration on the aggregation kinetics of two amyloid peptides. J. Phys. Chem. B. 2009;113:531–535. doi: 10.1021/jp809817s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okabayashi H., Ishida M., O’Connor C.J. Fourier transform IR study of aggregational behavior of N-acetyl-L- and N-butyloxycarbonyl-L-glutamic acid oligomeric benzyl esters in dioxane and benzene: β-turn→antiparallel β-sheet transition. Biopolymers. 2002;65:129–141. doi: 10.1002/bip.10186. [DOI] [PubMed] [Google Scholar]
- 40.Krimm S., Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 41.Mantsch H.H., Moffatt D.J., Casal H.L. Fourier-transform methods for spectral resolution enhancement. J. Mol. Struct. 1988;173:285–298. [Google Scholar]
- 42.Vass E., Kurz M., Konat R.K., Hollosi M. FTIR and CD spectroscopic studies on cyclic penta- and hexa-peptides. Detailed examination of hydrogen bonding in β- and γ-turns determined by NMR. Spectrochim. Acta A. 1998;54:773–786. [Google Scholar]
- 43.Williamson J.A., Miranker A.D. Direct detection of transient α-helical states in islet amyloid polypeptide. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutchinson E.G., Thornton J.M. A revised set of potentials for β-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valéry C., Pouget E., Paternostre M. Molecular origin of the self-assembly of lanreotide into nanotubes: a mutational approach. Biophys. J. 2008;94:1782–1795. doi: 10.1529/biophysj.107.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishihara Y., Kimura S. Nanofiber formation of amphiphilic cyclic tri-β-peptide. J. Pept. Sci. 2010;16:110–114. doi: 10.1002/psc.1206. [DOI] [PubMed] [Google Scholar]
- 48.Joshi K.B., Verma S. Ordered self-assembly of a glycine-rich linear and cyclic hexapeptide: contrasting ultrastructural morphologies of fiber growth. Supramol. Chem. 2006;18:405–414. [Google Scholar]
- 49.Clark T.D., Buriak J.M., Ghadiri M.R. Cylindrical β-sheet peptide assemblies. J. Am. Chem. Soc. 1998;120:8949–8962. [Google Scholar]
- 50.Kobayashi K., Granja J.R., Ghadiri M.R. β-Sheet peptide architecture: measuring the relative stability of parallel vs. antiparallel β-sheets. Angew. Chem. Int. Ed. Engl. 1995;34:95–98. [Google Scholar]
- 51.Gibbs A.C., Kondejewski L.H., Wishart D.S. Unusual β-sheet periodicity in small cyclic peptides. Nat. Struct. Biol. 1998;5:284–288. doi: 10.1038/nsb0498-284. [DOI] [PubMed] [Google Scholar]
- 52.Khakshoor O., Nowick J.S. Use of disulfide “staples” to stabilize β-sheet quaternary structure. Org. Lett. 2009;11:3000–3003. doi: 10.1021/ol901015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shim S.H., Gupta R., Zanni M.T. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc. Natl. Acad. Sci. USA. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marek P., Mukherjee S., Raleigh D.P. Residue-specific, real-time characterization of lag-phase species and fibril growth during amyloid formation: a combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J. Mol. Biol. 2010;400:878–888. doi: 10.1016/j.jmb.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng F.L., Raleigh D.P., Abedini A. Combination of kinetically selected inhibitors in trans leads to highly effective inhibition of amyloid formation. J. Am. Chem. Soc. 2010;132:14340–14342. doi: 10.1021/ja1046186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abedini A., Meng F.L., Raleigh D.P. A single-point mutation converts the highly amyloidogenic human islet amyloid polypeptide into a potent fibrillization inhibitor. J. Am. Chem. Soc. 2007;129:11300–11301. doi: 10.1021/ja072157y. [DOI] [PubMed] [Google Scholar]
- 57.Dupuis N.F., Wu C., Bowers M.T. The amyloid formation mechanism in human IAPP: dimers have β-strand monomer-monomer interfaces. J. Am. Chem. Soc. 2011;133:7240–7243. doi: 10.1021/ja1081537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hay D.L., Christopoulos G., Sexton P.M. Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol. Pharmacol. 2005;67:1655–1665. doi: 10.1124/mol.104.008615. [DOI] [PubMed] [Google Scholar]
- 59.Poyner D. Pharmacology of receptors for calcitonin gene-related peptide and amylin. Trends Pharmacol. Sci. 1995;16:424–428. doi: 10.1016/s0165-6147(00)89093-8. [DOI] [PubMed] [Google Scholar]
- 60.Conner A.C., Simms J., Poyner D.R. Ligand binding and activation of the CGRP receptor. Biochem. Soc. Trans. 2007;35:729–732. doi: 10.1042/BST0350729. [DOI] [PubMed] [Google Scholar]
- 61.Saha S., Waugh D.J.J., Smith D.D. Role of conformational constraints of position 7 of the disulphide bridge of h-α-CGRP derivatives in their agonist versus antagonist properties. J. Pept. Res. 1998;52:112–120. doi: 10.1111/j.1399-3011.1998.tb01365.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


