Abstract
Background
Oligonucleotides (ONs) have shown great promise as therapeutic agents for various diseases. It is necessary to provide a protocol for preparation of ON-loaded lipid nanoparticles (LNPs) in a reproducible manner on a laboratory scale.
Materials and Methods
A 3-inlet microfluidic (MF) chip-based device was used to synthesize LNPs at the lipid/ON ratio of 10/1 (w/w) and at flow rates ranging from 50 to 1100 µl/min. A series of LNPs containing either antisense oligodeoxyribonucleotide (AS-ODN) or small-interfering RNA (siRNA) were synthesized. Bulk mixing was used as control.
Results
The MF method was shown to be particularly useful for synthesis of LNPs loaded with AS-ODN. The optimal range of flow rates for AS-ODN LNPs was found to be 100 to 200 µl/min. MF synthesis produced LNPs with lower polydispersity values. However, the MF was less effective in preparing LNPs loaded with siRNA, which may have been due to greater rigidity of double-stranded siRNA comparing to single-stranded AS-ODN.
Conclusion
MF technology is a simple, affordable and reproducible method for production of ON-LNPs.
Keywords: Microfluidics, oligonucleotide, lipid nanoparticles
Oligonucleotides (ONs) are able to specifically silence gene expressions and hold great promise as therapeutic agents. Substantial efforts have been made on the synthesis and evaluation of therapeutic ONs including antisense oligodeoxynucleotides (AS-ODN) and small-interfering RNA (siRNA) (1–4). The development of safe and effective delivery systems for in vivo application is of central importance to realize the therapeutic potential of ONs (3, 4). Lipid nanoparticles (LNPs) have been recognized as one of the most promising delivery systems mainly due to their biocompatibility and the ease of large-scale production (4–9).
Several technologies have been used in the production of ON LNPs, including detergent dialysis (9), ethanol dilution (10), freeze-thawing (11) and thin-film hydration (12). In conventional methods, i.e., bulk-mixing (BM), lipids and ONs are spontaneously assembled into heterogeneous nanostructures in a bulk phase, which is not precisely controlled and typically produces LNPs with high polydispersity. Post-processing by sonication and extrusion is frequently required. Microfluidics (MF) is a versatile technology platform that is able to provide a well-controlled mixing environment (14, 15). MF has found applications in chemical synthesis and biological analysis, as well as in nanoparticle synthesis (5, 16–18). For example, MF has been used for the synthesis of CdSe quantum dots (QDs) (19, 20), titanium dioxide (21), and lipid- or polymer-based nanoparticles (16, 18, 22, 23).
In this study, LNPs were synthesized by simple mixing of ONs with empty liposomes in the MF channel. Both ASODN and siRNA were evaluated as cargos.
Materials and Methods
Materials
1,2-Dioleoyl-3-trimethylammonium-propane (DOTAP), and N-palmitoyl-sphingosine-1-succinyl [methoxy(polyethylene glycol) 2000] (Ceramide-PEG2000) were obtained from Avanti Polar Lipids, Inc (Alabaster, AL, USA). Egg phosphatidylcholine (egg PC) was obtained from Lipoid (Newark, NJ, USA). Cholesterol (Chol) and other chemicals and reagents were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). All tissue culture media and supplies were obtained from Invitrogen (Carlsbad, CA, USA). Phosphorothioate oligo G3139 with the sequence 5’-TCT CCC AGC GTG CGC CAT- 3’ was custom synthesized by Alpha DNA, Inc (Montreal, Canada). Luciferase-targeted and negative control siRNA were obtained from Applied Biosystems (Austin, TX, USA).
MF device
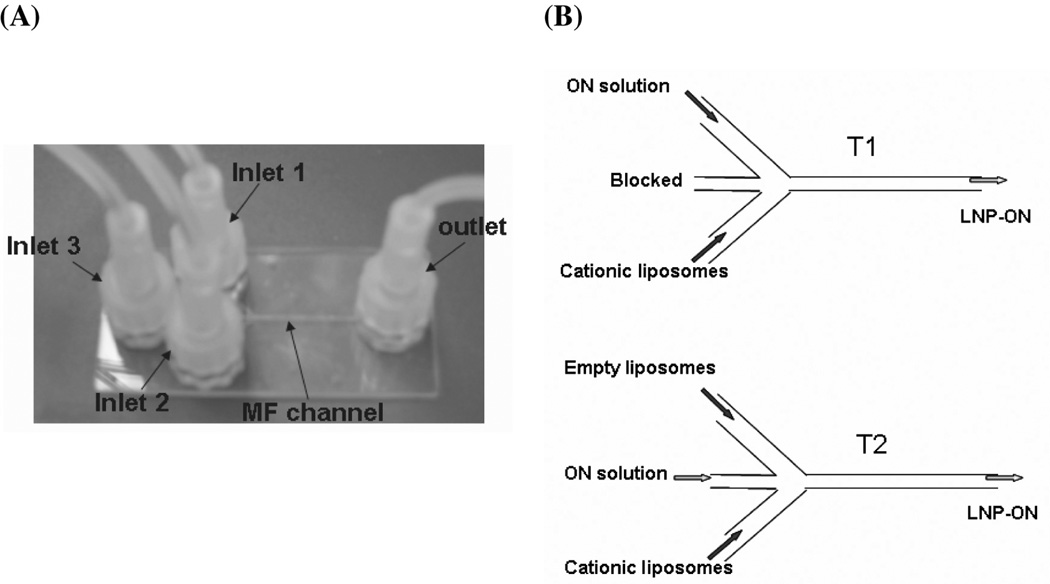
The MF chip used in this study is shown in Figure 1A. It was made of fused silica glass with microchannels 300 µm in width and 100 µm in depth, and with a length of 5 cm (Translume Inc., Ann Arbor, MI, USA). The device consisted of three inlets and one outlet (Figure 1A). The inlets were connected to two sterile syringes containing either liposomes or ON in HEPES (pH=7.4). These syringes were mounted on a single syringe infusion pump (model 975; Harvard Apparatus, Holliston, MA, USA).
Figure 1.
Preparation of ON-loaded LNPs using an MF chip. (A) Photographs of the MF chip. The device consisted of three inlet ports and one outlet port. The inlet ports were connected to sterile syringes containing either cationic liposomes or ON buffer solution. (B) Schematics for flow patterns T1 and T2. In the case of T1, the ON solution and cationic liposomes were injected at the same flow rate from inlets 1 and 2, respectively. Inlet 3 was blocked. In the case of T2, the ON solution was injected from inlet 3 and two streams of cationic liposomes were injected through inlets 1 and 2. The flow rates of the three streams were the same.
Preparation of ON-loaded LNPs
As depicted in Figure 1B, the MF preparation of ON-loaded LNPs was performed by constant-rate infusion of cationic liposomes and ON (ODN or siRNA) in HEPES buffer (20 mM HEPES, pH 7.4) through the inlets. The empty cationic liposomes were first prepared by ethanol injection as described previously (15) with minor modification. Briefly, an ethanolic lipid solution (composed of DOTAP/egg PC/Chol/PEGCeramide at 45:18:35:2, mol/mol) was injected into a stirring HEPES solution (20 mM HEPES, pH 7.4) at room temperature. Ethanol was removed by dialysis using a MWCO 10,000 Dalton Float-A-Lyzer (Spectrum Laboratories Inc., Ranco Dominguez, CA, USA) against HEPES buffer (20 mM HEPES, pH 7.4) for 2 h at room temperature. The resulting liposomes were then sterilized using a 0.22-µm filter (Fisher Scientific, Pittsburgh, PA, USA).
Two flow patterns were evaluated (Figure 1B). In the flow pattern of T1, the ON solution (50 µg/ml) and the cationic liposomes (500 µg/ml) were injected at the same flow rate in inlets 1 and 2, respectively. Inlet 3 was blocked. The resulting ON-LNPs were collected at the outlet port. In the flow pattern of T2, the ON solution (50 µg/ml) was injected at inlet 3. Meanwhile, cationic liposomes (250 µg/ml) were injected through inlets 1 and 2. The flow rates of three streams were identical. The resulting ON-LNPs were collected at the outlet port. The effect of shear stress on MF preparation of ON-LNPs was investigated by varying the flow rate from 50 to 1100 µl/min. The synthesis was repeated at least twice under each set of conditions.
Size and zeta potential measurements
The particle size of LNPs was determined by dynamic light scattering using a particle sizer BI-200SM (Brookhaven Instruments Corp., Holtsville, NY, USA) in an intensity-weighted mode. Prior to measurement, the samples were diluted with de-ionized water to 1 ml. The zeta potential of ON-loaded LNPs was measured following dilution in PBS and then determined by a PALS zeta potential analyzer (Brookhaven Instrument Corp.).
Cell transfection study
SK Hep-1 cells, stably expressing the luciferase gene, were plated at a concentration of 2×104 cells per well in 96-well plates and grown to 60–70% confluent prior to transfection. Luciferase-specific and negative control siRNAs were formulated into LNPs by MF or BM. Cells were treated with various siRNA-LNPs at siRNA concentration of 100 nM and were incubated for 24 h at 37°C. The cells were washed with PBS and lysed. The luciferase activity for each well was determined using luciferase reagent from Promega (Madison, WI, USA). The luminescence for each sample was measured using a Berthold MicroLumatPlus LB96V plate luminometer (Huntsville, Alabama, USA) and was normalized for the amount of protein measured using the Micro BCA assay kit (Pierce, Rockford, IL, USA). Luciferase gene down-regulation was then determined for each preparation. Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) was used as a positive control.
Statistical analysis
Data were represented as mean±standard deviation (SD) and were analyzed by two-tailed Student’s t-test using the MiniTAB Program (Minitab Inc., State College, PA, USA). A difference with a p-value of <0.05 was considered statistically significant.
Results
Production of AS-ODN loaded LNPs in MF device
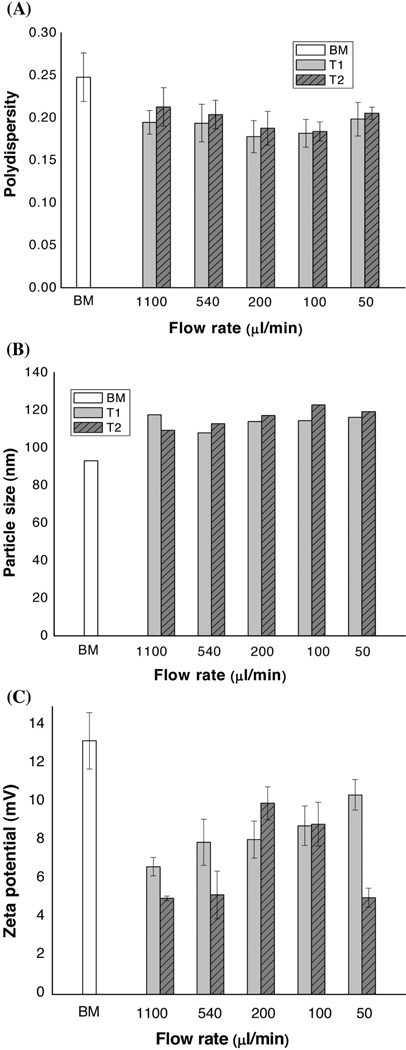
An MF device was used to produce the LNPs. G3139, an AS-ODN against human Bcl-2, was used as a model agent. The difference between LNP-G3139 produced by the MF and BM methods was evaluated. As demonstrated in Figure 1B, the MF approach was implemented with a pair of syringes and a glass MF chip with a ‘Y’-channel design. The effects of flow pattern (T1 and T2) and flow rate on particle size were examined. Figure 2A shows that narrower distribution (lower polydispersity) was obtained using either T1 or T2 flow patterns compared to the value obtained for the BM method. In particular, the polydispersity reached a minimum when MF synthesis was carried out at the flow rate range between 100 and 200 µl/min. More importantly, the SD values of LNP-G3139 obtained by the MF method were lower than when using the BM method, resulting in a more reproducible preparation of LNP-AS-ODN by MF.
Figure 2.
Effects of the flow rate on physical properties of G3139-LNPs prepared by the MF method. (A) The relationship between the polydispersity of LNPs and the flow rate. (B) The relationship between the particle size of LNPs and the flow rate. (C) The relationship between the zeta potential of LNPs and the flow rate. The LNPs prepared by the BM method were used as a reference control. The formulation composition was as follows: DOTAP/egg PC/Chol/Ceramide-PEG=45/18/35/2 (mol/mol); Lipids/G3139=10.0/1 (w/w). Results are presented as mean±SD of three independent experiments.
Figure 2B shows the effect of flow rate on the particle size of LNP-G3139 synthesized by MF. LNPs produced by MF were slightly larger in size than those produced by the BM method. For the MF T2 flow pattern, when the flow rate was increased from 50 to 1100 µl/min, the mean particle size decreased from approximately120nm to approximately 110 nm. A similar trend was observed in the MF synthesis using the flow pattern T1. In addition, as seen in Figure 2C, the zeta potential of LNPs prepared by the MF method was slightly lower than that prepared by the BM method. Figure 2 suggests that the flow pattern (T1 and T2) did not significantly affect the properties of the resulting LNPs.
Production of siRNA-loaded LNPs by MF
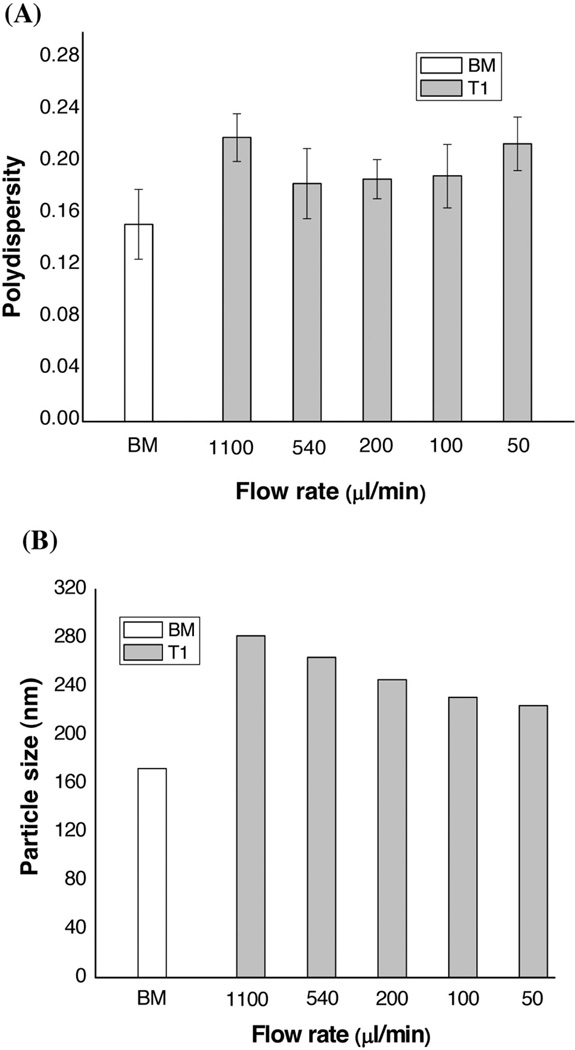
In contrast to the single-stranded AS-ODN, siRNA is double-stranded RNA with 19–23 nt in length. This study further examined the effects of preparation method on the particle size of siRNA-LNPs, using the flow pattern T1. The weight ratio of lipids/siRNA (10/1) was the same as that used with AS-ODN. As shown in Figure 3, siRNA-LNPs prepared by MF showed not only larger particle size but also greater polydispersity compared to LNPs made by the BM method. However, the reproducibility of the MF method was better than that of the BM method (Figure 3A). Figure 3B indicates that the siRNA-LNPs by MF had larger particle sizes (220– 280 nm versus approximately 170 nm for BM). Interestingly, when the flow rate was increased from 50 to 1100 µl/min in MF, the mean particle size of LNPs increased from approximately 220 nm to approximately 280 nm. Additionally, the zeta potentials of the siRNA-LNPs prepared by the MF and BM methods did not differ significantly. Therefore, the cargo molecule (siRNA versus ODN) had a profound effect on the synthetic method (MF versus BM). To the Authors’ best knowledge, this is the first time such an observation has been made.
Figure 3.
Effects of the flow rate on the physical properties of siRNA-LNPs prepared by the MF method. (A) The relationship between the polydispersity of LNPs and the flow rate. (B) The relationship between the particle size of LNPs and the flow rate. The LNPs prepared by the BM method were used as a reference control. The formulation composition was as follows: DOTAP/egg PC/Chol/Ceramide-PEG=45/18/35/2 (mol/mol); Lipids/G3139=10.0/1 (w/w). Results are presented as mean±SD of three independent experiments.
In vitro evaluation of siRNA-LNPs prepared by MF
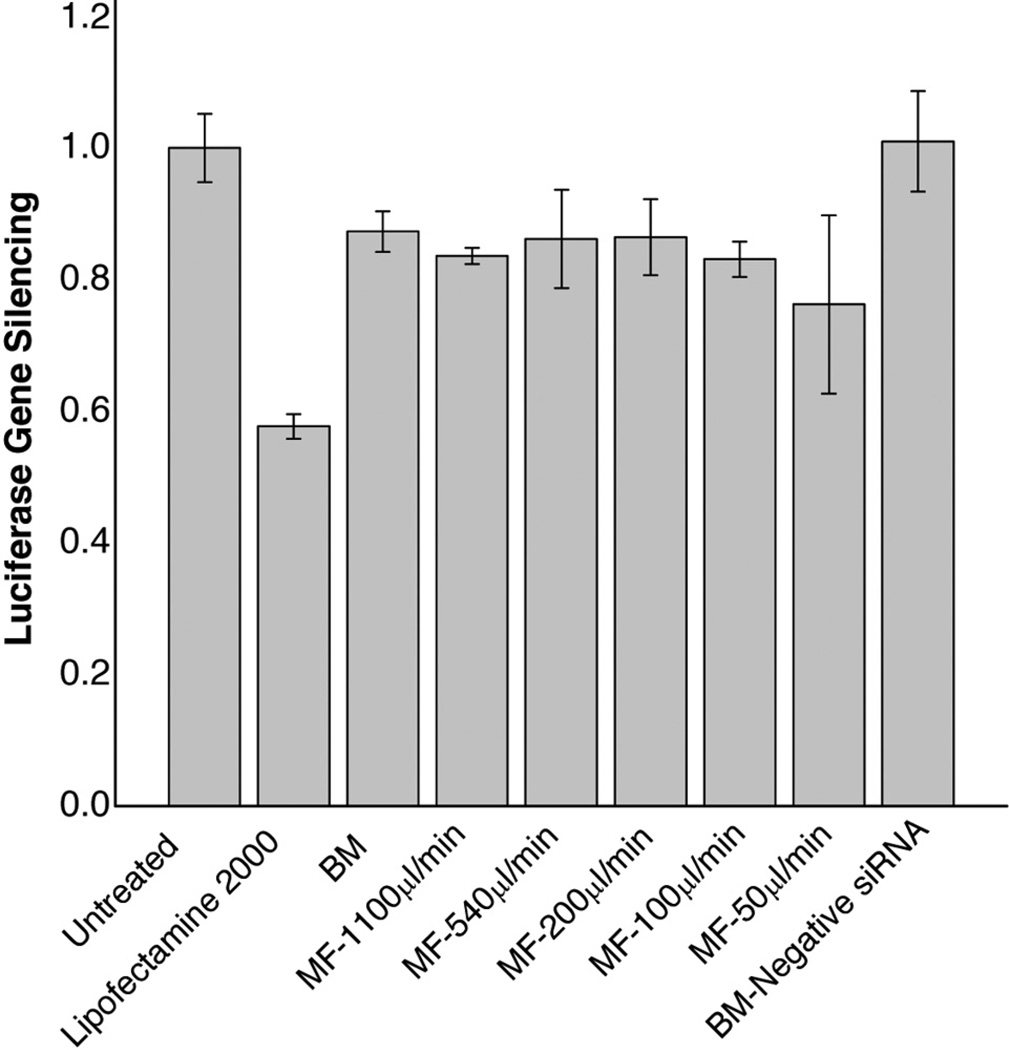
siRNA-LNPs were evaluated for transfection efficiency in SK Hep-1 cells expressing a luciferase reporter gene (Figure 4). Lipofectamine 2000, a common transfection agent, was used as a positive control. Figure 4 shows that the luciferase level, relative to the untreated control, was reduced by 20–30% when transfected using the siRNA-LNPs prepared by either the MF or BM method. There was not a significant difference in the down-regulation of luciferase between the two methods. Furthermore, the siRNA-LNPs produced at different flow rates by MF also did not show a significant effect on luciferase silencing. Combined with the results in Figure 3, these results suggested that the larger particle size and broader size distribution of siRNA-LNPs synthesized by MF did not influence their transfection efficiency, which was essentially the same as that of the LNPs produced by the BM method.
Figure 4.
Silencing of luciferase in SK Hep-1 cells by siRNA-LNPs prepared by the MF method. Cells were treated with various formulations containing 100nM siRNA at 24 h and the luciferase expression was analyzed. The siRNA-LNPs prepared by the BM method were used as a control. Lipofectamine was used as a positive control for transfection. The formulation composition was: DOTAP/egg PC/Chol/Ceramide-PEG=45/18/35/2 (mol/mol); Lipids/siRNA=10.0/1 (w/w). Results are presented as the mean±SD of three independent experiments.
Discussion
Clinical application of ON-based therapeutics has been hampered by the lack of safe and efficient delivery systems. LNPs are widely recognized as one of the most promising delivery systems (4–9). The in vivo delivery of ONs is affected by the physical chemical properties of LNPs, including composition, particle size, surface charge and morphology (6, 7, 9). In conventional BM methods, lipids and ONs are spontaneously assembled into heterogeneous nanostructures in a bulk phase. This tends to suffer from irreproducibility of properties of the nanoparticles from batch to batch. It is also a challenge to scale up the production procedures (5, 7, 24, 25). This study described a MF chip-based approach for preparation of ON loaded LNPs.
MF is a relatively novel technology for producing nano-sized lipid nanoparticles (5, 16, 22, 25). The characteristics of laminar flow and tunable mixing in MF have distinctive advantages in the formation of LNPs (5, 17). The present study examined the effects of the MF technology on the production of ODN and siRNA LNPs in the absence of ethanol (Figure 1). To implement MF in a laboratory setting, Yu et al: Microfludic Assembly of Lipid-based Oligonucleotide Nanoparticles syringe pump and a glass MF chip were used. In general, an MF chip is fabricated in a poly(methyl methacrylate) (PMMA) or PDMS plate (5, 17, 24, 25), which requires costly microfabrication equipment and advanced manipulation skills. This study, however, used a commercial glass-based chip, which was chemically stable and readily reusable. For both patterns (T1 and T2), the flow of ONs aqueous solution met with the flow of cationic liposomes to form LNPs in the MF channel (Figure 1B). Unlike the ethanol dilution method, the direct mixing of liposomes and ON in MF avoided the need for removal of ethanol from the product, resulting in reduced production time. In fact, in this study formation of a precipitation was often observed when the ethanol dilution method was used to produce ON-LNPs in conjunction with MF (data not shown).
In this study, MF-produced G3139-LNPs were found to have similar sizes (approximately 110 nm) and narrower size distribution compared to BM-produced G3139-LNPs (Figures 2A and 2B). Importantly, the MF method was more reproducible than the BM method. MF is characterized by laminar flow (Re<2000) (5, 14, 26). In the chip used in this study, as the flow rate was increased from 50 to 1100 µl/min, the calculated Re values were 3.6 to 58.4, which indicated laminar flow. The optimal range of flow rates for preparing ODN-loaded LNPs were further determined to be 100 to 200 µl/min (corresponding to Re from 7 to 14), which resulted in reduced polydispersity.
Interestingly, siRNA-LNPs prepared by MF did not appear to have any advantages over LNPs prepared by BM, showing larger particle size and similar gene silencing activity. This may have been due to the double-stranded structure of the siRNA and the increased rigidity of its structure, which in turn affects its interactions with the cationic liposomes.
The findings of this study suggest that MF provides a new platform for the development and optimization of ON-loaded LNPs. Reproducible control of particle size and size distribution can be implemented in continuous MF flow systems. This method does not require a highly specialized and expensive instrument and it can be easily carried out in a typical biomedical or pharmaceutical laboratory. Synthesis of ON-LNPs with small size and narrow-size distribution can be optimized by varying flow rate, flow ratio, concentration of lipids solution and the characteristic length of the MF channel.
Conclusion
A novel method of MF synthesis of ON-LNPs is reported. The method improved LNP synthesis by improving its reproducibility. It also produced LNPs with narrower size distribution with AS-ODN but not with siRNA. Further studies are warranted to characterize the MF LNPs in vivo.
Acknowledgements
This work was supported by the National Institute of Health Grants R01 CA135243 and R21 CA131832.
References
- 1.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 2.Dykxhoorn DM, Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126:231–235. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu B, Zhao X, Lee LJ, Lee RJ. Targeted delivery systems for oligonucleotide therapeutics. AAPS J. 2009;11:195–203. doi: 10.1208/s12248-009-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayburn ER, Zhang R. Antisense, RNAi, gene silencing strategies for therapy Mission possible or impossible? Drug Discov Today. 2008;13:513–521. doi: 10.1016/j.drudis.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, Lee RJ, Lee LJ. Microfluidic methods for production of liposomes. Methods Enzymol. 2009;465:129–141. doi: 10.1016/S0076-6879(09)65007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu SY, McMillan NA. Lipidic systems for in vivo siRNA delivery. AAPS J. 2009;11:639–652. doi: 10.1208/s12248-009-9140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semple SC, Akinc A, Chen J, Hope MJ. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Szoka FC. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 10.Maurer N, Wong KF, Stark H, Louie L, McIntosh D, Wong T, Scherrer P, Semple SC, Cullis PR. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys J. 2001;80:2310–2326. doi: 10.1016/S0006-3495(01)76202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada Y, Kogure K, Nakamura Y, Inoue K, Akita H, Nagatsugi F, Sasaki S, Suhara T, Harashima H. Development of efficient packaging method of oligodeoxynucleotides by a condensed nanoparticle in lipid envelope structure. Biol Pharm Bull. 2005;28:1939–1942. doi: 10.1248/bpb.28.1939. [DOI] [PubMed] [Google Scholar]
- 12.Podesta JE, Kostarelos K. Chapter 17 - Engineering cationic liposome siRNA complexes for in vitro and in vivo delivery. Methods Enzymol. 2009;464:343–354. doi: 10.1016/S0076-6879(09)64017-9. [DOI] [PubMed] [Google Scholar]
- 13.Semple SC, Klimuk SK, Harasym TO, Dos Santos N, Ansell SM, Wong KF, Maurer N, Stark H, Cullis PR, Hope MJ, Scherrer P. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim Biophys Acta. 2001;1510:152–166. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 14.Stone HA, Stroock AD, Ajdari A. Engineering flows in small devices Microfluidics toward a lab-on-a-chip. Ann Rev Fluid Mech. 2004;36:381–411. [Google Scholar]
- 15.Whitesides GM. The origins the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 16.Jahn A, Vreeland WN, DeVoe DL, Locascio LE, Gaitan M. Microfluidic-directed formation of liposomes of controlled size. Langmuir. 2007;23:6289–6293. doi: 10.1021/la070051a. [DOI] [PubMed] [Google Scholar]
- 17.Jahn A, Reiner JE, Vreeland WN, DeVoe DL, Locascio LE, Gaitan M. Preparation of nanoparticles by continuous-flow microfluidics. J Nanopart Res. 2008;10:925–934. [Google Scholar]
- 18.Karnik R, Gu F, Basto P, Cannizzaro C, Dean L, Kyei-Manu W, Langer R, Farokhzad OC. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Lett. 2008;8(9):2906–2912. doi: 10.1021/nl801736q. [DOI] [PubMed] [Google Scholar]
- 19.Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WC. Convergence of quantum dot barcodes with microfluidics signal processing for multiplexed high-throughput infectious disease diagnostics. Nano Lett. 2007;7(9):2812–2818. doi: 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- 20.Valencia PM, Basto PA, Zhang L, Rhee M, Langer R, Farokhzad OC, Karnik R. Single-step assembly of homogenous lipid-polymeric lipid-quantum dot nanoparticles enabled by microfluidic rapid mixing. ACS Nano. 2010;4(3):1671–1679. doi: 10.1021/nn901433u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiba K, Ogawa M. Microfluidic syntheses of well-defined sub-micron nanoporous titania spherical particles. Chem Comm (Camb) 2009;44:6851–6853. doi: 10.1039/b914322j. [DOI] [PubMed] [Google Scholar]
- 22.Jahn A, Stavis SM, Hong JS, Vreeland WN, DeVoe DL, Gaitan M. Microfluidic mixing the formation of nanoscale lipid vesicles. ACS Nano. 2010;4:2077–2087. doi: 10.1021/nn901676x. [DOI] [PubMed] [Google Scholar]
- 23.Thiele J, Steinhauser D, Pfohl T, Forster S. Preparation of monodisperse block copolymer vesicles via flow focusing in microfluidics. Langmuir. 2010;26:6860–6863. doi: 10.1021/la904163v. [DOI] [PubMed] [Google Scholar]
- 24.Koh CG, Kang X, Xie Y, Fei Z, Guan J, Yu B, Zhang X, Lee LJ. Delivery of polyethylenimine (PEI)/DNA complexes assembled in a microfluidics device. Mol Pharm. 2009;6(5):1333–1342. doi: 10.1021/mp900016q. [DOI] [PubMed] [Google Scholar]
- 25.Koh CG, Zhang X, Liu S, Golan S, Yu B, Yang X, Guan J, Jin Y, Talmon Y, Muthusamy N, Chan KK, Byrd JC, Lee RJ, Marcucci G, Lee LJ. Delivery of antisense oligodeoxyribonucleotide lipopolyplex nanoparticles assembled by microfluidic hydrodynamic focusing. J Control Release. 2010;141(1):62–69. doi: 10.1016/j.jconrel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradhan P, Guan J, Lu D, Wang PG, Lee LJ, Lee RJ. A facile microfluidic method for production of liposomes. Anticancer Res. 2008;28:943–948. [PubMed] [Google Scholar]