Abstract
Abnormal patterns of meiotic recombination (i.e., crossing-over) are believed to increase the risk of chromosome nondisjunction in human oocytes. To date, information on recombination has been obtained using indirect, genetic methods. Here we use an immunocytological approach, based on detection of foci of a DNA mismatch-repair protein, MLH1, on synaptonemal complexes at prophase I of meiosis, to provide the first direct estimate of the frequency of meiotic recombination in human oocytes. At pachytene, the stage of maximum homologous chromosome pairing, we found a mean of 70.3 foci (i.e., crossovers) per oocyte, with considerable intercell variability (range 48–102 foci). This mean equates to a genetic-map length of 3,515 cM. The numbers and positions of foci were determined for chromosomes 21, 18, 13, and X. These chromosomes yielded means of 1.23 foci (61.5 cM), 2.36 foci (118 cM), 2.5 foci (125 cM), and 3.22 foci (161 cM), respectively. The foci were almost invariably located interstitially and were only occasionally located close to chromosome ends. These data confirm the large difference, in recombination frequency, between human oocytes and spermatocytes and demonstrate a clear intersex variation in distribution of crossovers. In a few cells, chromosomes 21 and 18 did not have any foci (i.e., were presumptively noncrossover); however, configurations that lacked foci were not observed for chromosomes 13 and X. For the latter two chromosome pairs, the only instances of absence of foci were observed in abnormal cells that showed chromosome-pairing errors affecting these chromosomes. We speculate that these abnormal fetal oocytes may be the source of the nonrecombinant chromosomes 13 and X suggested, by genetic studies, to be associated with maternally derived chromosome nondisjunction.
Introduction
Meiotic recombination (i.e., crossing-over) not only generates genetic variation but also is vital for the correct segregation (i.e., disjunction) of homologous chromosomes at the first meiotic division. In humans, meiotic chromosome nondisjunction, resulting in aneuploid conception, is a main cause of miscarriage and of trisomies at birth. Variations in the patterns of maternal recombination have been identified as a risk factor for meiotic chromosome nondisjunction (Robinson et al. 1993; Sherman et al. 1994; Fisher et al. 1995; Hassold et al. 1995; Lamb et al. 1996; Nicolaidis and Petersen 1998; Brown et al. 2000). However, direct investigation of meiotic recombination in human adult oocytes has proved difficult, for technical reasons. To date, the only information on female recombination has been derived indirectly from genetic-linkage studies. This approach entails the determination of the patterns of inheritance of DNA polymorphisms in children, to estimate the rates of meiotic recombination in the germ cells of their parents; it is also necessary to know the grandparental distribution of DNA polymorphisms, to assign recombination events to maternal or paternal germ cells. Most of these analyses have been performed in the CEPH families that have the necessary three-generation information (e.g., see Broman et al. 1998). Through this approach, it has been possible to determine that the human female genetic-map length is ∼4,400 cM. Estimates have similarly been made of the rates of recombination for each chromosome pair and also for the positions of recombination events along the chromosome arms (Broman et al. 1998). These family studies have identified three particular crossover categories that may influence the risk of maternal chromosome nondisjunction: (1) failure of crossing-over; (2) a reduction in frequency, together with more distal placement of a crossover; and (3) an increased rate of recombination, together with a very proximally placed crossover (Robinson et al. 1993; Sherman et al. 1994; Fisher et al. 1995; Hassold et al. 1995; Lamb et al. 1996; Nicolaidis and Petersen 1998; Brown et al. 2000). However, it is also clear that the influence that these variant recombination patterns have on the risk of nondisjunction differs between chromosome pairs. Thus, for example, chromosome 21 is affected by all of the variant patterns, whereas only failure of reduced recombination appears to be correlated with nondisjunction risk for chromosome X (e.g., see Lamb et al. 1996; Thomas et al. 2001).
In contrast to the situation in human female germ cells, meiotic recombination in human male germ cells has been investigated through both indirect and direct means. The indirect method is identical to that outlined above for female germ cells. Classically, the direct approach has made use of chiasmata (i.e., the cytologically visible consequences of crossing-over) to determine the numbers and distributions of crossovers at diakinesis/metaphase I (Hultén 1974; Laurie and Hultén 1985). Recently, an alternative approach has been developed, and this method exploits the remarkable behavior of the DNA mismatch-repair protein MLH1, which forms discrete foci along the axes of paired homologous chromosomes in germ cells at prophase I of meiosis (Baker et al. 1996). The number and distribution of foci in pachytene spermatocytes mirror those of chiasmata at metaphase I in mouse spermatocytes (Anderson et al. 1999). The crucial role that the MLH1 protein plays in crossing-over is additionally supported by the observation that chiasma formation is essentially abolished in mice with a knockout of the Mlh1 gene (Edelman et al. 1996; Woods et al. 1999). The immunocytological approach to the identification of crossover positions at pachytene has also been applied to human spermatocytes, and, as in the mouse, the numbers and distributions of MLH1 foci at pachytene match those of chiasmata at metaphase I (Barlow and Hultén 1998). A preliminary study showed that MLH1 foci could likewise be identified in human fetal oocytes at prophase I (Barlow and Hultén 1998), the developmental stage when meiotic recombination occurs in female germ cells. This observation indicates this approach's applicability to the direct and detailed analysis of meiotic recombination in human oocytes. We report here the first fully analyzed case, and we provide a description of the patterns of crossing-over in human fetal oocytes, for both the whole genome and selected chromosome pairs. These observations provide novel information on meiotic recombination in human fetal oocytes and enable a direct search for the particular recombination patterns that are predicted, by indirect linkage studies, to influence the risk of chromosome nondisjunction. Through this approach, it should be possible (1) to verify whether such recombination patterns can be detected in fetal oocytes, (2) to estimate the rate of their occurrence, and (3) to demonstrate whether there is the predicted interchromosome variability in their frequency. These comparisons should thereby provide further insight into the role that meiotic recombination plays in the etiology of chromosome nondisjunction in human oocytes.
Material and Methods
Cell Preparation
Meiosis begins during the second trimester of fetal gestation, and prophase I stages from leptotene to diplotene are present in 16–24-wk fetuses (Speed 1985). The ovarian tissue used in this investigation was obtained from a second-trimester fetus (approximate gestational age 19 wk, on the basis of the mother's last menstrual period), after prostaglandin-induced termination of pregnancy. The use of the tissue in this project was approved by the Department of Health, Coventry Research Ethics Committee, and the East Birmingham Research and Ethics Committee. Informed consent, in accordance with the recommendations of the Polkinghorne report (Polkinghorne 1989), was received from the patient undergoing the termination of pregnancy. Ovaries were removed at ⩽12 h postmortem and were placed in Liebovitz medium (Life Technology) containing 0.3% BSA (Sigma).
A full description of the cell-preparation and protein-detection protocols has recently been provided elsewhere (Hultén et al. 2001). Two primary antibodies were applied to the cells: rabbit polyclonal anti-SCP3 and mouse monoclonal anti-MLH1 (Pharmingen). The cells were incubated overnight at room temperature and were washed with PBS/0.1% Tween 20, and appropriate, differentially labeled secondary antibodies were applied. The cells were mounted in Vectashield/4′,6-diamidino-2-phenylindole (Vector Laboratories).
FISH
Repeat-sequence DNA probes for the α-satellite regions of the centromeric heterochromatin were used to identify specific chromosomes. Chromosomes 21 and 13 were identified using a DNA probe common to both (Appligene and Oncor), and the chromosomes were distinguished on the bases of size and arm ratios. Chromosomes 18 and X were identified using differentially labeled probes (Vysis). All the probes used were directly labeled with fluorochromes. FISH was performed according to the manufacturers’ protocols. After FISH, the staining of synaptonemal complex (SC) was refreshed by application of an anti-rabbit antibody, conjugated with Texas Red, in the same manner as used for the initial immunocytology.
Analysis
The cells were viewed using a Zeiss Axioskop fluorescence microscope that was equipped with both a cooled CCD camera (Photometrics) and a SmartCapture image-acquisition and -analysis system (Vysis/Applied Imaging). The anti-SCP3 antibody binds to a protein component of axial elements (AEs) in unpaired chromosomes and to the lateral elements of SCs in paired chromosomes (Schalk et al. 1998). The meiotic stage of the oocytes was determined using AE/SC formation, as described elsewhere (Hartshorne et al. 1999). Only cells with normal chromosome pairing/synapsis (fig. 1), determined using published criteria (Speed 1988; Hartshorne et al. 1999), are described in the present article.
Figure 1 .
Paired images of two pachytene oocytes. SCs are shown in red, and MLH1 foci are shown in yellow. a and b, Oocyte with normal chromosome synapsis. The cell is shown after detection of MLH1 (a) and after FISH to identify selected chromosome pairs (b). Chromosome 21 has 1 focus, chromosome 18 has 3 foci, chromosome 13 is obscured by overlaps, and chromosome X has 5 foci. c and d, Abnormal oocyte showing synaptic errors. The cell is shown after MLH1 detection (c) and subsequent FISH (d). Chromosome X displays synaptic failure and, as a result, is present as two separate AEs (leftward-pointing arrowheads); chromosome 18 shows partial synapsis (downward-pointing arrow). MLH1 foci are present along fully synapsed bivalents and on the synapsed segment of chromosome 18 (c); there are no foci on the asynapsed axes of chromosomes X.
Pachytene oocytes that were well spread and flattened (i.e., in the present instance, lying in no more than two focal planes) were selected for enumeration of MLH1 foci. Captured images were transferred, as PICT files, to the IPLab software, and the counting of foci was performed using this software. Only those foci that were unambiguously colocated with SCs were included. Faint (i.e., out-of-focus) signals were excluded. For the analysis of chromosome-specific numbers and distributions of MLH1 foci, the slides were reanalyzed, and images of cells with either peripherally located bivalents or dispersed SCs were captured. These cells were rescreened after FISH, to determine whether they contained analyzable examples of the target chromosome pairs. When there was ambiguity, because of overlaps or entanglements, in the location of either a focus or the axis of a chromosome pair, the bivalent was excluded. The positions of foci along the axis of a chromosome pair were measured (in pixels) from the q-arm telomere by tracing along the axis, with the IPLab software. Each chromosome pair was measured five times, and mean distances were used to estimate focal positions. Since the length of bivalents varied considerably between cells, the positions were transformed to percentages, to facilitate comparison. First, the position of the centromere was estimated for each bivalent, by use of the center point of the FISH signal. Second, the q arm of the bivalent was divided into 20 (5%) intervals, and the p arm was divided into intervals of the same size. The number of p-arm intervals thus varies between the different chromosome pairs that were selected for detailed analysis.
Results
The observations described here come from one fully analyzed case. Initial observations from other fetuses indicate that the recombination patterns described here are typical of events in human fetal oocytes. In the latter, analyses were limited, for technical reasons. Two principal problems were encountered: First, the oocytes from some ovaries failed to respond to the hypotonic treatment and remained small and round at spreading. Second, the cells could show considerable background MLH1 staining. We believe that both problems are a consequence of postmortem, degenerative changes to the germ cells (see Wallace and Hultén 1985). We are currently exploring other means of tissue preservation in a condition more optimal to this type of analysis.
Timing of MLH1 Appearance
MLH1 foci were not present along the axes of unpaired chromosomes at leptotene. The foci first appeared at early zygotene, although not in all cells (6/11 oocytes). It is unclear whether absence of foci in some early zygotene cells represented (a) variable expression of the protein or (b) failure of detection when the level of the antigen was low. Most oocytes contained foci by mid-to-late zygotene (10/13 and 16/17 cells, respectively) and, similarly, by pachytene (169/176 cells; fig. 1a). Again, it is unclear whether absence of foci was a technical artifact or represented cellwide failure of crossing-over in a small percentage of oocytes. Foci were observed on SCs until early diplotene. In diplotene cells, the foci often appeared to be present at the forks of desynapsing homologues, giving the impression that they were holding homologous axes together despite the SC breakdown elsewhere. At later diplotene, the proportions of cells with foci decreased (table 1), as would be expected if the MLH1 protein was lost from the bivalents at this stage.
Table 1.
Mean ± SD Number of MLH1 Foci in Late-Zygotene, Pachytene, and Early-Diplotene Oocytes
| Stage | Mean ± SD No. (Range) of Foci | No. ofCells |
| Late zygotene | 65.2 ± 20.4 (30–90) | 10 |
| Pachytene | 70.3 ± 10.5 (48–102) | 95 |
| Early diplotene | 58.3 ± 13.3 (35–89) | 24 |
The numbers of foci per cell at late zygotene and early diplotene largely fell within the range observed at pachytene (table 1). Two-tailed t tests showed that the mean number of foci did not differ significantly (P=.49) between late zygotene and pachytene but did differ significantly (P<.001) between pachytene and early diplotene. Overall, these analyses indicate constancy of numbers of foci through late zygotene and pachytene but a decline at diplotene, after the onset of desynapsis.
In total, 95 pachytene oocytes were selected for the assessment of numbers of MLH1 foci. These gave a mean of 70.3 foci per cell (table 1). The numbers of foci showed considerable variation between cells, with a range of 48–102 foci. Limited observations from three other fetuses showed similarly wide intercell variation (table 2).
Table 2.
Mean ± SD Number of MLH1 Foci in Pachytene Oocytes from Three Further Cases
| Fetus | Mean ± SD No. (Range) of Foci | No. ofCells |
| 1a | 95.0 ± 12.3 (81–104) | 3 |
| 2 | 77.3 ± 13.0 (62–94) | 6 |
| 3 | 71.6 ± 12.5 (53–87) | 5 |
Reported by Barlow and Hultén (1998).
Numbers and Positions of Foci along Selected Chromosomes
FISH was used to identify four chromosome pairs—namely, chromosomes 21, 18, 13, and X (fig. 1). The numbers of foci along the axes of these four chromosomes were determined, and their positions were measured (fig. 1). Only a small sample of X chromosomes could be analyzed, because of frequent entanglements and overlaps with other SCs, even in relatively well-spread cells (fig. 1a). The mean number of foci on each of the selected chromosome pairs increased as chromosome size increased (table 3). The lengths of the particular chromosome pairs showed considerable intercell variation. Some of this variation may be due to the cell-preparation technique. Interestingly, however, the mean lengths of each chromosome pair tended to be larger with increased numbers of foci (table 4).
Table 3.
Frequency of Bivalents with Different Numbers of Foci in the Chromosome Pairs Selected for Detailed Analysis
|
No. of BivalentsWhen No. of Foci Is |
||||||||
| Chromosome | 0 | 1 | 2 | 3 | 4 | 5 | Mean Foci/Bivalent | Total Bivalents |
| 21 | 3 | 60 | 23 | 0 | 0 | 0 | 1.23 | 86 |
| 18 | 1 | 4 | 35 | 24 | 3 | 0 | 2.36 | 67 |
| 13 | 0 | 2 | 31 | 28 | 3 | 0 | 2.5 | 64 |
| X | 0 | 0 | 6 | 9 | 5 | 3 | 3.22 | 23 |
Table 4.
Mean ± SD Lengths of Bivalents with Different Numbers of Foci
| No. of Foci | No. ofBivalents | Mean ± SD Lengtha |
| Chromosome 21: | ||
| 1 | 60 | 126.3 ± 13.3 |
| 2 | 23 | 145.2 ± 22.2 |
| Chromosome 18: | ||
| 1 | 4 | 182.7 ± 9.3 |
| 2 | 35 | 208.4 ± 31.7 |
| 3 | 24 | 217.1 ± 25.5 |
| 4 | 3 | 228.9 ± 13.9 |
| Chromosome 13: | ||
| 1 | 2 | 232.7 ± 27.2 |
| 2 | 31 | 264.7 ± 35.9 |
| 3 | 28 | 289.9 ± 32.1 |
| 4 | 3 | 263.1 ± 29.9 |
| Chromosome X: | ||
| 2 | 6 | 350.8 ± 28.5 |
| 3 | 10 | 381.8 ± 63.2 |
| 4 | 5 | 396.2 ± 23.0 |
| 5 | 3 | 438.3 ± 57.0 |
Measured (in pixels) from the q-arm telomere by tracing along the axes, with the IPLab software.
In general, foci were located interstitially along the chromosome arms and were only occasionally located close to either the centromere or a telomere. Adjacent foci were normally separated by a relatively large chromosomal segment; in chromosome 21 bivalents with 2 foci, for example, the foci were separated by a mean ± SD distance equal to ∼44%±10.3% of the chromosome axial length (table 5). A few exceptions to this generalization were found. One notable example was a chromosome 13 bivalent in which 2 foci were located within 4% of the chromosome arm: The mean distance between adjacent foci decreased as the number of foci on the bivalents increased (table 5).
Table 5.
Interfocus Distances for Bivalents of Each Chromosome Pair with Different Numbers of Foci
|
Mean ± SD Interfocus Distancea for Interfocus Interval(% of Chromosome Arm) |
|||||
| No. of Foci | 1–2 | 2–3 | 3–4 | 4–5 | No. ofBivalents |
| Chromosome 21: | |||||
| 2 | 44.1 ± 10.3 | … | … | … | 23 |
| Chromosome 18: | |||||
| 2 | 57.8 ± 11.1 | … | … | … | 35 |
| 3 | 33.9 ± 12.0 | 39.9 ± 11.4 |
… | … | 24 |
| 4 | 21.2 ± 11.5 | 29.4 ± 9.5 | 28.3 ± 5.2 |
… | 3 |
| Chromosome 13: | |||||
| 2 | 48.1 ± 10.9 | … | … | … | 31 |
| 3 | 33.7 ± 11.5 | 32.7 ± 12.2 | … | … | 28 |
| 4 | 24.7 ± 6.1 | 31.9 ± 9.0 | 18.4 ± 4.8 | … | 3 |
| Chromosome X: | |||||
| 2 | 68.0 ± 11.7 |
… | … | … | 6 |
| 3 | 37.2 ± 12.0 | 34.3 ± 15.0 | … | … | 9 |
| 4 | 25.1 ± 4.7 | 35.0 ± 7.5 | 21.3 ± 9.9 | … | 5 |
| 5 | 15.7 ± 3.9 | 15.3 ± 3.3 | 41.6 ± 4.0 |
15.1 ± 4.4 | 3 |
Measured (in pixels) from the q-arm telomere by tracing along the axes, with the IPLab software. The transcentromere distance is underlined; intervals with a mixture of intra- and interarm distances are boldface italic. Foci are numbered from the q-arm telomere, across the centromere, to the p-arm telomere.
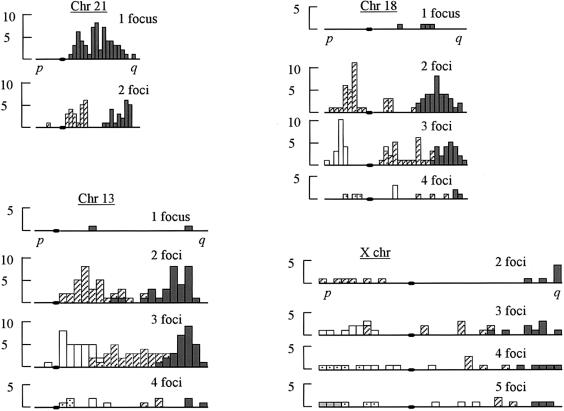
Chromosome 21, the smallest member of the human karyotype, had a mean of 1.23 foci (table 3), with a maximum of 2 foci per bivalent. In three cells, the chromosome 21 bivalent lacked an MLH1 focus, although foci were present on other bivalents. The distribution pattern of the foci along chromosome 21 is summarized in figure 2. A distally located focus (within 5% of the q-arm telomere) was not seen in any of the bivalents with 1 focus, nor were any bivalents found that had a highly proximal focus. In bivalents with 2 foci, three examples of a proximally located focus were found within 5% of the centromere.
Figure 2 .
Histograms showing the distributions of foci. Each chromosome is divided into 5% intervals (see “Material and Methods”), and the position of the centromere is indicated by a blackened oval on the X-axis. The positions of foci are shown separately (for bivalents with 1, 2, 3, etc., foci). For ease of presentation, the chromosome pairs are not shown to scale.
Chromosome 18 is a small submetacentric chromosome. A variable number of foci were recorded on this chromosome, with a mean of 2.36 per bivalent (table 3). Only one chromosome 18 bivalent was found that lacked a focus. When ⩾2 foci were present, one was generally positioned on the p arm (28/35 bivalents). One bivalent was identified with a focus positioned within 5% of the p-arm telomere, and two other bivalents had a focus within 5% of the q-arm telomere (fig. 2).
Chromosome 13 is a large acrocentric chromosome. All the bivalents screened carried ⩾1 focus (table 3). Only one bivalent had a focus on its p arm, the remainder being on the q arm. A mean of 2.5 foci were present on the chromosome, with a range of 1–4 per bivalent (table 3). The distribution pattern of the foci is summarized in figure 2. In both bivalents with 1 focus, the latter was positioned interstitially. In the remaining bivalents, only 2 had a focus within 5% of the q-arm telomere, and 11 were within 5% of the centromere (fig. 2).
The positions of foci were measured along a relatively small number of chromosome X bivalents, because of the problem of overlaps with other bivalents. However, in all cells examined, even when the chromosome X bivalent was eventually excluded from the analysis, there were foci along the chromosomal axis. Overall, the chromosome X bivalent had a mean of 3.22 foci (table 3). The minimum number of foci seen was 2, and the maximum was 5. In all the chromosomes X that were included in the analysis, both the p arm and the q arm had ⩾1 MLH1 focus. The positions of the MLH1 foci are summarized in figure 2. Of the 23 bivalents screened, 3 had a focus within 5% of the p-arm telomere, 4 had a focus within 5% of the q-arm telomere, and 1 had a focus within 5% of the centromere.
Chromosomes 18 and X both had a tendency toward larger interarm (i.e., across-the-centromere) distances between foci, compared to intra-arm distances (table 5).
Discussion
Timing of MLH1 Formation
We have found that MLH1 foci can first be detected at early zygotene in human fetal oocytes and that the number of these foci is relatively stable from late zygotene to pachytene but that they start to disappear at early diplotene. These characteristics distinguish the behavior of MLH1 foci in human oocytes from those in the mouse. Baker et al. (1996) similarly noted the presence of foci in mouse zygotene oocytes. However, they found that the numbers of foci were at a peak (65±12 per cell) at early pachytene and had subsided (31±2 per cell) by midpachytene. The number of foci further decreased at late pachytene, although some still remained in early-diplotene oocytes. A similar interspecies difference has also been described for male germ cells. Anderson et al. (1999) reported that MLH1 foci in mouse spermatocytes increased in number from early to midpachytene, before decreasing to 0 at late pachytene. In contrast, the number of MLH1 foci remains constant throughout pachytene in human spermatocytes (Barlow and Hultén 1998; C.T. and M.A.H., unpublished data). This interspecies disparity is intriguing and merits more-detailed investigation, since it may provide new insights into the mechanisms that control crossover numbers and distributions in mammalian germ cells.
Comparison of Numbers of MLH1 Foci in Male and Female Germ Cells
The study of MLH1 foci in human spermatocytes gave a mean of 50.6 foci per cell at pachytene (Barlow and Hultén 1998). Moreover, these foci were frequently located in the subterminal regions of the chromosomes, a distribution pattern that is in contrast to the generally more-interstitial location in oocytes (see fig. 1). Analyses of MLH1 foci enable direct demonstration of the pronounced intersex difference in both the numbers and distributions of meiotic crossovers in human germ cells, as suggested by indirect genetic-linkage studies (Broman et al. 1998). Our data indicate that the level of crossing-over in female germ cells is 1.4 times greater than that in male germ cells. This is in comparison to the recent human female germ cell:human male germ cell estimate of 1.6:1, from a study of genetic-map lengths (Broman et al. 1998). It is not yet clear whether crossover interference, a crucial factor in the determination of the numbers and distributions of crossovers, varies between human male meiosis and human female meiosis (e.g., see Broman and Weber 2000; Lynn et al. 2000). One possible explanation for the sex-related difference in recombination frequency is that the strength of interference is greater in spermatocytes, hence giving the reduced number of crossovers in spermatocytes, in comparison to that in oocytes. An alternative possibility can be envisaged from the observation that pachytene chromosomes in oocytes are approximately double the length of those in spermatocytes (Bojko 1983; Wallace and Hultén 1985)—that is, the female genome has a much longer physical platform for meiotic recombination than the male genome. An increased number of crossovers could therefore be accommodated along the longer SCs of female germ cells while the same physical intercrossover distance as in the male genome is retained. Comparison of interfocus distances on specific chromosomes from oocytes and spermatocytes should establish unequivocally whether there is any intersex variation in interference strength.
Distribution of MLH1 Foci along Chromosome Arms
Crossover (i.e., chiasma) positions are influenced not only by interference but also by preferential distribution (for review, see Jones 1984; Petes 2001). The latter implies that certain chromosomal segments are more favored for crossing-over than are others, a phenomenon that was first described in human spermatocytes by use of chiasma analysis (Hultén 1974) and that was subsequently elaborated by molecular-genetic characterization of recombination hotspots (for review, see Lichten and Goldman 1995; Jeffreys et al. 1999; Petes 2001). In the present article, the influence of interference is manifested by the large interfocus distances observed. However, it is also clear that interference is not the sole determinant of position, particularly in bivalents with fewer foci. First, the distance between adjacent foci decreased as the numbers of foci increased, and there is no reason to believe that the strength of interference varies with the number of recombination events. Second, the interfocus distance was generally larger across the centromere than within a chromosome arm. In many organisms, it has been noted that, close to the centromere, crossing-over generally occurs at a reduced rate (see Colombo and Jones 1997). One possible interpretation of the tendency toward larger interfocus distances over the centromere in chromosomes 18 and X is that it could be the result of the supplementation of interference, by an inhibitory effect of pericentromeric heterochromatin. However, full explanation of this phenomenon awaits a more complete understanding of the mechanism of interference.
The positions favored for crossing-over do not seem to be dependent simply on the DNA content of particular chromosomal regions. In human spermatocytes, for example, chiasma formation is favored in chromosomal segments that are adjacent to telomeres (Hultén 1974; Laurie and Hultén 1985). This distribution pattern is mirrored by MLH1 foci in pachytene spermatocytes (Barlow and Hultén 1998). In stark contrast, MLH1 foci rarely form in the distal regions of bivalents in pachytene oocytes (fig. 2). This considerable intersex difference in the placement of crossovers presumptively reflects control over their positions. Currently, it is not clear how such control may function.
Lamb et al. (1997) summarized the distribution of recombination events along chromosome 21, in bivalents with one or two crossovers. They divided the chromosome 21q arm into five equal segments and found that each segment had a similar likelihood of recombination when one crossover was present. When two crossovers were present, however, the proximal and distal segments had a higher rate of exchange. In our study, in bivalents with 1 MLH1 focus, the focus was very rarely in the final, distal segment of the chromosome arm. The proximal segment also showed a diminished potential for MLH1-focus formation. Thus, for bivalents with one exchange, our determination of crossover position is somewhat at odds with that estimated by Lamb et al. (1997). However, our observations in bivalents with 2 MLH1 foci coincide with those by Lamb et al. (1997), in finding (1) an enhanced number in the proximal and distal segments and (2) a relative dearth interstitially (fig. 2).
Estimated Genetic-Map Lengths
The human female genetic-map length was recently estimated as 4,435 cM (Broman et al. 1998). This study also reported the occurrence of significant interindividual variation between female genetic-map lengths. In eight women, autosomal-map lengths were found to have a range of 3,300–4,700 cM (Broman et al. 1998). We found a mean of 70.3 MLH1 foci per oocyte at pachytene. On the bases that each MLH1 focus represents one crossover and that one crossover is equal to a genetic distance of 50 cM, we then estimated a human female genetic-map length of 3,515 cM. The other three fetuses, from which very limited data were available (table 2), provided means of 95 foci (4,750 cM), 77.3 foci (3,815 cM), and 71.6 foci (3,580 cM) per cell. Overall, the estimates provided by indirect genetic-linkage studies and direct cytogenetic analyses are in reasonable agreement. This is all the more surprising when one considers the disparate nature of the germ cell populations examined. The cytogenetic approach uses a population of oocytes prior to the wave of atresia that eliminates >70% of germ cells during the late-second and third trimesters (Morita and Tilly 1999). In contrast, the genetic approach screens a highly selected population of adult germ cells (the survivors of atresia in both fetal and adult ovaries and of postzygotic development). Our genetic-map estimates correspond reasonably to those produced by the linkage approach. This conclusion encourages the use of our data to examine the relationship between meiotic recombination in fetal oocytes and chromosome nondisjunction in adult cells.
The data from the identification of MLH1 foci in specific chromosomes can be used to provide genetic-map estimates for these chromosomes, in a manner similar to that described above for the whole genome. These estimates are given in table 6, along with previously published figures obtained from genetic studies. Our estimated genetic length for chromosome 13 is consistent with the previously published information. However, the estimates for other chromosomes are generally smaller than expected. Since the overall genomic level of recombination was smaller in our material, it is unsurprising that this effect can also be seen for specific chromosomes. Possibly the most intriguing difference between the two methodologies was observed for chromosome 18: The genetic estimates indicate that chromosome 18 is more recombinogenic than chromosome 13 is, despite the smaller physical size of the former (85 Mb vs. 114 Mb, respectively; Morton 1991). In the pressent study, the two chromosomes have a similar genetic length, with chromosome 13 being slightly larger (table 6). It is also notable that genetic studies themselves may provide very disparate outcomes, as in the case of chromosome 21, for which recent estimates vary from 65 cM to 80.1 cM (Dib et al. 1996; Lamb et al. 1997; Broman et al. 1998; Lynn et al. 2000).
Table 6.
Estimates of Chromosome-Specific Genetic-Map Lengths in Human Female Germ Cells
Crossing-Over and Nondisjunction
As described in the “Introduction” section, there is evidence that the numbers and distributions of crossovers can influence the risk of chromosome nondisjunction in human oocytes. Thus, the lack of crossing-over and the placement of crossovers either very distally or very close to the centromere are associated with increased risk of nondisjunction. Interestingly, different chromosomes are affected to a different extent by each of these (Hassold and Hunt 2001). A reduction in recombination is associated with nondisjunction for all chromosomes; however, for chromosomes 18 and X, this would appear to mean failure of crossing-over, since there is no indication that the exact placement of the exchange correlates with the risk of nondisjunction. For chromosome 21, distal positioning of a single crossover increases the risk of malsegregation. Through our analyses of MLH1 foci, we can determine whether bivalents with “at-risk” configurations are actually seen in human fetal oocytes, and, if they are present, we can estimate their relative frequencies.
A few chromosomes (3/86 and 1/67, respectively) 21 and 18 lacked a focus; this apparent failure of recombination should result in these bivalents being achiasmate. We have therefore identified, in the fetal ovary, bivalents that, because they are presumptively achiasmate, will be at particular risk of nondisjunction when meiosis is completed in the adult ovary. No chromosome 21 bivalents were found that had a single, distally located MLH1 focus. Although this suspect configuration was not present, chromosome pairs with a highly proximal MLH1 focus (3/23 bivalents) were evident among those with 2 foci (fig. 2).
We did not find any instances of chromosome 13 (or chromosome X) without a focus, nor did we observe distal location of the focus in bivalents with a single focus—that is, two of the suspect configurations were not present in this sample. We did, however, identify bivalents with the third configuration—namely, a highly proximal MLH1 focus in a bivalent with 2 or 3 foci (11/59 chromosome 13 bivalents; fig. 2).
Overall, we have identified some of the configurations that are believed to increase the risk of nondisjunction, and we found that these configurations occur at low incidences in fetal oocytes. It is notable that the relative frequencies of these different types of configuration vary between the different chromosomes. This variability is consistent with the pattern, identified by genetic studies (Hassold and Hunt 2001), of interchromosome differences in susceptibility to recombination-mediated nondisjunction. The absence, among the two longer chromosomes examined here, of any bivalents that lack foci is puzzling in light of the evidence that achiasmate meiosis is important for malsegregation that affects chromosomes 13 and X (Hassold and Hunt 2001). One possible way to reconcile the present cytogenetic observations with the expectations of the genetic-linkage studies may be through the consideration of cells with abnormal synapsis. In addition to the cells with normal chromosome pairing that are described here, we found others with anomalous synapsis (fig. 1b). In human fetal oocytes, MLH1 foci are present only along synapsed chromosome regions and are not observed where synapsis either has yet to commence or has failed to occur. The only pachytene oocytes in which failure of MLH1-focus formation was observed for either chromosome 13 or chromosome X were those that were designated as “abnormal” because of partial or complete asynapsis of these chromosomes (fig. 1; authors' unpublished data). We suggest that these cells may be the source of the presumptively achiasmate bivalents that are implicated in nondisjunction of chromosomes 13 and X (Hassold and Hunt 2001; Thomas et al. 2001). (Note that occasional examples of similar synaptic errors that affect chromosomes 21 and 18—and, thereby, MLH1-focus formation—were also identified.) This proposal presumes that cells with asynapsis of some chromosome pairs are able to complete fetal development and contribute to the oocyte pool of the adult ovary. There is considerable evidence from yeast and from mammalian spermatocytes that cell checkpoints, which monitor chromosome synapsis and recombination, cause arrested development in cells with errors (for review, see Roeder and Bailis 2000). If our suggestion is valid, then we must further speculate that these checkpoints are, for some reason, less effective in human fetal oocytes: either some cells with pairing and recombination errors evade checkpoint arrest and continue development or such checkpoints are not active in female meiosis. There is some evidence to support this conjecture. Female mice homozygous for a null mutation of Mlh1 have only one or two crossovers per cell, and most chromosome pairs form univalents at metaphase I (Woods et al. 1999). Despite this failure of recombination in fetal germ cells, oocytes do survive and mature in the adult ovary. Other mouse-gene knockouts that cause disrupted meiotic chromosome synapsis—for example, Spo11 (Baudat et al. 2000; Romanienko and Camerini-Otero 2000) and Mei1 (Libby et al. 2002)—support the concept of sexual dimorphism as spermatogenesis arrests at pachytene, whereas some oocytes progress beyond this stage and survive to the adult-ovary stage. It remains to be determined whether a comparable tolerance to synaptic error in combination with failure of recombination is also present in human oocytes.
Acknowledgments
This study was supported by Wellbeing grant H1/98 and Wellcome Trust grant 061202/ZOOZ. We thank the Calthorpe Clinic, Birmingham, for the provision of tissue, and Prof. Christa Heyting, for kindly providing the anti-SCP3 antibody.
References
- Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13:336–342 [DOI] [PubMed] [Google Scholar]
- Barlow AL, Hultén MA (1998) Crossing over analysis at pachytene in man. Eur J Hum Genet 6:350–358 [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6:989–998 [DOI] [PubMed] [Google Scholar]
- Bojko M (1983) Human meiosis. VIII. Chromosome pairing and formation of the synaptonemal complex in oocytes. Carlsberg Res Commun 48:457–483 [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL (2000) Characterization of human crossover interference. Am J Hum Genet 66:1911–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Feingold E, Broman KW, Sherman SL (2000) Genome-wide variation in recombination in female meiosis: a risk factor for non-disjunction of chromosome 21. Hum Mol Genet 9:515–523 [DOI] [PubMed] [Google Scholar]
- Colombo PC, Jones GH (1997) Chiasma interference is blind to centromeres. Heredity 79:214–227 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, Pollard JW, Kolodner RD, Kucherlapati R (1996) Meiotic pachytene arrest in MLH1-deficient mice. Cell 85:1125–1134 [DOI] [PubMed] [Google Scholar]
- Fisher JM, Harvey JF, Morton NE, Jacobs PA (1995) Trisomy 18: studies of the parent and cell division of origin and the effect of aberrant recombination on nondisjunction. Am J Hum Genet 56:669–675 [PMC free article] [PubMed] [Google Scholar]
- Hartshorne GM, Barlow AL, Child TJ, Barlow DH, Hultén MA (1999) Immunocytogenetic detection of normal and abnormal oocytes in human fetal ovarian tissue in culture. Hum Reprod 14:172–182 [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2:280–291 [DOI] [PubMed] [Google Scholar]
- Hassold T, Merrill M, Adkins K, Freeman S, Sherman S (1995) Recombination and maternal age-dependent nondisjunction: molecular studies of trisomy 16. Am J Hum Genet 57:867–874 [PMC free article] [PubMed] [Google Scholar]
- Hultén M (1974) Chiasma distribution at diakinesis in the normal human male. Hereditas 76:55–78 [DOI] [PubMed] [Google Scholar]
- Hultén MA, Barlow AL, Tease C (2001) Meiotic studies in humans. In: Rooney DE (ed) Human cytogenetics: constitutional analysis, a practical approach, 3d ed. Oxford University Press, Oxford, pp 211–236 [Google Scholar]
- Jeffreys AJ, Barber R, Bois P, Buard J, Dubrova YE, Grant G, Hollies CRH, May CA, Neumann R, Panayi M, Ritchie AE, Shone AC, Signer E, Stead JDH, Tamaki K (1999) Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis 20:1665–1675 [DOI] [PubMed] [Google Scholar]
- Jones GH (1984) The control of chiasma distribution. Symp Soc Exp Biol 38:293–320 [PubMed] [Google Scholar]
- Lamb NE, Feingold E, Sherman SL (1997) Estimating meiotic exchange patterns from recombination data: an application to humans. Genetics 146:1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, Gu Y, Shen J, Saker D, May KM, Avramopoulos D, Petersen MB, Hallberg A, Mikkelsen M, Hassold TJ, Sherman SL (1996) Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet 14:400–405 [DOI] [PubMed] [Google Scholar]
- Laurie DA, Hultén MA (1985) Further studies on bivalent chiasma frequency in human males with normal karyotypes. Ann Hum Genet 49:189–201 [DOI] [PubMed] [Google Scholar]
- Libby BJ, de la Fuente R, O’Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC (2002) The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol 242:174–187 [DOI] [PubMed] [Google Scholar]
- Lichten M, Goldman ASH (1995) Meiotic recombination hotspots. Annu Rev Genet 29:423–444 [DOI] [PubMed] [Google Scholar]
- Lynn A, Kashuk C, Petersen MB, Bailey JA, Cox DR, Antonarakis SE, Chakravarti A (2000) Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res 10:1319–1332 [DOI] [PubMed] [Google Scholar]
- Morita Y, Tilly JL (1999) Oocyte apoptosis: like sand through an hourglass. Dev Biol 213:1–17 [DOI] [PubMed] [Google Scholar]
- Morton NE (1991) Parameters of the human genome. Proc Natl Acad Sci USA 88:7474–7476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaidis P, Petersen MB (1998) Origin and mechanisms of non-disjunction in human autosomal trisomies. Hum Reprod 13:313–319 [DOI] [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2:360–369 [DOI] [PubMed] [Google Scholar]
- Polkinghorne J (1989) Review of the guidance of the research use of fetuses and fetal material. Vol Cm762. Her Majesty’s Stationery Office, London [Google Scholar]
- Robinson WP, Bernasconi F, Mutirangura A, Ledbetter DH, Langlois S, Malcolm S, Morris MA, Schinzel AA (1993) Nondisjunction of chromosome 15: origin and recombination. Am J Hum Genet 53:740–751 [PMC free article] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM (2000) The pachytene checkpoint. Trends Genet 16:395–403 [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6:975–987 [DOI] [PubMed] [Google Scholar]
- Schalk JA, Dietrich AJ, Vink AC, Offenberg HH, van Aalderen M, Heyting C (1998) Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma 107:540–548 [DOI] [PubMed] [Google Scholar]
- Sherman SL, Petersen MB, Freeman SB, Hersey J, Pettay D, Taft L, Frantzen M, Mikkelsen M, Hassold TJ (1994) Non-disjunction of chromosome 21 in maternal meiosis. I. Evidence for a maternal age-dependent mechanism involving reduced recombination. Hum Mol Genet 3:1529–1535 [DOI] [PubMed] [Google Scholar]
- Speed RM (1985) The prophase I stages in human foetal oocytes studied by light and electron microscope. Hum Genet 69:69–75 [DOI] [PubMed] [Google Scholar]
- ——— (1988) The possible role of meiotic pairing anomalies in the atresia of human fetal oocytes. Hum Genet 78:260–266 [DOI] [PubMed] [Google Scholar]
- Thomas NM, Ennis S, Sharp AJ, Durkie M, Hassold TJ, Collins AR, Jacobs PA (2001) Maternal sex chromosome non-disjunction: evidence for X chromosome-specific factors. Hum Mol Genet 10:243–250 [DOI] [PubMed] [Google Scholar]
- Wallace BMN, Hultén MA (1985) Meiotic chromosome pairing in the normal human female. Ann Hum Genet 49:215–226 [DOI] [PubMed] [Google Scholar]
- Woods LM, Hodges CA, Baart E, Baker SM, Liskay M, Hunt PA (1999) Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J Cell Biol 145:1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]