Abstract
Although the predisposition to morbid obesity is heritable, the identities of the disease-causing genes are largely unknown. Therefore, we have conducted a genomewide search with 628 markers, using multigenerational Utah pedigrees to identify genes involved in predisposition to obesity. In the genomewide search, we identified a highly significant linkage to high body-mass index in female patients, at D4S2632, with a multipoint heterogeneity LOD (HLOD) score of 6.1 and a nonparametric linkage (NPL) score of 5.3. To further delineate the linkage, we increased both the marker density around D4S2632 and the size of our pedigree data set. As a result, the linkage evidence increased to a multipoint HLOD score of 9.2 (at D4S3350) and an NPL score of 11.3. Evidence from almost half of the families in this analysis support this linkage, and therefore the gene in this region might account for a significant percentage of the genetic predisposition to severe obesity in females. However, further studies are necessary to clarify the effect that this gene has in males and in the general population.
Introduction
The incidence of obesity ([MIM 601665]) has reached epidemic levels (Flegal et al. 1998; Mokdad et al. 1999). Patients with severe obesity (typically defined as BMI [in kg/m2] >35) suffer not only from the direct physical and psychological consequences of being overweight but also from increased risk for heart disease, diabetes, hypertension, and some types of cancer (Drenick et al. 1980; Manson et al. 1995; Troiano et al. 1996). Unfortunately, current obesity therapies have proven largely ineffective (Van der Ploeg 2000). Therefore, there is an urgent need to define the etiology of this disease and to use that information to initiate rational drug development.
Although it is clear that predisposition to severe obesity is a heritable trait (Bouchard 1987; Hunt et al. 1989; Price et al. 1990), identifying the genes that confer this risk has been difficult. Many potential obesity-causing genes have been identified in studies of humans and animals. However, variation in these genes has not been shown to account for a significant amount of severe obesity in the human population (Pérusse et al. 2001). Unfortunately, linkage studies to identify obesity-causing genes also have been difficult. Numerous genomewide searches for obesity genes have been published, but, with a few notable exceptions (e.g., see Comuzzie et al. 1997; Hager et al. 1998; Feitosa et al. 2002), these studies have, at best, produced equivocal results.
The difficulty with genetic studies of obesity presumably stems from the genetic complexity underlying the predisposition to the disease. This is particularly true for linkage analysis, which is very sensitive to the degree of genetic complexity (Risch 1990; Greenberg 1993; Greenberg et al. 1998). Therefore, in this linkage study, we employed several strategies designed to ameliorate some of these complexities. First, we focused on patients with very severe obesity. In general, linkage analysis is best at detecting genes that have strong effects on the phenotype of interest. Also, by focusing on a severe phenotype (i.e., BMI ⩾40), we hoped to reduce the underlying genetic heterogeneity. Second, we applied strict selection criteria to the pedigrees included in this study, in a manner that was intended to maximize our ability to detect genetic linkage—that is, we selected pedigrees to enrich for genes with large effects on BMI, to enrich for genes with high penetrances, and to reduce the frequency of intrafamilial genetic heterogeneity. Finally—and, again, to reduce genetic heterogeneity—we employed sex-specific models and data sets. Various groups have detected sex-influenced effects on obesity-related phenotypes and on obesity-prone genotypes (Borecki et al. 1993; Comuzzie et al. 1995; Gallagher et al. 1996; Elbers et al. 1999; Lee et al. 1999; Garenc et al. 2002; Martin et al. 2002), and, importantly for the present study, these include effects on BMI. Here we present the results of a genomewide search using the strategies described above, in conjunction with multigenerational Utah pedigrees to identify genes involved in predisposition to obesity.
Subjects and Methods
Pedigree Ascertainment
Families were ascertained from two sources. The majority of families were selected from the Utah population-based Health Family Tree Program (Hunt et al. 1986; Williams et al. 1988). Families who indicated on this high school–based family-history questionnaire that they had two or more first-degree relatives of the students’ parents who were ⩾100 lbs overweight were contacted, to assess their willingness to participate in this study and to further validate the reported weights. All family members who reported that they had a BMI ⩾32 were asked to participate in a clinical examination. Spouses were also invited to the clinic. Initially, normal-weight subjects were examined, but, as the study progressed, fewer normal-weight subjects were recruited. Pedigrees were expanded by following branches with reported severe obesity. The second source of pedigrees was a consecutive series of over 8,000 patients with gastric bypass, in a local registry begun in 1980 by a group of surgeons (Smith et al. 1995). As part of an annual follow-up of the health status of these subjects, we ascertained their family history of obesity and their willingness to participate in this research study. All subjects from both methods of ascertainment signed a consent form, and this study was approved by the University of Utah Institutional Review Board.
Pedigree Selection
It is difficult for linkage studies to detect (a) genes that have weak to moderate effects on the phenotype of interest, (b) genes with a low penetrance, and (c) the effect that a single gene has in the presence of multiple disease-causing genes within a family (i.e., intrafamilial genetic heterogeneity). Therefore, for this linkage study, we selected a set of families in a manner that was meant to reduce genetic complexity—and, thus, to increase our ability to detect linkage. First, to enrich for genes with both strong effects on BMI (i.e., high expressivity) and high penetrance, we selected pedigrees that contained at least three members with an extreme (i.e., ⩾40) BMI. The three family members could have been either (a) first-degree relatives (i.e., siblings, or parents and children) or (b) two first-degree relatives and at least one second- or third-degree relative (e.g., a grandparent or first cousin). Such a group of closely related individuals with a BMI ⩾40 were referred to as an “affected cluster” (fig. 1). Second, to reduce the potential for intrafamilial genetic heterogeneity, we limited the size of our selected pedigrees. For any common disease, the probability that intrafamilial genetic heterogeneity will be observed increases as the size of the family increases. Therefore, we chose pedigree founders in order to define subpedigrees (subsequently referred to as “clustered pedigrees”), such that the clustered pedigree contained the affected cluster and any other family member with a BMI ⩾40, who was related to a member of the affected cluster by no more than three generations (e.g., first cousins). The final clustered pedigrees were formed by defining the appropriate founders, followed by a unilineal descent to include all of their ascertained posterity (fig. 2). In some cases, the originally ascertained pedigree contained multiple affected clusters. If the shortest genetic distance between affected clusters exceeded three generations, each of the clusters was considered as a separate pedigree. We applied these selection criteria to a total data set of >435 pedigrees of European descent that have obesity, to create a data set of clustered pedigrees. On average, the clustered pedigrees contained 27 subjects, 10 of whom had a BMI ⩾35. Descriptive statistics of the clustered pedigrees are given in table 1. These selection rules were chosen without regard to genotype information and to no extent did they bias the results of the linkage analysis.
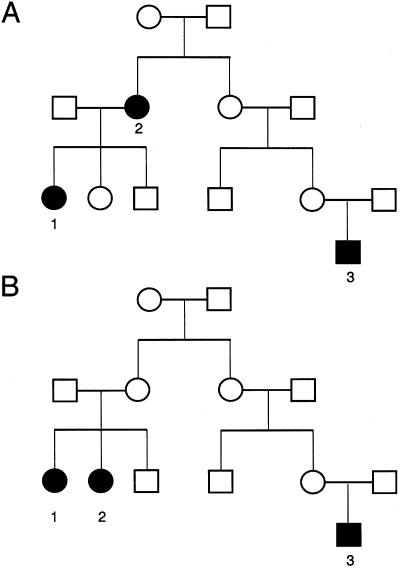
Figure 1.
Pedigrees illustrating requirements to contain an affected cluster. Blackened symbols denote subjects with BMI ⩾40. A, Pedigree with an affected cluster. B, Pedigree similar to that in panel A, but without a cluster. In both pedigrees, individuals 1 and 2 are first-degree relatives, but, in the pedigree in panel B, the genetic distance from individual 3 to either individual 1 or individual 2 is more than three generations.
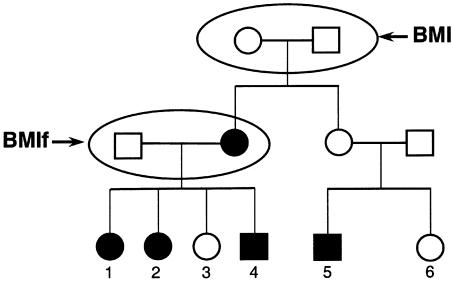
Figure 2.
Illustration of effect that phenotype has on pedigree structure used in linkage analysis. Founder pairs for the BMI phenotype and the BMIf phenotype are indicated by the oval. Blackened symbols denote subjects with BMI ⩾40. The final pedigree structures were formed by defining the appropriate founders, followed by a unilineal descent to include all of their ascertained posterity. Note that individual 4 would be included within the BMIf pedigree but that his phenotype would be unknown. Also, the entire pedigree would be excluded from analyses of the BMIm phenotype.
Table 1.
Descriptive Statistics for Pedigrees
|
Mean BMI ± SD |
|||
| Pedigree Type (No.) | Average Size (Range) | Females | Males |
| BMI clusters (64) | 26 (4–104) | 35.5±8.6 | 34.7±8.0 |
| BMIf clusters (37) | 27 (4–104) | 36.7±9.2 | 34.5±8.0 |
| BMIm clusters (14) | 34 (4–87) | 34.3±9.2 | 36.4±8.4 |
| Total data set (435) | 17 (2–435) | 32.7±8.5 | 31.7±7.9 |
Phenotypes
Height was measured, by a stadiometer, with the individual in bare feet and with the head in the Frankfort Plane and with examiner prompts to maintain a straight back and posture. Weight was measured by an electronic scale with a maximum capacity of 800 lbs (Scaletronic; Sharp). For subjects who had undergone gastric-bypass surgery, the greatest measured or reported BMI was used instead of current BMI. Subjects <15 years of age, as well as deceased pedigree members, were defined as having an unknown phenotype. When sex-specific phenotypes were analyzed, the excluded sex was defined as having an unknown phenotype.
Genetic Markers and Genotyping
DNA was extracted from blood buffy coats by PUREGENE DNA isolation kits (Gentra Systems). For the genomewide search, genotyping was performed by use of 628 fluorescent dye–labeled microsatellite markers (di-, tri-, and tetranucleotide repeats) covering the entire human genome. The mean heterozygosity index for these genomic-search markers was 75%. All dinucleotide-repeat markers contained GTTT extensions at the 5′ ends of the non–dye-coupled PCR primers, to reduce the variability of addition of nontemplated nucleotides at the 3′ ends of the labeled products (Brownstein et al. 1996). PCR products were analyzed on ABI 377 fluorescent sequencers. The average spacing between genomic-search markers was 5.8 cM. The genetic map used for all analyses was generated internally by the CRI-MAP program (Lander and Green 1987), on 3,916 meioses. After identification of significant linkage, 73 additional markers (di-, tri-, and tetranucleotide repeats) were developed within a 60-cM interval centered on the peak LOD score. This decreased the average marker spacing to 0.75 cM. The relative order of the markers within this interval was determined by use of the UCSC Human Genome Project Working Draft (also see International Human Genome Sequencing Consortium 2001), and the genetic distances between markers were calculated by the CRI-MAP program. Inheritance of alleles was verified by the PedCheck program (O'Connell and Weeks 1998). Samples with incompatible calls were generally regenotyped. The marker data for that family were set to missing if the incompatibility could not be resolved. The average completeness of genotyping was 95% after incompatibilities were set to zero.
Statistical Analysis
Prior to linkage analysis, BMI was adjusted, by linear regression, for sex and for age within each sex. In this study, we employed robust multipoint linkage statistics as proposed by Göring and Terwilliger (2000). Linkage analysis was performed by MCLINK, a program developed to perform multipoint analysis on very large pedigrees with any number of markers (Thomas et al. 2000). Three models (dominant, codominant, and recessive) were used in linkage analysis. A disease-gene frequency of 0.003 was chosen for the dominant and codominant models, and 0.0775 was chosen for the recessive model, yielding a disease prevalence, due to a single locus, of ∼0.6%. This disease frequency would be equivalent to assuming the existence of 10 major genes with similar effects on BMI, resulting in heritable severe obesity being present in 6% of the general population. A logistic distribution was used for the BMI-dependent ratio of the sporadic rate to the penetrance (Ott 1991). The ratio of sporadic rate to penetrance for a genotype with two obesity-related mutations for all models and with one obesity-related mutation for the dominant model is described by the formula
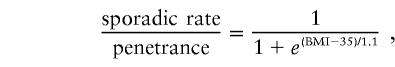 |
and the ratio of sporadic rate to penetrance for a genotype with one obesity-related mutation for the codominant model is described by the formula
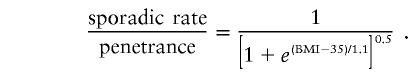 |
These two ratios change gradually from 1 to 0 as BMI increases. Individuals with low BMI are largely uninformative, whereas individuals with high BMI are almost fully informative. In practical terms, only phenotypes from severely obese individuals (BMI >35) are informative.
Since the genetic model underlying predisposition to obesity is unknown, we also performed analyses using a model-independent method. The haplotype solutions generated by MCLINK (Thomas et al. 2000) can also be used for nonparametric linkage (NPL) statistics (Camp et al. 2001). The method is similar to the method developed by Kruglyak et al. (1996) but is extended to the analysis of quantitative traits. The main difference between these two methods is in the scoring function. Our scoring function is extended for analysis of quantitative traits and is defined for each pedigree, at each marker position, by the formula 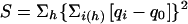 , where the first sum is taken over all haplotypes h, the second sum is taken over individuals i carrying haplotype h, qi is the quantitative-trait value for individual i, and q0 is a value close to the population mean or median for the trait. In our NPL analysis, we used BMI as the quantitative-trait value and set q0 equal to 27. The value of q0 is the mean BMI of the Utah population, as derived from 270 randomly ascertained adults.
, where the first sum is taken over all haplotypes h, the second sum is taken over individuals i carrying haplotype h, qi is the quantitative-trait value for individual i, and q0 is a value close to the population mean or median for the trait. In our NPL analysis, we used BMI as the quantitative-trait value and set q0 equal to 27. The value of q0 is the mean BMI of the Utah population, as derived from 270 randomly ascertained adults.
The NPL scoring function is large when individuals sharing the same haplotype have their quantitative-trait values deviating in the same direction from q0, and it is small when the quantitative-trait values do not correlate with haplotype sharing. Thus, NPL statistics can detect genes with causal mutations that segregate among individuals with either low or high BMI values. NPL analysis not only circumvents the problem of model misspecifications but also has the ability to recognize the effects of multiple mutations, even if their penetrances, phenotypic effects, and modes of inheritance are quite different.
To be able to compare NPL and LOD scores directly, we calculated NPL and LOD scores in 20 independent genomic scans of the same set of pedigrees but with randomly generated haplotypes (data not shown). Subsequently, an asymptotic formula was derived to adjust NPL scores, such that equal LOD and NPL scores translated to the same P values.
Results
In this study, we considered three different BMI-based phenotypes: a sex-nonspecific phenotype (BMI), a female-only phenotype (BMIf), and a male-only phenotype (BMIm). In the sex-specific phenotypes, the opposite sex is considered completely uninformative. The pedigree-selection criteria employed in this study (see the “Pedigree Selection” subsection, above) depended on the affected status of pedigree members. As a consequence, the set of selected clustered pedigrees changed as the phenotype definition changed. This was achieved by altering, in a phenotype-specific manner, the selection of pedigree founders (fig. 2). Therefore, although there was some overlap, in general we analyzed a different set of clustered pedigrees for each of the three BMI phenotypes. As a result, we included 64 pedigrees with 1,687 individuals in the analysis of BMI, 37 pedigrees with 994 individuals in the analysis of BMIf, and 14 pedigrees with 479 individuals in the analysis of BMIm (for additional descriptive statistics, see table 1).
We considered three inheritance models: dominant, codominant, and recessive. In all of these models, BMI was used to determine the likelihood that a subject carried a disease allele. In general, results for individuals with a high BMI were informative, and these individuals were considered likely to be disease-allele carriers, whereas results for individuals with a low BMI were uninformative.
Genomewide Search
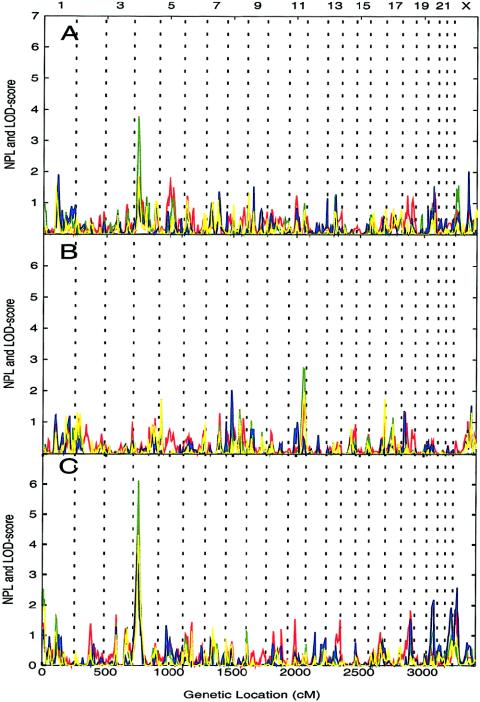
The clustered pedigrees were genotyped with 628 genetic markers covering the human genome, at an average spacing of 5.8 cM. Marker-segregation patterns were determined by MCLINK (Thomas et al. 2000), and the haplotype solutions were subsequently used to generate model-based multipoint heterogeneity LOD (HLOD) scores and NPL scores (Camp et al. 2001) scores (fig. 3). The NPL scores reported in the present study were adjusted such that NPL scores and HLOD scores of equal magnitude correspond to the same P value. Linkage for the BMIf phenotype was detected at D4S2632 (4p15-p14) (fig. 3); the HLOD scores were 6.1 for the codominant model, 4.3 for the recessive model, and 3.7 for the dominant model; and the NPL score was 5.3. The results of the genomewide search are summarized in table 2 (linkage scores [HLOD or NPL] >2.0 are shown).
Figure 3.
Results of genomic search by four analyses: NPL (red), codominant model (green), dominant model (blue), and recessive model (yellow). The HLOD or NPL score is plotted on the Y-axis, and marker positions (in cM) are plotted on the X-axis. Vertical dashed lines delimit the chromosomes; chromosome numbers (odd numbers only) are indicated at the top of the figure. A, HLOD or NPL scores from BMI phenotype. B, HLOD or NPL scores from BMIm phenotype. C, HLOD or NPL scores from BMIf phenotype.
Table 2.
Significant and Interesting Multipoint HLOD Scores from the Genomewide Search
| Marker | Position(cM) | HLOD Score | Phenotype | Model |
| D1S468 | 7.7 | 2.5 | BMIf | Codominant |
| D1S508 | 18.4 | 2.2 | BMIf | Codominant |
| D4S2639 | 31.8 | 2.2 | BMI | Codominant |
| D4S2289 | 32.2 | 2.6 | BMIf | Dominant |
| D4S2397 | 40.5 | 4.1 | BMIf | Codominant |
| D4S2632 | 49.9 | 6.1 | BMIf | Codominant |
| D4S1627 | 59.5 | 3.4 | BMIf | Codominant |
| D4S3019 | 68.1 | 2.1 | BMIf | Codominant |
| D4S3248 | 70.6 | 2.0 | BMIf | Codominant |
| D8S282 | 41.8 | 2.0 | BMIm | Dominant |
| D11S4464 | 114.9 | 2.8 | BMIm | Codominant |
| D11S934 | 118.9 | 2.6 | BMIm | Codominant |
| D11S912 | 123.9 | 2.7 | BMIm | Codominant |
| D20S478 | 50.8 | 2.0 | BMIf | Dominant |
| D20S438 | 51.2 | 2.0 | BMIf | Dominant |
| D20S465 | 52 | 2.0 | BMIf | Dominant |
| D20S481 | 59.1 | 2.2 | BMIf | Dominant |
| DXS8099 | 37.3 | 2.6 | BMIf | Dominant |
| DXS1059 | 115.2 | 2.0 | BMI | Dominant |
According to the guidelines proposed by Lander and Kruglyak (1995), a LOD score >4.9 represents highly significant evidence for linkage (“highly significant” is their most significant category). However, the present study employed three phenotypes, three models, and an NPL analysis for each of the three phenotypes. Therefore, we adjusted our highly significant threshold for 12 multiple tests. To be conservative, we assumed that the models and phenotypes were independent, requiring an adjustment of 1.1 (log1012) LOD-score units. Accordingly, we adjusted our genomewide highly significant threshold to 6.0. The linkage evidence at 4p15-p14 remained highly significant.
Linkage Analyses of the 4p15-p14 Interval
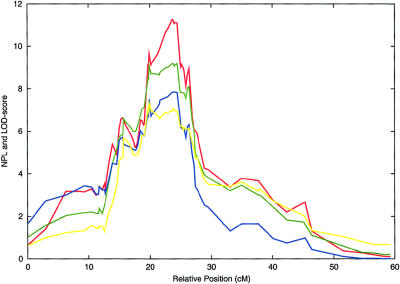
To further delineate the linkage on chromosome 4, we developed 73 additional genetic markers in a 60-cM interval centered on D4S2632 (the genetic markers within 10 cM of D4S2632 are shown in table 3). These markers (as well as the chromosome 4 markers in our genomewide search) were typed in our clustered pedigrees. As a result, the HLOD score for the codominant model increased from 6.1 to 7.2. Also, we increased the size of our pedigree set, in two ways. First, we expanded the initial 37 female-clustered pedigrees. Second, we added 14 newly ascertained pedigrees, all of which contained clusters of severely affected females. These families were collected in the same way and had the same average BMI as our genomewide search pedigrees. They had an average size of 13 subjects, and, in total, the expanded data set contained 1,182 subjects. The genotyping data were analyzed by use of the BMIf phenotype (fig. 4). After the analysis of the expanded data set, the codominant model generated a peak HLOD score of 9.2 at marker D4S3350. The heterogeneity fraction was 0.46. All of the model-based HLOD scores were also highly significant (table 3), and, importantly, the peak linkage scores for all analyses occurred at approximately the same genetic location. The highest linkage scores were generated by the NPL analysis, with a peak score of 11.3 at marker D4S3350. The families with LOD or NPL scores ⩾1 at D4S3350 are shown in table 4.
Table 3.
Markers and Linkage Scores for Regions Flanking D4S2632, When BMIf Phenotype Is Used
|
HLOD Score |
|||||
| Markera | Relative Positionb (cM) | NPL Score | Codominant | Dominant | Recessive |
| D4S2397 | 15 | 6.1 | 5.7 | 5.5 | 4.6 |
| D4S1609 | 15.3 | 6.6 | 5.8 | 5.7 | 4.7 |
| D4S391 | 15.6 | 6.6 | 5.8 | 5.7 | 4.8 |
| 4-MYR0306 | 15.7 | 6.5 | 6.6 | 5.4 | 5.7 |
| 4-MYR0310 | 16.1 | 6.3 | 6.4 | 5.3 | 5.6 |
| 4-MYR0305 | 16.9 | 5.9 | 6.2 | 5.3 | 5.3 |
| 4-MYR0301 | 17.3 | 5.6 | 6.0 | 5.2 | 4.9 |
| D4S418 | 17.6 | 5.3 | 6.0 | 5.1 | 4.9 |
| D4S2282 | 17.7 | 5.2 | 6.0 | 5.1 | 4.8 |
| 4-MYR0307 | 18.0 | 5.5 | 6.2 | 5.3 | 4.9 |
| 4-MYR0304 | 18.5 | 6.5 | 6.8 | 6.0 | 5.4 |
| D4S1643 | 18.7 | 6.5 | 7.0 | 5.9 | 5.8 |
| 4-MYR0263 | 19.0 | 6.3 | 7.1 | 5.8 | 5.8 |
| 4-MYR0264 | 19.1 | 6.3 | 7.1 | 5.8 | 5.8 |
| D4S2968 | 19.7 | 9.1 | 8.5 | 6.9 | 7.1 |
| D4S2408 | 19.8 | 9.1 | 8.5 | 6.8 | 6.9 |
| 4-MYR0251 | 19.9 | 9.7 | 9.1 | 7.5 | 7.3 |
| 4-MYR0250 | 20.2 | 9.1 | 8.7 | 6.7 | 7.0 |
| 4-MYR0233 | 20.3 | 9.2 | 8.7 | 6.8 | 7.0 |
| D4S2359 | 21.3 | 9.6 | 8.7 | 7.1 | 6.7 |
| D4S3027 | 21.6 | 9.9 | 8.7 | 7.3 | 6.6 |
| 4-MYR0249 | 21.7 | 9.9 | 8.8 | 7.5 | 6.6 |
| D4S3001 | 22.6 | 10.7 | 9.0 | 7.6 | 6.9 |
| D4S188 | 22.7 | 10.7 | 9.0 | 7.6 | 6.9 |
| 4-MYR0220 | 22.8 | 10.8 | 9.0 | 7.6 | 6.9 |
| 4-MYR0246 | 22.9 | 10.8 | 9.0 | 7.6 | 6.9 |
| AFMa070ta9 | 23.6 | 11.2 | 9.2 | 7.8 | 7.0 |
| D4S3350c | 23.7 | 11.3 | 9.2 | 7.8 | 7.0 |
| 4-MYR0234 | 23.8 | 11.2 | 9.2 | 7.9 | 7.1 |
| 4-MYR0235 | 23.9 | 11.2 | 9.1 | 7.8 | 7.0 |
| D4S2415 | 24.0 | 11.1 | 9.1 | 7.8 | 7.0 |
| 4-MYR0248 | 24.1 | 11.1 | 9.1 | 7.8 | 7.0 |
| 4-MYR0247 | 24.5 | 11.1 | 9.2 | 7.8 | 7.0 |
| D4S1587 | 25.0 | 9.2 | 7.8 | 6.6 | 6.2 |
| 4-MYR0262 | 25.1 | 9.0 | 7.8 | 6.2 | 6.2 |
| D4S2632 | 25.2 | 9.1 | 7.8 | 6.2 | 6.3 |
| D4S2400 | 25.3 | 9.1 | 7.8 | 6.1 | 6.2 |
| D4S483 | 25.4 | 9.0 | 7.8 | 6.1 | 6.1 |
| 4-MYR0267 | 25.5 | 9.0 | 7.8 | 6.1 | 6.1 |
| D4S2995 | 25.6 | 8.9 | 7.8 | 6.1 | 6.2 |
| D4S2955 | 25.9 | 8.1 | 7.6 | 5.7 | 6.1 |
| 4-MYR0266 | 26.1 | 8.6 | 7.9 | 6.1 | 6.1 |
| 4-MYR0265 | 26.2 | 8.7 | 8.1 | 6.2 | 6.1 |
| 4-MYR0302 | 26.4 | 9.0 | 8.1 | 6.3 | 6.1 |
| 4-MYR0300 | 26.8 | 7.3 | 6.8 | 5.2 | 5.6 |
| 4-MYR0303 | 26.9 | 6.7 | 6.5 | 4.9 | 5.4 |
| D4S3040 | 27.2 | 6.1 | 6.0 | 4.4 | 5.1 |
| D4S1581 | 27.4 | 6.0 | 5.0 | 3.4 | 4.7 |
| D4S617 | 27.7 | 5.9 | 4.8 | 3.3 | 4.8 |
| D4S2382 | 28.9 | 4.3 | 4.0 | 2.5 | 3.7 |
| D4S405 | 29.5 | 4.2 | 3.8 | 2.4 | 3.5 |
| D4S1627 | 33.1 | 3.4 | 3.2 | 1.3 | 3.4 |
| D4S1536 | 34.9 | 3.8 | 3.5 | 1.7 | 3.6 |
Markers denoted “4-MYR” were developed at Myriad Genetics. Primer sequences are available on request.
Starting 15 cM from D4S403, the first marker developed to investigate this linkage.
Located at 49.5 cM on the Marshfield sex-averaged linkage map.
Figure 4.
Results of adding additional markers and individuals to linkage analyses of the 4p15-p14 interval, in four analyses: NPL (red), codominant model (green), dominant model (blue), and recessive model (yellow). The highest linkage score is at marker D4S3350, which is located at 49.5 cM on the Marshfield sex-averaged linkage map. The phenotype is BMIf. Linkage scores are plotted on the Y-axis, and relative marker positions are plotted on the X-axis.
Table 4.
Families with Multipoint Linkage Scores of ⩾1.0 at D4S3350, When BMIf Phenotype Is Used
|
No. of Individuals |
|||||||
| Females |
HLOD Score |
||||||
| Family | Total | BMI ⩾35a | BMI ⩾40 | NPL Score | Codominant | Dominant | Recessive |
| 43601 | 81 | 18 | 11 | 2.6 | 4.9 | 3.4 | 3.1 |
| 708201 | 13 | 5 | 3 | 2.3 | 1.7 | 1.3 | 1.6 |
| 604401 | 58 | 11 | 7 | 2.0 | 1.7 | 2.0 | 1.1 |
| 7444 | 13 | 7 | 3 | 1.9 | 1.2 | 1.4 | .5 |
| 736201 | 13 | 4 | 4 | 1.3 | 1.1 | .2 | 1.0 |
| 738001 | 63 | 7 | 4 | 1.1 | .8 | 1.4 | .7 |
| 7135101 | 65 | 14 | 5 | 1.1 | .8 | 1.0 | .9 |
| 722801 | 34 | 12 | 6 | 1.0 | .5 | .7 | .2 |
| 7158201 | 15 | 5 | 4 | .7 | 1.3 | .2 | 1.2 |
| 1105501 | 19 | 8 | 3 | .3 | .8 | .0 | 1.2 |
Includes females with BMI ⩾40.
Discussion
In the present study we have conducted a genomewide search for genes involved in obesity predisposition and have reported on a region, at 4p15-p14, that generated highly significant evidence for an obesity-predisposition gene. No other significant linkages were detected; however, other interesting regions are shown in table 2. One of the interesting regions (the second-highest-scoring region in the genome) was on chromosome 11. These markers have been linked to high BMI values in Pima Indians, and markers in the same genomic region have been associated with weight in a mostly white population (Hanson et al. 1998; Norman et al. 1998; Gu et al. 2002). We also detected a weak linkage to chromosome 20. Numerous groups have previously detected significant linkage of obesity to chromosome 20 (Borecki et al. 1994; Lembertas et al. 1997; Bottini and Gloria-Bottini 1999; Lee et al. 1999); in addition, we have previously reported such linkage in Utah pedigrees (Hunt et al. 2001). Presumably, this linkage signal was diminished because many of the pedigrees from our previous study (including some of the chromosome 20–linked pedigrees) did not meet the strict selection criteria of the present study and, therefore, were excluded from this analysis. To our knowledge, none of the other regions listed in table 2 correspond to previously detected linkages.
Given the magnitude of the linkage evidence presented here, it is notable that this region has not been previously identified (Pérusse et al. 2001). In particular, several studies have generated highly significant evidence for predisposition to obesity, but none of the studies detected this linkage (Comuzzie et al. 1997; Hager et al. 1998; Feitosa et al. 2002). However, there are significant differences between this and previous studies. For example, we employed several strategies that were intended to reduce the underlying genetic complexity of obesity. We collected a large set of multigenerational obesity pedigrees and, from these, selected pedigrees that were favorable for linkage analysis. We also applied sex-specific phenotypes and phenotype-specific pedigree sets and focused our analyses on a severe phenotype. The combination of these strategies is likely to have an impact on the results of linkage analysis. For example, prior to the implementation of these analytic strategies, the highest HLOD that we observed in the 4p15-p14 region was 3.0 (data not shown). It is worth noting that a recent abstract (Arya et al. 2001) indicates that the 4p15-p14 region has been linked to high BMI in Mexican Americans. Although the data are preliminary, the results seem to confirm the present linkage.
Both parametric linkage analysis and NPL analysis detected the linkage to 4p15-p14; however, the most significant linkage statistic was generated by NPL analysis. One of the strengths of NPL is its ability to detect linkage even in the presence of genetic complexity. In particular, NPL analyses can simultaneously detect different patterns of haplotype sharing in affected pedigree members. Here, the data suggest that the actual disease inheritance contains components of dominant, codominant, and recessive models. If any of these models had been grossly misspecified, they would not have generated the observed HLOD scores (see fig. 4). Perhaps the actual disease inheritance involves multiple causal variants, each with different penetrances, phenotypic effects, and modes of inheritance, and, obviously, it would be difficult to reproduce that amount genetic complexity in any parametric analysis.
The linkage evidence at 4p15-p14 was sensitive to phenotype definition. The BMIf phenotype was the only phenotype that generated highly significant evidence for an obesity linkage at 4p15-p14 (fig. 3). Analysis of the BMIm phenotype generated no evidence for linkage to this region, and analysis of the BMI phenotype generated only suggestive evidence for linkage. In addition, none of the linked pedigrees (table 4) generated higher LOD scores when the BMI phenotype (data not shown) was used. This suggests that affected males within the linked pedigrees either did not share the presumptive disease haplotype(s) or, at least, did not share to the same extent as did affected females. For example, if we consider the affected subjects (BMI ⩾35) in the three families that generated the highest linkage scores (table 4), 29 of 32 females share the disease haplotype(s), compared to 13 of 23 males. One explanation for these data is that the obesity-predisposition gene described in the present study does not cause male obesity. Various lines of evidence suggest that sex-specific obesity genes may exist. As stated previously, sex-specific effects on obesity have been described, and several of these studies have specifically described these effects on BMI (Borecki et al. 1993; Gallagher et al. 1996; Lee et al. 1999; Garenc et al. 2002). However, even moderately complex relationships between genotypes and phenotypes can severely suppress the resultant LOD scores. Therefore, it is often difficult to infer biology from the results of linkage analysis. For example, the gene detected in the present study may have an effect on males, but that effect could simply be weaker than its effect in females. It would be difficult to create a single linkage analysis that could simultaneously detect linkage in males and in females if the genetic effect varied by sex. Obesity-causing alleles of melanocortin 4 receptor (a gene implicated in 2%–5% of early-onset morbid obesity) offer a clear example of this scenario. Although these alleles affect both males and females, the effect on females is more profound (Sina et al. 1999). Alternatively, it is possible that the 4p15-p14 gene affects both sexes equally but that there is an elevated sporadic rate of high BMI values in males. For example, the environment could have a larger impact on high BMI values in males than it does in females, or there might be male-specific obesity genes that effect some of the male members of the pedigrees that we studied. In either case, including males in our analysis would significantly reduce the linkage scores. In fact, there are myriad possible scenarios that are consistent with the observed results, and therefore the final assessment of the role that this gene plays in male obesity awaits the discovery of the disease-causing variation.
The human draft genomic sequence around D4S3350 contains no obvious obesity-related candidate genes in the region (International Human Genome Sequencing Consortium 2001; Venter et al. 2001). Of the genes that previously have been implicated in obesity, CCKAR (cholecystokinin A receptor) (Ulrich et al. 1993; Miller et al. 1995; Inoue et al. 1997; Funakoshi et al. 2000) is the one closest to this region. However, in light of the position of this gene, we do not think that it will explain this linkage. CCKAR is located 1.5 cM upstream of the first marker listed in table 2, a location that places it outside the 4-LOD-unit support interval for this linkage. The region near D4S3350 appears to be gene poor, which should aid in the subsequent identification of the disease gene and, ultimately, should provide significant information regarding the genetics of predisposition to severe obesity.
Acknowledgments
We thank the families that participated in the study. This work was funded by National Institutes of Health grant DK44655 and the Bayer Corporation.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for AHC [MIM 601665])
- UCSC Human Genome Project Working Draft, http://genome.ucsc.edu/
References
- Arya R, Blangero J, Almasy L, O'Connell P, Stern MP (2001) A major locus for body mass index (BMI) on chromosome 4p in Mexican Americans. Obes Res 9 Suppl 3:70S [Google Scholar]
- Borecki IB, Bonney GE, Rice T, Bouchard C, Rao DC (1993) Influence of genotype-dependent effects of covariates on the outcome of segregation analysis of the body mass index. Am J Hum Genet 53:676–687 [PMC free article] [PubMed] [Google Scholar]
- Borecki IB, Rice T, Perusse L, Bouchard C, Rao DC (1994) An exploratory investigation of genetic linkage with body composition and fatness phenotypes: the Quebec Family Study. Obes Res 2:213–219 [DOI] [PubMed] [Google Scholar]
- Bottini E, Gloria-Bottini F (1999) Adenosine deaminase and body mass index in non-insulin-dependent diabetes mellitus. Metabolism 48:949–951 [DOI] [PubMed] [Google Scholar]
- Bouchard C (1987) Genetics and human obesity. Ann Behav 9:9–14 [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1006, 1008–1010 [DOI] [PubMed] [Google Scholar]
- Camp NJ, Gutin A, Abkevich V, Farnham JM, Cannon-Albright L, Thomas A (2001) A new nonparametric linkage statistic for mapping both qualitative and quantitative trait loci. Genet Epidemiol 21 Suppl 1:S461–S466 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Blangero J, Mahaney MC, Mitchell BD, Hixson JE, Samollow PB, Stern MP, MacCluer JW (1995) Major gene with sex-specific effects influences fat mass in Mexican Americans. Genet Epidemiol 12:475–488 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Hixson JE, Almasy L, Mitchell BD, Mahaney MC, Dyer TD, Stern MP, MacCluer JW, Blangero J (1997) A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat Genet 15:273–275 [DOI] [PubMed] [Google Scholar]
- Drenick EJ, Bale GS, Seltzer F, Johnson DG (1980) Excessive mortality and causes of death in morbidly obese men. JAMA 243:443–445 [PubMed] [Google Scholar]
- Elbers JM, Asscheman H, Seidell JC, Gooren LJ (1999) Effects of sex steroid hormones on regional fat depots as assessed by magnetic resonance imaging in transsexuals. Am J Physiol 276:E317–325 [DOI] [PubMed] [Google Scholar]
- Feitosa MF, Borecki IB, Rich SS, Arnett DK, Sholinsky P, Myers RH, Leppert M, Province MA (2002) Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: the National Heart, Lung, and Blood Institute Family Heart Study. Am J Hum Genet 70:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kuczmarski RJ, Johnson CL (1998) Overweight and obesity in the United States: prevalence and trends, 1960–1994. Int J Obes Relat Metab Disord 22:39–47 [DOI] [PubMed] [Google Scholar]
- Funakoshi A, Miyasaka K, Matsumoto H, Yamamori S, Takiguchi S, Kataoka K, Takata Y, Matsusue K, Kono A, Shimokata H (2000) Gene structure of human cholecystokinin (CCK) type-A receptor: body fat content is related to CCK type-A receptor gene promoter polymorphism. FEBS Lett 466(2–3): 264–266 [DOI] [PubMed] [Google Scholar]
- Gallagher D, Visser M, Sepulveda D, Pierson RN, Harris T, Heymsfield SB (1996) How useful is body mass index for comparison of body fatness across age, sex, and ethnic groups? Am J Epidemiol 143:228–239 [DOI] [PubMed] [Google Scholar]
- Garenc C, Perusse L, Chagnon YC, Rankinen T, Gagnon J, Borecki IB, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C (2002) The hormone-sensitive lipase gene and body composition: the HERITAGE Family Study. Int J Obes Relat Metab Disord 26:220–227 [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD (2000) Linkage analysis in the presence of errors I: complex-valued recombination fractions and complex phenotypes. Am J Hum Genet 66:1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA (1993) Linkage analysis of “necessary” disease loci versus “susceptibility” loci. Am J Hum Genet 52:135–143 [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D, O'Dell SD, Chen XH, Miller GJ, Day IM (2002) Evidence of multiple causal sites affecting weight in the IGF2-INS-TH region of human chromosome 11. Hum Genet 110:173–181 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SC, Abkevich V, Hensel CH, Gutin A, Neff CD, Russell DL, Tran T, Hong X, Jammulapati S, Riley R, Weaver-Feldhaus J, Macalma T, Richards MM, Gress R, Francis M, Thomas A, Frech GC, Adams TD, Shattuck D, Stone S (2001) Linkage of body mass index to chromosome 20 in Utah pedigrees. Hum Genet 109:279–285 [DOI] [PubMed] [Google Scholar]
- Hunt SC, Hasstedt SJ, Kuida H, Stults BM, Hopkins PH, Williams RR (1989) Genetic heritability and common environmental components of resting and stressed blood pressures, lipids, and body mass index in Utah pedigrees and twins. Am J Epidemiol 129:625–638 [DOI] [PubMed] [Google Scholar]
- Hunt SC, Williams RR, Barlow GK (1986) A comparison of positive family history definitions for defining risk of future disease. J Chronic Dis 39:809–821 [DOI] [PubMed] [Google Scholar]
- Inoue H, Iannotti CA, Welling CM, Veile R, Donis-Keller H, Permutt MA (1997) Human cholecystokinin type A receptor gene: cytogenetic localization, physical mapping, and identification of two missense variants in patients with obesity and non-insulin-dependent diabetes mellitus (NIDDM). Genomics 42:331–335 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lee JH, Reed DR, Li W-D, Xu W, Joo E-J, Kilker RL, Nanthakumar E, North M, Sakul H, Bell C, Price RA (1999) Genome scan for human obesity and linkage to markers in 20q13. Am J Hum Genet 64:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembertas AV, Perusse L, Chagnon YC, Fisler JS, Warden CH, Purcell-Huynh DA, Dionne FT, Gagnon J, Nadeau A, Lusis AJ, Bouchard C (1997) Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J Clin Invest 100:1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson JE, Willett WC, Stampfer MJ, Colditz GA, Hunter DJ, Hankinson SE, Hennekens CH, Speizer FE (1995) Body weight and mortality among women. N Engl J Med 333:677–685 [DOI] [PubMed] [Google Scholar]
- Martin LJ, Mahaney MC, Almasy L, MacCluer JW, Blangero J, Jaquish CE, Comuzzie AG (2002) Leptin's sexual dimorphism results from genotype by sex interactions mediated by testosterone. Obes Res 10:14–21 [DOI] [PubMed] [Google Scholar]
- Miller LJ, Holicky EL, Ulrich CD, Wieben ED (1995) Abnormal processing of the human cholecystokinin receptor gene in association with gallstones and obesity. Gastroenterology 109:1375–1380 [DOI] [PubMed] [Google Scholar]
- Mokdad AH, Serdula MK, Dietz WH, Bowman BA, Marks JS, Koplan JP (1999) The spread of the obesity epidemic in the United States, 1991–1998. JAMA 282:1519–1522 [DOI] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1991) Analysis of human genetic linkage. Johns Hopkins University Press, Baltimore [Google Scholar]
- Pérusse L, Chagnon YC, Weisnagel SJ, Rankinen T, Snyder E, Sands J, Bouchard C (2001) The human obesity gene map: the 2000 update. Obes Res 9:135–169 [DOI] [PubMed] [Google Scholar]
- Price RA, Ness R, Laskarzewski P (1990) Common major gene inheritance of extreme overweight. Hum Biol 62:747–765 [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Sina M, Hinney A, Ziegler A, Neupert T, Mayer H, Siegfried W, Blum WF, Remschmidt H, Hebebrand J (1999) Phenotypes in three pedigrees with autosomal dominant obesity caused by haploinsufficiency mutations in the melanocortin-4 receptor gene. Am J Hum Genet 65:1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Goodman GN, Edwards CB (1995) Roux-en-Y gastric bypass: a 7-year retrospective review of 3,855 patients. Obes Surg 5:314–318 [DOI] [PubMed] [Google Scholar]
- Thomas A, Gutin A, Abkevich V, Bansal A (2000) Multilocus linkage analysis by blocked Gibbs sampling. Stat Comput 10:259–269 [Google Scholar]
- Troiano RP, Frongillo EA Jr, Sobal J, Levitsky DA (1996) The relationship between body weight and mortality: a quantitative analysis of combined information from existing studies. Int J Obes Relat Metab Disord 20:63–75 [PubMed] [Google Scholar]
- Ulrich CD, Ferber I, Holicky E, Hadac E, Buell G, Miller LJ (1993) Molecular cloning and functional expression of the human gallbladder cholecystokinin A receptor. Biochem Biophys Res Commun 193:204–211 [DOI] [PubMed] [Google Scholar]
- Van der Ploeg LH (2000) Obesity: an epidemic in need of therapeutics. Curr Opin Chem Biol 4:452–460 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- Williams RR, Hunt SC, Barlow GK, Chamberlain RM, Weinberg AD, Cooper HP, Carbonari JP, Gotto AM (1988) Health family trees: a tool for finding and helping young members of coronary and cancer prone pedigrees in Texas and Utah. Am J Public Health 78:1283–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]