Abstract
Fabry disease is an inborn error of glycosphingolipid catabolism, resulting from deficient activity of lysosomal α-galactosidase A (α-Gal A). A rare alternative splicing that introduces a 57-nucleotide (nt) intronic sequence to the α-Gal A transcript from intron 4 of the gene has been identified. In addition, a novel midintronic base substitution that results in substantially increased alternative splicing has been identified in a patient with Fabry disease who has the cardiac variant phenotype. The sequence of the patient’s intron 4 contains a single G→A transversion at genomic nt 9331 (IVS4+919G→A), located at the −4 position of the 3′ end of the intronic insertion (nts 9278–9334 in the genomic sequence). Minigene constructs containing the entire intron 4 sequence with G, A, C, or T at nt 9331 within an α-Gal A complementary DNA expression vector were prepared and expressed in COS-1 cells. Whereas transfection of the G or T minigenes transcribed predominantly normal-sized transcripts, the transfection of the A or C minigenes produced a large amount of the alternatively spliced transcript. These results suggest that the G→A mutation, within an A/C-rich domain, results in increased recognition of the alternative splicing by an A/C-rich enhancer–type exonic splicing enhancer. The intronic mutation was not observed in 100 unrelated unaffected men but was present in 6 unrelated patients with cardiac Fabry disease. Reverse-transcriptase polymerase chain reaction of total RNA of various normal human tissues revealed that the alternatively spliced transcript was present in all of the samples, and especially at a higher ratio in the lung and muscle. The normal transcript was present in the patients’ lymphoblasts and resulted in ∼10% residual enzyme activity, leading to a cardiac phenotype of Fabry disease.
Fabry disease (MIM 301500) is an X-linked recessive inborn error of glycosphingolipid catabolism, caused by lysosomal α-galactosidase A (α-Gal A, or GLA; Enzyme Commission [EC] number 3.2.1.22) deficiency. The enzymatic defect results in the accumulation of neutral glycosphingolipids with terminal α-galactosyl residues—predominantly globotriaosylceramide—in body fluids and in tissue lysosomes (Desnick et al. 2001). Classically affected men with no α-Gal A activity have angiokeratoma, acroparesthesias, hypohidrosis, and characteristic corneal and lenticular opacities. In contrast, those afflicted with a milder form have residual α-Gal A activity and do not have the classical manifestations but typically present with left-ventricular hypertrophy (Nakao et al. 1995) and have late-onset cardiomyopathy (Elleder et al. 1990; Ogawa et al. 1990; von Scheidt et al. 1991).
To date, >190 α-Gal A mutations that cause Fabry disease have been described (Human Gene Mutation Database Web site), and all of these mutations are located within the coding or the flanking intronic sequences. Mutations that cause the classic phenotype include missense and nonsense mutations, large and small insertions or deletions, and splicing defects (Desnick et al. 2001). In contrast, mutations that cause cardiac Fabry disease include missense mutations, a small in-frame deletion, and decreased α-Gal A mRNA (Desnick et al. 2001). Among the mutations reported, all of the splicing mutations have been found in the flanking regions of exons, resulting in exon skipping or in misreading of the splice site (Yokoi et al. 1991; Sakuraba et al. 1992; Eng et al. 1993, 1997; Germain and Poenaru 1996, 1999; Okumiya et al. 1996; Matsumura et al. 1998; Topaloglu et al. 1999).
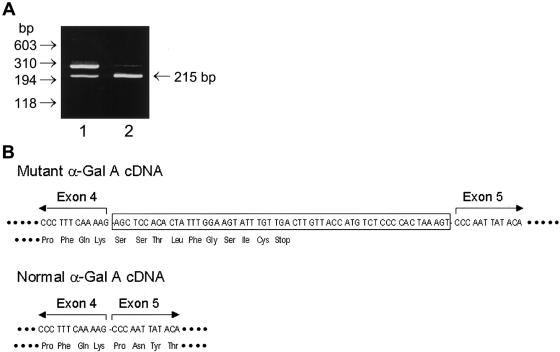
During the course of mutant analysis of a patient with cardiac Fabry disease (proband 1) who had residual enzyme activity of 8.7 U/mg protein (9.1% of normal) in lymphoblasts, we were unable to identify any mutation in the exonic or flanking intronic regions of the α-Gal A gene. Therefore, we directed our efforts toward the determination of the responsible mutation, by means of RT-PCR of the RNA from the cardiac variant. Direct sequencing of the RT-PCR product revealed an insertion between exons 4 and 5 (data not shown). To further characterize the abnormal splicing, RT-PCR of the region that includes exons 4 and 5 was performed. Two bands were visualized on gel electrophoresis: the expected wild-type 215-bp fragment and a larger fragment, ∼270 base pairs in length (fig. 1A). This abnormal DNA fragment was also found in the normal cells, at a very low level. From the intensity of the DNA bands, it was estimated that the abnormal α-Gal A mRNA in the patient’s lymphoblasts was ⩾70% of the total α-Gal A mRNA, whereas it was <5% of the total in normal lymphoblasts. These results suggested that the abnormal DNA fragment was generated by a rare splicing event within the α-Gal A gene.
Figure 1.
Identification of the mutant α-Gal A cDNA. A, To determine the sequence of the cDNA at the junction between exons 4 and 5, RT-PCR amplification of 0.1 μg of total RNA, prepared from the patient's lymphoblasts and from normal lymphoblasts, was performed using primers 5′-GTCCTTGGCCCTGAATAG-3′ and 5′-GTCCAGCAACATCAACAATT-3′, and the results were analyzed by agarose gel electrophoresis. Lane 1, patient lymphoblasts; lane 2, normal lymphoblasts. B, To analyze the sequence of mutant α-Gal A cDNA, the RT-PCR product amplified with the above primers was subcloned into the TA cloning pCR vector (Invitrogen), and the mutant cDNA fragment was sequenced by an automated sequencer.
To determine the sequence of the abnormal fragment, the RT-PCR product generated from the patient’s lymphoblast RNA through use of primer set 5′-GTCCTTGGCCCTGAATAG-3′ and 5′-GTCCAGCAACATCAACAATT-3′, which amplified a fragment that included exons 4 and 5, was subcloned into the TA cloning pCR vector (Invitrogen). Sequencing of the RT-PCR product revealed a 57-nucleotide (nt) insertion at the junction between exons 4 and 5 of the α-Gal A cDNA (fig. 1B), consistent with the observed size of the RT-PCR product from the patient’s lymphoblasts (fig. 1A). This insertion corresponded to an intronic sequence in the gene at nts 9278–9334, which is located in the middle of intron 4. This in-frame insertion caused a premature termination at the 27th nucleotide downstream from exon 4. Therefore, the predicted product of the mutant α-Gal A mRNA was a truncated protein of 222 amino acid residues. To examine whether the truncated protein exhibited enzyme activity, an expression construct (pBaIns57) containing the mutant insertion between exons 4 and 5 was prepared and expressed in COS-1 cells. A truncated protein of 30 kD was detected by western blot analysis (fig. 2A). The enzyme activities in the mock-transfected COS-1 cells and COS-1 cells transfected with pBaIns57 were 153±3 U/mg and 147±3 U/mg, respectively, compared with 2,670±200 U/mg in cells transfected with the wild-type α-Gal A cDNA (pBaN; fig. 2B). These results indicated that the C-terminal truncated protein had no enzyme activity.
Figure 2.
Transient expression of mutant α-Gal A cDNA in COS-1 cells. COS-1 cells were transfected with 1 μg of plasmid DNA and 8 μl of LipofectAMINE Reagent (Life Technologies). A, Western blot analysis was performed using a rabbit anti-human α-Gal A antibody, as described elsewhere (Ishii et al. 1994). Lane 1, mock transfection (pBactE vector; see Koide et al. 1990); lane 2, pBaIns57, an α-Gal A cDNA containing the 57-nt intronic insertion; lane 3, pBaN, a wild-type α-Gal A expression construct (Ishii et al. 1992). Approximately 50 μg of protein was loaded in each well for SDS-PAGE analysis. The truncated protein is indicated by an arrow. B, α-Gal A activity was assayed with 4-methylumbelliferyl α-D-galactoside, as described by Fan et al. (1999). Bars 1, 2, and 3 in the graph correspond to lanes 1, 2, and 3, respectively, in panel A. The enzyme activity is expressed as the mean±SD of triplicate experiments.
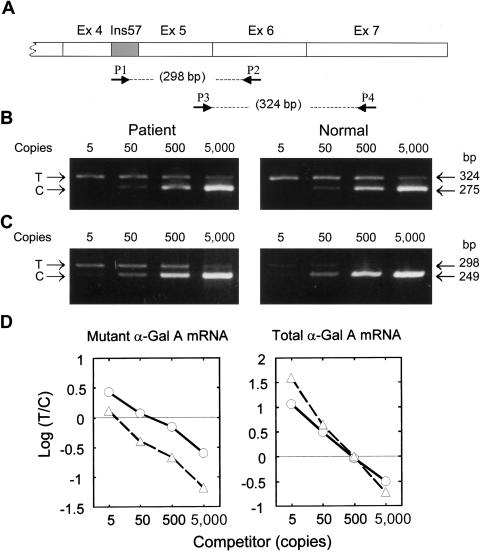
To determine the ratio of mutant and total (both mutant and normal) α-Gal A mRNA in the patient’s lymphoblasts, a quantitative competitive RT-PCR was performed. DNA competitors were prepared by PCR amplification of a λ-DNA provided by the Competitive DNA Construction Kit (Takara Shuzo), through use of sense and antisense primers 5′-GCTCCACACTATTTGGAAGTACGGTCATCATCTGACAC-3′ and 5′-AAAGGAGCAGCCATGATAGAACTATCGACATCATTACGC-3′, for specific determination of the mutant α-Gal A mRNA, or sense and antisense primers 5′-GGTTGGAATGACCCAGATAGTACGGTCATCATCTGACAC-3′ and 5′-TGCGATGGTATAAGAGCGCAGTTAATCGAACAAGAC-3′, for specific determination of total α-Gal A mRNA. A 298-bp PCR product or a 324-bp PCR product was amplified in the presence of the respective competitors to detect either mutant α-Gal A mRNA or both normal and mutant α-Gal A mRNAs, respectively (fig. 3A). The total α-Gal A mRNA amount was essentially the same in both the normal cells and the patient's cells (fig. 3B). On the other hand, the 298-bp mutant fragment was the predominant product in the patient’s cells, whereas this fragment was present at a very low level in the normal cells (fig. 3C). On the basis of the intensity of the DNA bands, the mutant α-Gal A mRNA was estimated to be present at ∼115 copies, compared with the DNA competitor; this amount was ∼10-fold higher than that in the normal cells (10 copies) (fig. 3D).
Figure 3.
Quantitative analysis of the mutant α-Gal A mRNA. The relative amounts of normal and mutant α-Gal A mRNA were determined by a competitive RT-PCR method (Becker-Andre and Hahlbrock 1989). The concentration of total RNA prepared from the lymphoblasts was determined by OD260 and was adjusted using the human β-actin Competitive PCR Set (Takara Shuzo). RT was performed with 0.6 μg of total RNA in 60 μl of the reaction mixture, through use of the RNA PCR Kit (Takara Shuzo). Aliquots (5 μl) of the RT reaction were used for PCR amplifications of mutant or total α-Gal A mRNAs. A, Diagram of mutant α-Gal A mRNA and the location of the primers. In the assay of mutant α-Gal A mRNA, target (298 bp) and competitor (249 bp) DNA fragments were amplified with primer set P1 (5′-GCTCCACACTATTTGGAAG-3′) and P2 (5′-AAAGGAGCAGCCATGATAG-3′) and DNA competitors prepared specifically for the mutant α-Gal A mRNA (see text) at various concentrations. In the assay for the total α-Gal A mRNA, target (324 bp) and competitor (275 bp) DNA fragments were amplified with primers P3 (5′-GGTTGGAATGACCCAGATA-3′) and P4 (5′-TGCGATGGTATAAGAGCG-3′) and DNA competitors prepared specifically for the total α-Gal A mRNA (both mutant and normal) at various concentrations. The relative amounts of the total α-Gal A mRNA (B) and the mutant α-Gal A mRNA (C) in both the patient's lymphoblasts and the normal lymphoblasts were determined by comparison with the competitors’ concentration, indicated at the top of the photos. D, After agarose gel electrophoresis, the concentrations of the PCR products were determined using an image processing system, according the method reported elsewhere (Minamikawa-Tachino et al. 1993). Circles = patient; triangles = unaffected subject; Ex = exon; T = target; C = competitor.
To determine whether the alternative splicing occurred in human tissues, RT-PCR between exons 4 and 5 was performed using total tissue RNA (fig. 4). The alternatively spliced transcript (the 272-bp fragment) was observed in a trace amount (<1% of total α-Gal A mRNA) in heart and small intestine, and a small amount (<10%) was observed in kidney, spleen, and liver. However, the ratio of the abnormal transcript to the total α-Gal A mRNA was higher in muscle and lung. These results clearly indicate that this abnormal transcript is indeed an alternative splicing product of the human α-Gal A gene.
Figure 4.
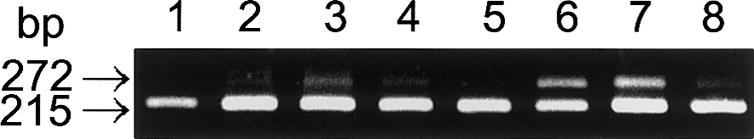
Alternative splicing in human tissues. RT-PCR was performed with 0.1 μg of total human tissue RNA obtained from OriGene, as described in the legend to figure 1. Lane 1, heart; lane 2, kidney; lane 3, spleen; lane 4, liver; lane 5, small intestine; lane 6, muscle; lane 7, lung; lane 8, lymphoblasts.
Intron 4 (nts 8413–10130) of the α-Gal A gene was sequenced using genomic DNA prepared from the patient’s lymphoblasts. Four differences were observed when it was compared with the sequence reported by Kornreich et al. (1989): a G→A transition was present at nt 9331, an A→G transition was present at nt 9401, a G insertion was present at nt 9828, and a CG→GC transversion was present at nt 9876. Because the substitution at nt 9401, the insertion at nt 9828, and the transversions at nt 9876 were detected in genomic DNA of an unaffected Japanese male (data not shown), only the G→A transition at nt 9331 (IVS4+919G→A) was considered to be patient-specific. To determine whether the G→A transition at nt 9331 was a polymorphism, genomic DNA from 100 unaffected Japanese males was amplified and analyzed for the G→A substitution by BfaI digestion, since the G→A transversion at nt 9331 disrupts a BfaI cleavage site. All of the PCR products were completely cleaved by the enzyme (data not shown), indicating that this transversion was not present in 100 normal alleles.
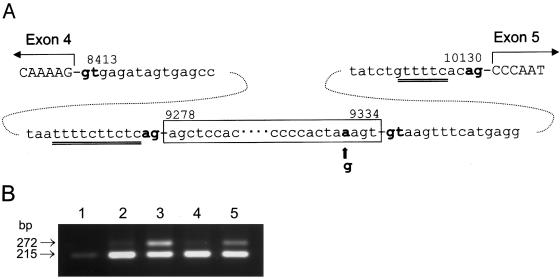
The 57-nt sequence inserted in the mutant α-Gal A mRNA was identical to the genomic sequence at nts 9278–9334 in the patient (fig. 5A). The invariant GT and AG dinucleotides involved in the splicing were present at both sites of the 57-nt sequence, and a 10-nt polypyrimidine tract (nts 9265–9275) was observed upstream of the 3′ AG acceptor site. It is known that efficient splicing of mammalian RNAs involves four sequence elements in introns: (1) a conserved GT dinucleotide at the 5′ splice junction, (2) an invariant AG dinucleotide at the 3′ splice junction, (3) a weakly conserved sequence at the site of lariat formation with an adenine at the branch point, and (4) a pyrimidine-rich region of variable length between the branch point and the 3′ splice junction, referred to as the polypyrimidine tract (Padgett et al. 1986; Shapiro and Senapathy 1987). All of the structural characters surrounding the insertion site indicate that the 57-nt sequence could be an additional exon in the α-Gal A gene. Studies (Dominski and Kole 1991) on the effects that internal exon length has on splice-site selection have revealed that internal exons >87 nts in length were included in the splice product, whereas complete skipping of an internal exon was observed when the exons were <33 nt in length. A 51-nt exon was processed in vitro to result in both skipped products and included products, and the importance of these elements has been shown experimentally in studies employing pre-mRNAs that contain point mutations or deletions within an existing sequence (Aebi et al. 1986). In the present case, a slightly shorter sequence, generated by deletion of 3 bp from the 57-nt sequence, was exclusively skipped from the transcript (data not shown), suggesting that the length of 57-nt probably limits the inclusion of this sequence. This may be the reason that only low-level splicing of this intronic sequence (∼5% of α-Gal A mRNA) occurred in normal lymphoblasts and tissues.
Figure 5.
Contribution of nucleotide at nt 9331 to alternative splicing. A, Sequence of α-Gal A intron 4 from the patient. The 57-nt sequence is enclosed in the box. The G→A transition at nt 9331 is indicated by an arrow. The polypyrimidine tract is indicated by double underlining. B, Transient expression of minigene constructs in COS-1 cells. Minigene constructs containing the entire normal or mutant intron 4 were prepared as follows: fragment A (nts 8324–9450) was amplified, from genomic DNA of an unaffected subject, with primers 5′-ATAAGCACATGTCCTTGGC-3′ and 5′-TGCAGAGATAATGAGGGAAT-3′ and then was digested with AflIII and SacI; fragment B (nts 9112–10282) was amplified from normal or mutant genomic DNA with primers 5′-CTGCCTAGAACAGTTCTTC-3′ and 5′-TCATTCCAACCCCCTGGT-3′, followed by digestion with SacI and BstXI. The 951-bp AflIII-SacI–digested fragment A was ligated to the 992-bp SacI-BstXI–digested fragment B, to generate a 1,943-bp AflIII-BstXI fragment that was inserted into the AflIII-BstXI of the pCXN2Gal expression construct (Ishii et al. 1993). The constructs containing normal and patient intron 4 were designated as “pCXN2Gal/Int4N” and “pCXN2Gal/Int4P,” respectively. To generate the G→C and G→T conversions at the nt 9331 mutation site, site-directed mutagenesis by Pfu-polymerase–based PCR using the QuickChange kit (Stratagene) was performed in a small fragment (SacI-XmaI, nts 9282–9587) of α-Gal A intron 4. The complete sequence of the mutated fragment was confirmed by DNA sequencing. The fragment with the desired mutation was subcloned into its original position in a minigene construct using two steps of subcloning. First it was inserted into a vector containing NheI-XmaI (exons 2–4 [nts 358–639 of cDNA] and intron 4 [nts 8413–9587 of genomic DNA]). Next, it was transferred to the pCXN2 expression vector that contains the full-length α-Gal A cDNA and intron 4. The constructs containing the G→C or G→T conversion are designated “pCXN2Gal/Int4C” or “pCXN2Gal/Int4T,” respectively. COS-1 cells transfected with 1 μg of plasmid DNA were grown for 2 d, and RT-PCR analysis was performed using primer pair 5′-GTCCTTGGCCCTGAATAG-3′ and 5′-GTCCAGCAACATCAACAATT-3′ and total RNA prepared from COS-1 cells transfected with each plasmid construct. Lane 1, mock transfection, pCXN2 vector (Niwa et al. 1991); lane 2, pCXN2Gal/Int4N; lane 3, pCXN2Gal/Int4P; lane 4, pCXN2Gal/Int4T; lane 5, pCXN2Gal/Int4C.
To evaluate whether the G→A transition at nt 9331 was responsible for the mutant α-Gal A mRNA in the patient’s cells, minigene constructs containing the entire normal intron 4 sequence (pCXN2Gal/int4N) or the patient’s intron 4 sequence (pCXN2Gal/int4P) were prepared and then expressed in COS-1 cells. To confirm that the pCXN2Gal/int4P construct contained the G→A transition, the minigene construct was digested with BfaI. Presence of a 427-bp fragment (nts 9116–9543) after the enzyme digestion indicated the presence of the mutation in pCXN2Gal/int4P (data not shown), since this fragment would be cleaved into two smaller fragments by the enzyme if the wild-type sequence was present. α-Gal A activity in COS-1 cells transfected with both normal and mutant constructs was detected, although the enzyme activity was substantially lower in the cells transfected with pCXN2Gal/int4P, suggesting that the minigene constructs were transcribed, spliced, and translated in the COS-1 cells. α-Gal A mRNA was analyzed by RT-PCR of total RNA isolated from the transfected COS-1 cells. A large amount of the 272-bp RT-PCR fragment was obtained from the COS-1 cells transfected with pCXN2Gal/int4P, indicating that alternative splicing of the α-Gal A transcript occurred in COS-1 cells (fig. 5B). This indicated that the IVS4+919G→A mutation is responsible for the drastic increase of the alternative splicing in intron 4.
Intronic mutations have been found to cause many human diseases, including β-thalassemia (Treisman et al. 1983), ornithine δ-aminotransferase deficiency (Mitchell et al. 1991), and ataxia-telangiectasia (Teraoka et al. 1999). These mutations often exist in consensus AG/GT dinucleotides, the branch point, or the polypyrimidine tract of a cryptic exon, resulting in inclusion of the cryptic exon or in changing of the normal splicing site upstream of the cryptic exon. In the present case, the IVS4+919G→A mutation that drastically changes the splicing of the 57-nt sequence is located within the alternative splicing exon. To assess the mechanism of the alternative splicing, additional minigenes with mutations at the −4 position and its surrounding region were generated by site-directed mutagenesis, and COS-1 cells were transfected with minigenes of α-Gal A cDNA containing intron 4 with IVS4+919G→C (pCXN2Gal/int4C) or IVS4+919G→T (pCXN2Gal/int4T) transversions. Only the IVS4+919G→C transversion, and not the G→T transversion, was shown to increase the inclusion of the 57-nt sequence (fig. 5B), indicating that an A or C at the position activates the alternative splicing.
Exonic splicing enhancers (ESEs) that direct the specific recognition of splicing sites during constitutive and alternative splicing are found within both coding and noncoding exons (Blencowe 2000). The most common feature of ESEs is that most are purine rich (Eldridge et al. 1999). Sequences other than purine-rich repeats have also been characterized as exon enhancers. For example, novel A/C-rich enhancers (ACEs) have been identified both in natural genes and in experiments designed to select exon enhancers. The ACEs were reported from human calcitonin exon 4 (van Oers et al. 1994), chicken cTNT exon 16 (Wang et al. 1995), and Drosophila doublesex exon 4 (Ryner and Baker 1991; Lynch and Maniatis 1995). Disruption of an ESE in the coding sequence can cause inappropriate exon skipping in vitro (Liu et al. 2001). In the 57-nt sequence, an A/C-rich sequence (nts 9323–9332: CCCCACUAGA) was observed in the alternative splicing exon within intron 4 of the α-Gal A gene. The IVS4+919G→A/C substitutions could further enrich the A/C predominance for the sequence and, thus, could increase the recognition of the alternative splice site. Therefore, the ACE recognition mechanism may be involved in the alternative splicing of the α-Gal A gene. ACE-type ESEs are not as common as purine-rich–type ESEs. Recently, the RNA-binding protein YB-1 was reported to bind ACEs and to stimulate the alternative splicing of the CD44 exon v4 (Stickeler et al. 2001), although the detailed structural mechanism was not revealed. Our finding of the increased alternative splicing in α-Gal A gene could be valuable for the elucidation of the mechanism of the ACE enhancement.
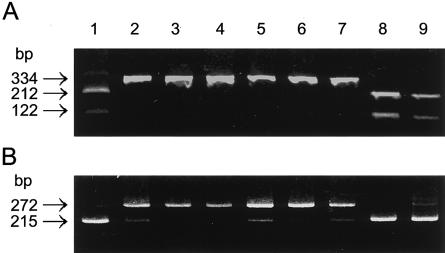
Mutations in another patient with Fabry disease (proband 2) with the cardiac variant and in four patients with Fabry disease of the cardiac phenotype (or atypical Fabry disease) previously characterized as expressing a decreased level of α-Gal A mRNA (Nakao et al. 1995) were analyzed in the present study. Proband 2 had residual lymphoblast α-Gal A activity that was 10.6% of normal. The residual enzyme activities in other patients were 8%–15% of normal. BfaI digestion of the genomic fragment containing the substitution site did not cleave the fragment from these patients (fig. 6A), clearly indicating the presence of the G→A substitution. These results were confirmed by sequencing of the appropriate PCR products (data not shown). RT-PCR amplification of total RNA from these patients’ lymphoblasts revealed a high level of mutant α-Gal A mRNA, detected as a 272-bp fragment (fig. 6B). The percentage of mutant α-Gal A mRNA was 64%–88%. In contrast, only 5%–8% of the alternatively spliced mRNA was detected in the lymphoblasts from unaffected subjects or from patients with Fabry disease who had missense mutations. These results indicated that the increased alternative splicing in these patients with cardiac Fabry disease was caused by the G→A substitution at nt 9331 and that the deficient enzyme activity was due to a decreased level of normal α-Gal A transcript. Accordingly, these patients should appropriately be classified as having the alternative splicing mutations.
Figure 6.
Detection of the mutation in unrelated patients with cardiac Fabry disease. The mutation can be readily detected by restriction enzyme analysis, because the G→A transition at nt 9331 disrupts a BfaI cleavage site. The mutant 334-bp fragment was not digested, whereas the normal fragment was cleaved into fragments of 212 bp and 122 bp. A, PCR fragment (334 bp; nts 9117–9450) amplified from genomic DNA with primers 5′-TAGAACAGTTCTTCCCCAA-3′ and 5′-TGCAGAGATAATGAGGGAAT-3′, digested with BfaI (New England Biolabs) and analyzed by agarose gel electrophoresis. B, RT-PCR analyses performed as described in the legend to figure 1, through use of total RNA from normal lymphoblasts or patient lymphoblasts. Lane 1, unaffected; lanes 2 and 3, probands 1 and 2; lanes 4, 5, 6, and 7, patients 2, 3, 6, and 7, respectively, all with Fabry disease and reported previously as having decreased α-Gal A mRNA content (Nakao et al. 1995); lane 8, patient with Fabry disease who has the M296I mutation (Nakao et al. 1995); lane 9, patient with Fabry disease who has the R301Q mutation (Sakuraba et al. 1990).
The cardiac variant of Fabry disease is usually associated with residual α-Gal A activity at 5%–10% of normal level. The residual enzyme activities in the patients with cardiac Fabry disease in the present study were determined to be ∼10% of normal. On the basis of RT-PCR amplification, the amount of mutant α-Gal A mRNA in the patients’ lymphoblasts was 64%–88% of the total α-Gal A mRNA, and the product of the mutant α-Gal A mRNA was a truncated polypeptide of 222 amino acid residues that had no detectable enzymatic activity (fig. 2B). The remainder of the normal α-Gal A mRNA was transcribed to result in the 10% residual α-Gal A activity and cause the cardiac variant phenotype. The truncated α-Gal A polypeptide may be also translated from the small amount of the abnormal α-Gal A mRNA in normal cells. However, it may be rapidly degraded in situ, since we were unable to detect the truncated protein in the patient’s lymphoblasts, although it was barely detected in COS-1 cells transfected with pBaIns57 (fig. 3A). Combining six unrelated patients who had cardiac Fabry disease retained the same mutation; this mutation may be considered to be a major mutation in Fabry disease, particularly in the patients who have no mutation in the coding and flanking regions. Until now, all splicing mutations reported in Fabry disease have been nucleotide substitutions or a single base deletion at or near the invariant dinucleotides of an intron, resulting in exon skipping or misreading of the splicing site (Yokoi et al. 1991; Sakuraba et al. 1992; Eng et al. 1993, 1997; Germain and Poenaru 1996, 1999; Okumiya et al. 1996; Matsumura et al. 1998; Topaloglu et al. 1999). The IVS4+919G→A mutation was unique because it occurred in the middle of an intron, and it increased the recognition of a normally weak splice site, resulting in the insertion of an additional sequence into the α-Gal A transcript, which leads to the cardiac phenotype of Fabry disease.
In summary, we report for the first time that there is a normally weakly regulated alternative splice site in the human α-Gal A gene. In addition, we determined that a novel intronic mutation causes a remarkable increase in the alternatively spliced α-Gal A transcript and, consequently, results in the cardiac phenotype of Fabry disease.
Acknowledgments
We thank Dr. E. Nanba (Gene Research Center, Tottori University, Japan) for supplying the DNA samples from unaffected Japanese men, and Mr. K. Inoue for excellent technical assistance. This work was supported, in part, by Ministry of Health and Welfare of Japan, Mizutani Foundation for Glycoscience grant 010092 and by American Heart Association grant AHA 0130522T.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Human Gene Mutation Database (HGMD), Cardiff, http://archive.uwcm.ac.uk/uwcm/mg/search/119272.html (for the α-Gal A gene)
- Online Mendelian Inheritance of Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Fabry disease [MIM 301500])
References
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C (1986) Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47:555–565 [DOI] [PubMed] [Google Scholar]
- Becker-Andre M, Hahlbrock K (1989) Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res 17:9437–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci 25:106–110 [DOI] [PubMed] [Google Scholar]
- Desnick RJ, Ioannou YA, Eng CM (2001) Fabry disease: α-galactosidase A deficiency. In: Scriver C, Beaudet A, Sly W, Valle D (eds) The metabolic and molecular bases of inherited disease, 7th ed. McGraw-Hill, New York, pp 3733–3774 [Google Scholar]
- Dominski Z, Kole R (1991) Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol 11:6075–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge AG, Li Y, Sharp PA, Blencowe BJ (1999) The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc Natl Acad Sci USA 96:6125–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleder M, Bradova V, Smid F, Budesinsky M, Harzer K, Kustermann-Kuhn B, Ledvinova J (1990) Cardiocyte storage and hypertrophy as a sole manifestation of Fabry’s disease: report on a case simulating hypertrophic non-obstructive cardiomyopathy. Virchows Arch A Pathol Anat Histopathol 417:449–455 [DOI] [PubMed] [Google Scholar]
- Eng CM, Ashley GA, Burgert TS, Enriquez AL, D'Souza M, Desnick RJ (1997) Fabry disease: thirty-five mutations in the α-galactosidase A gene in patients with classic and variant phenotypes. Mol Med 3:174–182 [PMC free article] [PubMed] [Google Scholar]
- Eng CM, Resnick-Silverman LA, Niehaus DJ, Astrin KH, Desnick RJ (1993) Nature and frequency of mutations in the α-galactosidase A gene that cause Fabry disease. Am J Hum Genet 53:1186–1197 [PMC free article] [PubMed] [Google Scholar]
- Fan JQ, Ishii S, Asano N, Suzuki Y (1999) Accelerated transport and maturation of lysosomal α-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat Med 5:112–115 [DOI] [PubMed] [Google Scholar]
- Germain D, Biasotto M, Tosi M, Meo T, Kahn A, Poenaru L (1996) Fluorescence-assisted mismatch analysis (FAMA) for exhaustive screening of the α-galactosidase A gene and detection of carriers in Fabry disease. Hum Genet 98:719–726 [DOI] [PubMed] [Google Scholar]
- Germain DP, Poenaru L (1999) Fabry disease: identification of novel α-galactosidase A mutations and molecular carrier detection by use of fluorescent chemical cleavage of mismatches. Biochem Biophys Res Commun 257:708–713 [DOI] [PubMed] [Google Scholar]
- Ishii S, Kase R, Sakuraba H, Fujita S, Sugimoto M, Tomita K, Semba T, Suzuki Y (1994) Human α-galactosidase gene expression: significance of two peptide regions encoded by exons 1-2 and 6. Biochim Biophys Acta 1204:265–270 [DOI] [PubMed] [Google Scholar]
- Ishii S, Kase R, Sakuraba H, Suzuki Y (1993) Characterization of a mutant α-galactosidase gene product for the late-onset cardiac form of Fabry disease. Biochem Biophys Res Commun 197:1585–1589 [DOI] [PubMed] [Google Scholar]
- Ishii S, Sakuraba H, Suzuki Y (1992) Point mutations in the upstream region of the α-galactosidase A gene exon 6 in an atypical variant of Fabry disease. Hum Genet 89:29–32 [DOI] [PubMed] [Google Scholar]
- Koide T, Ishiura M, Iwai K, Inoue M, Kaneda Y, Okada Y, Uchida T (1990) A case of Fabry's disease in a patient with no alpha-galactosidase A activity caused by a single amino acid substitution of Pro-40 by Ser. FEBS Lett 259:353–356 [DOI] [PubMed] [Google Scholar]
- Kornreich R, Desnick RJ, Bishop DF (1989) Nucleotide sequence of the human α-galactosidase A gene. Nucleic Acids Res 17:3301–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Cartegni L, Zhang MQ, Krainer AR (2001) A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet 27:55–58 [DOI] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T (1995) Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev 9:284–293 [DOI] [PubMed] [Google Scholar]
- Matsumura T, Osaka H, Sugiyama N, Kawanishi C, Maruyama Y, Suzuki K, Onishi H, Yamada Y, Morita M, Aoki M, Kosaka K (1998) Novel acceptor splice site mutation in the invariant AG of intron 6 of α-galactosidase A gene, causing Fabry disease: mutation in brief no. 146. Online. Hum Mutat 11:483 [DOI] [PubMed] [Google Scholar]
- Minamikawa-Tachino R, Ishii K, Sakuraba H, Suzuki Y, Kaminuma T (1993) Quantitative analysis of dystrophin gene amplification products using a PC-based image analysis system. Int J Biomed Comput 33:277–286 [DOI] [PubMed] [Google Scholar]
- Mitchell GA, Labuda D, Fontaine G, Saudubray JM, Bonnefont JP, Lyonnet S, Brody LC, Steel G, Obie C, Valle D (1991) Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc Natl Acad Sci USA 88:815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao S, Takenaka T, Maeda M, Kodama C, Tanaka A, Tahara M, Yoshida A, Kuriyama M, Hayashibe H, Sakuraba H, Tanaka H (1995) An atypical variant of Fabry’s disease in men with left ventricular hypertrophy. N Engl J Med 333:288–293 [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- Ogawa K, Sugamata K, Funamoto N, Abe T, Sato T, Nagashima K, Ohkawa S-I (1990) Restricted accumulation of globotriaocylceramide in the hearts of atypical cases of Fabry’s disease. Hum Pathol 21:1067–1073 [DOI] [PubMed] [Google Scholar]
- Okumiya T, Takenaka T, Ishii S, Kase R, Kamei S, Sakuraba H (1996) Two novel mutations in the α-galactosidase gene in Japanese classical hemizygotes with Fabry disease. Jpn J Hum Genet 41:313–321 [DOI] [PubMed] [Google Scholar]
- Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA (1986) Splicing of messenger RNA precursors. Annu Rev Biochem 55:1119–1150 [DOI] [PubMed] [Google Scholar]
- Ryner LC, Baker BS (1991) Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev 5:2071–2085 [DOI] [PubMed] [Google Scholar]
- Sakuraba H, Eng CM, Desnick RJ, Bishop DF (1992) Invariant exon skipping in the human α-galactosidase A pre-mRNA: Ag+1 to t substitution in a 5′-splice site causing Fabry disease. Genomics 12:643–650 [DOI] [PubMed] [Google Scholar]
- Sakuraba H, Oshima A, Fukuhara Y, Shimmoto M, Nagao Y, Bishop DF, Desnick RJ, Suzuki Y (1990) Identification of point mutations in the α-galactosidase A gene in classical and atypical hemizygotes with Fabry disease. Am J Hum Genet 47:784–789 [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA (2001) The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J 20:3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, Concannon P (1999) Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet 64:1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu AK, Ashley GA, Tong B, Shabbeer J, Astrin KH, Eng CM, Desnick RJ (1999) Twenty novel mutations in the alpha-galactosidase A gene causing Fabry disease. Mol Med 5:806–811 [PMC free article] [PubMed] [Google Scholar]
- Treisman R, Orkin SH, Maniatis T (1983) Specific transcription and RNA splicing defects in five cloned β-thalassaemia genes. Nature 302:591–596 [DOI] [PubMed] [Google Scholar]
- van Oers CC, Adema GJ, Zandberg H, Moen TC, Baas PD (1994) Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol Cell Biol 14:951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Scheidt W, Eng CM, Fitzmaurice TF, Erdmann E, Hubner G, Olsen EGJ, Christomanou H, Kandolf R, Bishop DF, Desnick RJ (1991) An atypical variant of Fabry’s disease with manifestations confined to the myocardium. N Engl J Med 324:395–399 [DOI] [PubMed] [Google Scholar]
- Wang Z, Hoffmann HM, Grabowski PJ (1995) Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA 1:21–35 [PMC free article] [PubMed] [Google Scholar]
- Yokoi T, Shinoda K, Ohno I, Kato K, Miyawaki T, Taniguchi N (1991) A 3′ splice site consensus sequence mutation in the intron 3 of the α-galactosidase A gene in a patient with Fabry disease. Jinrui Idengaku Zasshi 36:245–250 [DOI] [PubMed] [Google Scholar]