Abstract
To identify genes influencing age at onset (AAO) in two common neurodegenerative diseases, a genomic screen was performed for AAO in families with Alzheimer disease (AD; n=449) and Parkinson disease (PD; n=174). Heritabilities between 40%–60% were found in both the AD and PD data sets. For PD, significant evidence for linkage to AAO was found on chromosome 1p (LOD = 3.41). For AD, the AAO effect of APOE (LOD = 3.28) was confirmed. In addition, evidence for AAO linkage on chromosomes 6 and 10 was identified independently in both the AD and PD data sets. Subsequent unified analyses of these regions identified a single peak on chromosome 10q between D10S1239 and D10S1237, with a maximum LOD score of 2.62. These data suggest that a common gene affects AAO in these two common complex neurodegenerative diseases.
Introduction
Genetic studies of common complex neurodegenerative diseases, such as Alzheimer disease (AD [MIM 104300]) and Parkinson disease (PD [MIM 168600]), have focused on the identification of risk genes as targets for development of new treatments and improved diagnoses. This approach has identified the amyloid precursor protein (APP) (Goate et al. 1991), presenilin 1 (PS1) (Sherrington et al. 1995), presenilin 2 (PS2) (Levy-Lahad et al. 1995; Rogaev et al. 1995), and apolipoprotein E (APOE) (Corder et al. 1993) genes as contributing to risk in AD. APP, PS1, and PS2 cause rare early-onset autosomal dominant AD (5% of AD cases), whereas APOE is associated with both risk and age at onset (AAO) in late-onset familial AD, as well as in late- and early-onset sporadic AD. Similarly, three genes have been identified to associate with risk in PD: α-synuclein (Polymeropoulos et al. 1996) for rare autosomal dominant early-onset PD, Parkin (Abbas et al. 1999) for rare autosomal recessive juvenile parkinsonism and autosomal recessive early-onset PD, and tau (Martin et al. 2001) for classic PD. Genomic screens in both PD (Destefano et al. 2001; Scott et al. 2001) and AD (Kehoe et al. 1999; Pericak-Vance et al. 2000) have recently localized additional—but, as yet, unknown—risk genes.
However, risk is only one mode of genetic expression; AAO of disease may also be genetically influenced and may have an effect equivalent to that seen for the known risk genes (Daw et al. 2000). Identification of such genes would open new avenues of research with the potential to delay onset beyond the natural life span. Present knowledge about genes contributing to AAO in neurodegenerative diseases clearly lags behind the understanding of genes contributing to risk. Recently, there has been growing interest in using AAO information as a quantitative trait, to identify genes that influence onset of disease (Daw et al. 1999, 2000; Duggirala et al. 1999). Rapid development of methods of mapping quantitative trait loci (QTLs) for general pedigrees (Goldgar 1990; Amos 1994; Blangero and Almasy 1997) has now made the search for novel genes affecting AAO feasible.
In this study, the variance-component procedure in SOLAR (Blangero and Almasy 1997) was used to perform genomewide scans on the quantitative trait AAO for AD and PD to map QTLs influencing AAO. This method is less penetrance-model dependent than the classical segregation/linkage-mapping technique, and it can take into account covariate or random effects. The common regions showing evidence of linkage from independent analyses of AD and PD data sets were analyzed further by use of the combined AD and PD data set. Because AD and PD share some common clinical and pathological findings, we hypothesized that a gene or genes common to both disorders control AAO and could be localized by use of this genomic-screening approach.
Material and Methods
Family Ascertainment
In this study, we used a total of 449 families affected with AD and 174 families affected with PD. Ascertainment of data from families with AD and PD was independently conducted by various research centers. The data from families with AD were ascertained by the following centers: the Duke Center for Human Genetics (CHG); the Joseph and Kathleen Bryan Alzheimer’s Disease Research Center (Bryan ADRC); the Indiana Alzheimer’s Disease Research Center National Cell Repository (IADRC); the National Institute of Mental Health (NIMH); and Vanderbilt University (VAN). In all data sets, affected individuals were classified in accordance with the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Associations (NINCDS-ADRDA) clinical diagnostic criteria (McKhann et al. 1984). The reported AAO of patients with AD was defined as the age at which the caregiver, family, and/or individual first noted cognitive problems (most often short-term memory loss and, more rarely, other problems, such as dysphasia or disorientation to time or place, followed closely by memory change) sufficient to interfere with independent daily activities. The data from families with PD were ascertained by 13 centers in the United States and Australia (see Scott et al. 2001). Diagnostic and exclusion criteria, based on previously published diagnostic criteria for PD (Ward and Gibb 1990; Hughes et al. 1992a, 1992b), were adopted by all participating clinicians before beginning selection of families. Affected individuals were defined by having at least two cardinal signs of PD (e.g., rest tremor, bradykinesia, and rigidity) and no atypical clinical features or other causes of parkinsonism. The reported AAO was defined as the age at which an affected individual first noticed one of the cardinal signs of PD. In both AD and PD ascertainment, the reported AAE was recorded as the age at which study personnel clinically examined a participant. The data description is summarized in table 1. Overall, for AD, we have 1,121 affected individuals with reported AAO and 746 unaffected individuals with reported AAE, and, for PD, we have 378 affected individuals with reported AAO and 470 unaffected individuals with reported AAE. Average reported AAO ± SD was 72.8 ± 6.8 years for AD and 60.1 ± 12.7 years for PD. Average reported AAE for unaffected individuals was 70.2 ± 13.0 years for AD and 68.4 ± 12.7 years for PD. All participants or their legal representatives gave informed consent prior to joining the study, and data were collected according to protocols approved by each contributing group’s institutional review board.
Table 1.
Description of Data Sets
|
Value for Data Set |
|||
| Characteristic | AD | PD | ADPD |
| No. of families | 449 | 174 | 623 |
| No. of affected individuals with reported AAO information | 1,121 | 378 | 1,499 |
| No. of unaffected individuals with reported AAE information | 746 | 470 | 1,216 |
| Mean ± SD reported AAO, in years (range) | 72.8 ± 6.8 (49–97) | 60.1 ± 12.7 (12–90) | 69.6 ± 10.3 (12–97) |
| Mean ± SD reported AAE, in years, from unaffected individuals (range) | 70.2 ± 13.0 (29–105) | 68.4 ± 12.7 (34–98) | 69.6 ± 12.9 (29–105) |
Modeling AAO Data
We modeled AAO data as described by Duggirala et al. (1999), where AAO was suggested as a right-truncated quantitative trait in affected individuals (i.e., reported AAO is less than or equal to reported AAE) and a left-truncated quantitative trait in unaffected individuals (i.e., reported AAO is greater than reported AAE). That is, the AAO data was comprised of reported AAO for affected individuals and reported AAE for unaffected individuals. Outliers and normality of the data were examined for agreement with the assumption of QTL analysis. Four outliers that were 4 SD below the mean AAO were excluded in the AD data set.
Linkage Analysis
We used the variance-components procedure in SOLAR (Almasy and Blangero 1998) for linkage analysis. In theory, the quantitative phenotype AAO (y) was defined as a linear function of the n QTLs (ri) that influence the trait:
 |
where X is a matrix of covariates and β is the regression coefficient matrix associated with the covariates. The phenotype is assumed to follow a normal distribution. The likelihood function of y includes the identical by descent (IBD) probability at a marker that is linked to a QTL, the additive genetic variance attributed by an unobserved QTL (σ2q), and other variance components. SOLAR employs the likelihood-ratio test to test a null hypothesis of σ2q=0 (no linkage) and generates a LOD score that is the equivalent of the classical LOD score of linkage analysis. This technique can be applied to detect the evidence of linkage to an individual marker for two-point analysis or to an imputed chromosomal position in multipoint analysis. Locus-specific IBD information for pairs of relatives was obtained prior to computation of the likelihood function. The multipoint mapping strategy in SOLAR is an extension of the method of Fulker et al. (1995). It requires the map distance between the markers to create the IBD information of a pair of relatives at a QTL that is linked to a marker. We used a Kosambi sex-averaged map obtained from Map-O-Mat (Lander and Green 1987; Matise and Gitlin 1999). The linkage analysis method implemented in SOLAR does not require specification of disease-allele frequency, penetrance, or mode of inheritance, which differs from the classical linkage mapping procedure.
The initial genomic screens were performed on an AD data set of 449 families with a total of 4,316 relative pairs (sib pairs, cousin pairs, avuncular pairs, etc.) and a PD data set of 174 families with 2,256 relative pairs. A total of 323 (AD) and 330 (PD) microsatellite markers, with an average spacing of 10 cM, were analyzed (Vance and Ben Othmane 1998). Since AAO was modeled as a truncated quantitative trait, the overall distribution of AAO was thus considered as a mixture of two truncated normal distributions. We included sex and affection status as covariates in the polygenic model (model 1), in which affection status was used for adjustment of the contribution of reported AAO and reported AAE to the quantitative trait AAO.
The common interesting (see below) regions identified by the initial linkage analyses of AD and PD data sets were followed up by use of the combined AD and PD (ADPD) data set for linkage analysis. There are many reasons to consider the hypothesis of similar genetic mechanisms leading to AAO in AD and PD. Both AD and PD are neurodegenerative, late-AAO disorders. Clinically, a significant number of patients with AD develop signs of parkinsonism, including bradykinesia, rigidity, and gait abnormalities (Wilson et al. 2000). Conversely, dementia is a major factor in PD, and the two disorders both exhibit degeneration of cholinergic neurons in the nucleus basalis of Meynert (Korczyn 2001). Another pathological similarity is the presence of similar staining α-synuclein Lewy bodies (LB) in both disorders, supporting the premise that the two diseases may share a common pathway leading to LB formation (Lippa et al. 2001). APOE, a well-proven risk factor for AD, has been suggested, in some studies (e.g., Harhangi et al. 2000), to be involved in the risk for PD as well, although others have failed to confirm these findings (Khan et al. 2001). In addition, frontotemporal dementia-17, caused by mutations in the tau gene, presents phenotypically with a dementia similar to AD coupled with parkinsonism (McKhann et al. 2001). The tau gene, a major element in the neurofibulary tangles of AD, has recently been shown to be associated with PD as well (Martin et al. 2001). These similarities are likely not derived from the biologic events that initiate the start of each disease but rather from overlapping neuropathic pathways involved in the progression of each disorder. It is these same pathways and modifier genes that are likely to contribute to phenotypic traits such as AAO, which is why we pooled these two neurodegenerative diseases for a combined analysis.
For the analysis of the ADPD data set, two polygenic models were considered, to incorporate two different scenarios:
Model 1
We assumed that AD and PD are the same disease, so sex and affection status were included as covariates as that in the initial linkage analysis.
Model 2
We considered that the distribution of the ADPD data set is a mixture of two normal distributions (AAO from AD and AAO from PD), so disease status was included as an additional covariate, to distinguish the different contribution of AAO between AD and PD.
Results
Polygenic Models Revealed Strong Heritability of AAO Genes
The analyses of polygenic models showed that sex, affection status, and disease were all significant covariates, for instance, with P values <.0001 for the ADPD data set. The proportion of variance contributed by all covariates included in the model ranged from 0.9% to 15.3% among different data sets (table 2). We found that the AD data set showed the smallest proportion of variance from all covariates (0.9%). This may be due to the larger sample size for AD than for PD or to the marginally significant effect of sex and affection status in AD (P=.03 for sex and P=.05 for affection status). The heritability of AAO after these covariates were controlled for was highly significant in each of the data sets (P<.0001), with heritabilities of 42% (AD), 62% (PD), 56% (ADPD, model 1), and 49% (ADPD, model 2) (table 2). These data strongly indicate that genes are important modulators of AAO.
Table 2.
Residual Heritability (SE) of the AAO for Each Data Set under Each Model[Note]
|
Residual Heritabilitya [SE](%) |
Proportion ofVariance Contributedby All Covariates |
|||
| DataSet | Model 1 | Model 2 | Model 1 | Model 2 |
| AD | 41.8% [.038] | … | 0.9% | … |
| PD | 61.3% [.044] | … | 15.3% | … |
| ADPD | 55.7% [.028] | 49.2% [.029] | 1.8% | 8.3% |
Note.— The polygenic models are: model 1, covariate at sex and affection status; and model 2, covariate at sex, affection status, and disease.
The residual heritability is the ratio of the residual variance (after removing the covariate effects) to the total phenotypic variance.
Disease-Specific Linkage Evidence
The initial linkage analyses were performed on the AD and PD data sets separately. A threshold of LOD >1.00 was used for declaring a region “interesting” (table 3) (Weeks et al. 2000) and warranting follow-up analyses. Of greatest interest for PD is the result on chromosome 1, near D1S2134 (78 cM; LOD=3.41). For AD, examination of APOE, a known modulator of AAO in AD (Corder et al. 1993), generated a LOD score of 3.28 in the AD data set, confirming its role as a modulator of AAO. In addition to APOE, strong AD-specific suggestive linkage regions on chromosome 4q at D4S1652 (208 cM; LOD=2.29) and chromosome 8q (150 cM, LOD=2.09) were also found. Interestingly, neither chromosome 4q nor chromosome 8q have been reported as linkage regions with AD risk genes. It is possible that chromosomes 4q and 8q harbor genes that exclusively modulate onset of AD.
Table 3.
Summary of the Peak Regions with LOD >1 from the Multipoint Analysis
| LOD (Distance) in Data Set |
|||||
| ADPD |
|||||
| Chromosome andMarker Region | Map Positiona(cM) | AD | PD | Model 1 | Model 2 |
| Chromosome 1: | |||||
| D1S2134 | 76 | ||||
| Peak | 78 | 3.41 | |||
| D1S200 | 82 | ||||
| Chromosome 4: | |||||
| Peak (D4S1652) | 208 | 2.29 | |||
| Chromosome 5: | |||||
| D5S1462 | 105 | ||||
| Peak | 108 | 1.65 | |||
| D5S1453 | 115 | ||||
| Chromosome 6: | |||||
| D6S2439 | 43 | ||||
| Peak | 51 | 1.17 | |||
| D6S2427 | 54 | ||||
| Peak (D6S1017) | 63 | 1.88 | |||
| GATA184A08 | 146 | ||||
| Peak | 154/156 | 1.96 (154 cM) | 1.81 (156 cM) | ||
| D6S1007 | 160 | ||||
| Chromosome 8: | |||||
| D8S1128 | 140 | ||||
| Peak | 150 | 2.09 | |||
| D8S373 | 165 | ||||
| Chromosome 10: | |||||
| Peak (D10S1239) | 132 | 1.55 | |||
| Peak | 133/135 | 2.33 (135 cM) | 2.62 (133 cM) | ||
| Peak (D10S1237) | 139 | 2.39 | |||
| Chromosome 13: | |||||
| Peak (D13S800) | 55 | 1.41 | |||
| Peak (D13S285) | 111 | 1.46 | |||
| Chromosome 17: | |||||
| Peak (D17S1303) | 25 | 1.93 | |||
| Chromosome 18: | |||||
| Peak (D18S877) | 54 | 1.33 | |||
| Chromosome 20: | |||||
| D20S851 | 25 | ||||
| Peak | 27 | 1.47 | |||
| D20S604 | 33 | ||||
| Chromosome 22: | |||||
| Peak (D22S683) | 37 | 1.32 | |||
The map positions are based on the sex-averaged distance, in Kosambi centimorgans, in Map-O-Mat.
Linkage Evidence for a Common AAO Gene on Chromosome 10q
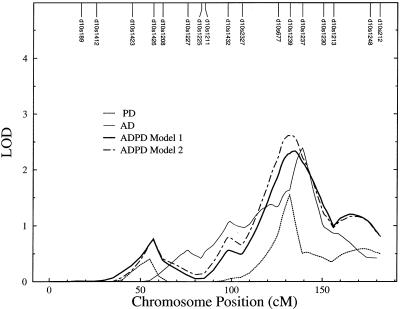
We found that, in the initial genomic screens, chromosomes 6 and 10 gave evidence for linkage to AAO in both the AD and PD data sets (table 3). The peaks were 12 cM apart on chromosome 6 (51 cM in AD and 63 cM in PD) and were 7 cM apart on chromosome 10 (132 cM in PD and 139 cM in AD). To decipher the role of these common interesting linkage regions, we performed linkage analyses on the combined ADPD data set for chromosomes 6 and 10. Analysis of the ADPD data set by use of models 1 and 2 did not result in a consistent area of interest for chromosome 6, as the combined data set gave peak LOD scores of 1.96 at 154 cM for model 1 and 1.81 at 156 cM for model 2. This peak from the combined ADPD data set is unlinked to both the independent AD (51 cM) and PD (63 cM) regions. However, analysis of the ADPD data set on chromosome 10 confirmed the findings from the analyses of the independent AD and PD data sets, resulting in a single peak region on chromosome 10q between D10S1239 and D10S1237, with LOD scores of 2.33 for model 1 at 133 cM and 2.62 for model 2 at 135 cM (table 3). Figure 1 summarizes the multipoint results of the four analyses on chromosome 10. As can be seen, four analyses revealed a similar pattern of LOD scores across chromosome 10, with a single peak region. Notably, inclusion of disease status as a covariate in model 2 slightly increases the LOD score in the ADPD data set. Our results strongly suggest that a common modulator of AAO for AD and PD may be located on chromosome 10q.
Figure 1.
Results of chromosome 10 multipoint linkage analyses. Marker distances were based on sex-averaged Kosambi centimorgans according to Map-O-Mat.
Discussion
Genomic screens have concentrated historically on identifying genes controlling the risk of developing a disease. However, risk is not the only important aspect of a disease. Onset of disease is also crucial, as understanding the regulation of onset could make it possible to delay onset beyond an individual’s normal life span. The results of this study, the first genomic screens for AAO in two major neurodegenerative diseases (AD and PD), demonstrate that AAO is highly heritable and that the search for AAO genes is possible. It should be noted that AAO data are very difficult to acquire reliably, and false-negative results may be produced. With this point in mind, this study followed published standards in ascertainment for definition of reported AAO for affected individuals and reported AAE for a participant. In addition, the large sample sizes assembled for both AD and PD should help to decrease the false-negative outcome. Although some limitations may exist, overall, we believe that our data set has significant power to detect genes modulating AAO.
Elsewhere, Daw et al. (2000) reported that at least four AAO genes with effect size possibly equal to or greater than that of APOE exist. However, the chromosomal locations linked to these putative AAO genes are still unknown. The present genomic screen for AAO in AD has identified six suggested linkage regions for AAO, in which chromosomes 4q, 8q, and 10q show the most promising results, with LOD scores >2. The APOE gene still yielded the strongest linkage effect among the newly identified regions in AD, and the role of APOE in controlling onset of AD was further confirmed. For PD, we identified a single peak with very strong linkage evidence (LOD=3.41) near D1S2134 (78 cM). Previously, Valente et al. (2001) and van Duijn et al. (2001) had localized genes for rare autosomal recessive early-onset PD to two independent regions on chromosome 1p (PARK6 and PARK7, respectively). The minimal candidate region (MCR) for PARK6 defined by the observed recombination in the family is between D1S483 (45.3 cM) and D1S247 (57.8 cM), whereas the MCR for PARK7 is between D1S468 (4.2 cM) and D1S214 (14 cM). In addition, the weak support for linkage on chromosome 1q (LOD=1.20 at 214 cM) to PD risk reported by DeStefano et al. (2001) is unlinked to our region.
Several recent reports have focused attention on chromosome 10q for AD (Bertram et al. 2000; Ertekin-Taner et al. 2000; Myers et al. 2000; Haines and Pericak-Vance 2001) but have been inconsistent in localization (fig. 2). Only the region identified by Bertram et al. (2000) by use of the NIMH family-sample data set maps near our region. To clarify our results, we further tested two subsets of data: a subset of the AD data, made up of all the NIMH families (∼60% of the overall AD data set), and a combined ADPD data set, without NIMH data, for chromosome 10. First, the NIMH AD subset generated a LOD score of 1.00 near D10S677, which is adjacent to D10S1239 (6 cM apart). The remaining non-NIMH data set thus contributed equally to the overall LOD score in AD. Second, the combined ADPD data set without NIMH data generated LOD scores of 1.48 for model 1 and 1.55 for model 2, at D10S1239 (132 cM). Figure 2 depicts the map positions of published linkage regions for risk genes on chromosome 10q and our linkage findings for AAO genes. It is clear that the linkage effect on chromosome 10q spans the different data sets used in the analyses. Three independent risk-gene studies (Ertekin-Taner et al. 2000; Myers et al. 2000; Pericak-Vance et al. 2000; Haines and Pericak-Vance 2001) identified an overlapping region (between D10S1225 and D10S1211) that is ∼47 cM proximal to our linkage region for AAO. Only NIMH data showed an overlapping linkage region for both risk and AAO genes. Our findings suggest three possible explanations for the previously reported inconsistent localizations of risk genes on chromosome 10 (Myers et al. 2000): (1) the same gene may affect both risk and onset in a subset of families, (2) onset and risk may be affected by separate genes within this region, or (3) the statistical methods may be detecting the genetic effects on onset when studying risk. Until the results can be replicated in different data sets, it is difficult to draw a conclusion at this stage to explain the inconsistent localization reported on chromosome 10. In addition, potential differences across ascertainment centers were investigated in our analysis by treating center as a random effect in the analyses. The center effect was not significant in PD but was significant in AD, with 1.9% heritability. After including the center effect for the AD data, the same peak region (near D10S1237) was found, with LOD = 2.03. These results confirmed that ascertainment through multiple centers is unlikely to significantly bias our findings on chromosome 10. For PD, no risk genes have yet been reported on chromosome 10 (Scott et al. 2001). Our study is the first to link chromosome 10q to PD.
Figure 2.
Map positions of reported linkage results for the risk and AAO genes on chromosome 10q.
AD and PD, although distinct clinical entities, share some common clinical and pathological features. Both have substantial variability in AAO. Although Lewy bodies are a cardinal feature of PD (Lippa et al. 2001), they can be found in individuals with autopsy-confirmed AD (Hulette et al. 2000; Scott et al. 2000). Dementia is the primary feature of AD, but a substantial number of patients with PD also develop dementia as their PD progresses (Scott et al. 2001). Thus, it is possible that AD and PD share common etiologic pathways. The use of combined AD and PD data sets for linkage analysis is therefore justified. The results presented here point to one or more genes on chromosome 10 as a common modulator of AAO in these neurodegenerative diseases.
Acknowledgments
We thank the patients with AD and PD and their families, whose help and participation made this work possible, as well as the clinical and research personnel of the Center for Human Genetics at Duke University Medical Center, the Program in Human Genetics at Vanderbilt University Medical Center, and the Joseph and the Kathleen Bryan ADRC. This work was supported by grants NS31153, AG05128, P50-AG-05128, AG13308, AG11268, AG10123, MH52453, P01 NS26630, and P50 NS39764 from the National Institutes of Health; grant M01 RR00865 from the U.S. Public Health Service; grant 95-23330 from the California Department of Health Services; grants IIRG94101 and IRG2-96044 from the Fran and Ray Stark Foundation Fund for Alzheimer’s Disease Research; a T. L. L. Temple Award (TLL-97-012); and Zenith Awards from the Alzheimer’s Disease and Related Disorders Association and GlaxoSmithKline, Inc.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD [MIM 104300] and PD [MIM 168600])
- Map-O-Mat, http://compgen.rutgers.edu/mapomat/
References
- Abbas N, Lucking CB, Ricard S, Durr A, Bonifati V, De Michele G, Bouley S, Vaughan JR, Gasser T, Marconi R, Brousolle E, Brefel-Courbon C, Harhangi S, Oostra B, Fabrizio E, Bohme GA, Pradier L, Wood NW, Filla A, Meco G, Denefle P, Agid Y, Brice A, The French Parkinson's Disease Study Group, The European Consortium on Genetic Susceptibility in Parknson's Disease (1999) A wide variety of mutations in the Parkin gene are responsible for autosomal recessive parkinsonism in Europe. Hum Mol Genet 8:567–574 [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis M, Go RCP, Vekrellis K, Selkoe DJ, Saunders A, Tanzi R (2000) Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science 290:2302–2305 [DOI] [PubMed] [Google Scholar]
- Blangero J, Almasy L (1997) Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol 14:959–964 [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923 [DOI] [PubMed] [Google Scholar]
- Daw EW, Heath SC, Wijsman EM (1999) Multipoint oligogenic analysis of age-at-onset data with applications to Alzheimer disease pedigrees. Am J Hum Genet 64:839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw EW, Payami H, Nemens E, Nochlin D, Bird TD (2000) The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet 66:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destefano AL, Golbe LI, Mark MH, Lazzarini AM, Maher NE, Saint-Hilaire M, Feldman RG, et al (2001) Genome-wide scan for Parkinson's disease: the GenePD Study. Neurology 57:1124–1126 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG (2000) Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer's disease pedigrees. Science 290:2303–2304 [DOI] [PubMed] [Google Scholar]
- Fulker DW, Cherny SS, Cardon LR (1995) Multipoint interval mapping of quantitative trait loci. Am J Hum Genet 56:1224–1233 [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance MA, Roses A, Williamson R, Rossor M, Owen M, Hardy J (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349:704–706 [DOI] [PubMed] [Google Scholar]
- Goldgar DE (1990) Multipoint analysis of human quantitative genetic variation. Am J Hum Genet 47:957–967 [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Pericak-Vance MA (2001) A genomic search for Alzheimer's disease genes. In: Iqbal K, Sisodia SS, Winblad B (eds) Alzheimer's disease: advances in etiology, pathogenesis and therapeutics. John Wiley & Sons, London, pp 33–43 [Google Scholar]
- Harhangi BS, de Rijk MC, Van Duijn CM, Van Broeckhoven C, Hofman A, Breteler MM (2000) APOE and the risk of PD with or without dementia in a population-based study. Neurology 54:1272–1276 [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Ben-Shlomo Y, Daniel SE, Lees AJ (1992a) What features improve the accuracy of clinical diagnosis in Parkinson's disease: a clinicopathologica study. Neurology 42:1142–1146 [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Daniel SE, Kilford L, Lees AJ (1992b) Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 55:181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulette CM, Rosenberg CR, Gaskell CP, Ervin JF, Saunders AM, Scott WK, Vance JM, Schmechel DE, Pericak-Vance MA (2000) Familial dementia with Lewy bodies. Paper presented at the Society for Neuroscience 30th Annual Meeting, New Orleans, November 4–9 [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ (1999) A full genome scan for late onset Alzheimer's disease. Hum Mol Genet 8:237–245 [DOI] [PubMed] [Google Scholar]
- Khan N, Graham E, Dixon P, Morris C, Mander A, Clayton D, Vaughan J, Quinn N, Lees A, Daniel S, Wood N, de Silva R (2001) Parkinson's disease is not associated with the combined alpha-synuclein/apolipoprotein E susceptibility genotype. Ann Neurol 49:665–668 [PubMed] [Google Scholar]
- Korczyn AD (2001) Dementia in Parkinson's disease. J Neurol Suppl 248:III1–III4 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu YH, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE (1995) Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science 269:973–977 [DOI] [PubMed] [Google Scholar]
- Lippa CF, Schmidt ML, Lee VM, Trojanowski JQ (2001) Alpha-synuclein in familial Alzheimer disease: epitope mapping parallels dementia with Lewy bodies and Parkinson disease. Arch Neurol 58:1817–1820 [DOI] [PubMed] [Google Scholar]
- Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, et al (2001) Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA 286:2245–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Gitlin JA (1999) MAP-O-MAT: marker-based linkage mapping on the World Wide Web. Am J Hum Genet Suppl 65:A435 [Google Scholar]
- McKhann G, Drachman D, Folstein M (1984) Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of the Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34:939–944 [DOI] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ (2001) Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol 58:1803–1809 [DOI] [PubMed] [Google Scholar]
- Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, Hamshere M, Abraham R, Tunstall N, Rice F, Carty S, Lillysstone S, Kehoe P, Rudrasingham V, Jones L, Lovestone S, Perez-Tur J, Williams J, Owen MJ, Hardy J, Goate AM (2000) Susceptibility locus for Alzheimer's disease on chromosome 10. Science 290:2304–2305 [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL (2000) Identification of novel genes in late-onset Alzheimer disease. Exp Gerontol 35:1343–1352 [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, Di Iorio G, Sanges G, Stenroose ES, Pho LT, Schaffer AA, Lazzarini AM, Nussbaum RL, Duvoisin RC (1996) Mapping of a gene for Parkinson's disease to chromosome 4q21-q23. Science 274:1197–1199 [DOI] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T (1995) Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature 376:775–778 [DOI] [PubMed] [Google Scholar]
- Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA (2000) Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet 66:922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, et al (2001) Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA 286:2239–2244 [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, et al (1995) Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature 375:754–760 [DOI] [PubMed] [Google Scholar]
- Valente EM, Bentivoglio AR, Dixon PH, Ferraris A, Ialongo T, Frontali M, Albanese A, Wood NW (2001) Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am J Hum Genet 68:895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duijn CM, Dekker MC, Bonifati V, Galjaard RJ, Houwing-Duistermaat JJ, Snijders PJ, Testers L, Breedveld GJ, Horstink M, Sandkuijl LA, Van Swieten JC, Oostra BA, Heutink P (2001) Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am J Hum Genet 69:629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JM, Ben Othmane K (1998) Methods of genotyping. In: Haines JL, Pericak-Vance MA (eds) Approaches to gene mapping in complex human diseases. Wiley-Liss, New York, pp 213–228 [Google Scholar]
- Ward CD, Gibb WR (1990) Research diagnostic criteria for Parkinson's disease. Adv Neurol 53:245–249 [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 [DOI] [PubMed] [Google Scholar]
- Wilson RS, Bennett DA, Gilley DW, Beckett LA, Schneider JA, Evans DA (2000) Progression of parkinsonism and loss of cognitive function in Alzheimer disease. Arch Neurol 57:855–860 [DOI] [PubMed] [Google Scholar]