Abstract
A total of 8,859 DNA sequences encompassing ∼1.9 million base pairs of the chimpanzee genome were sequenced and compared to corresponding human DNA sequences. Although the average sequence difference is low (1.24%), the extent of changes is markedly different among sites and types of substitutions. Whereas ∼15% of all CpG sites have experienced changes between humans and chimpanzees, owing to a 23-fold excess of transitions and a 7-fold excess of transversions, substitutions at other sites vary in frequency, between 0.1% and 0.5%. If the nucleotide diversity in the common ancestral species of humans and chimpanzees is assumed to have been about fourfold higher than in contemporary humans, all possible comparisons between autosomes and X and Y chromosomes result in estimates of the ratio between male and female mutation rates of ∼3. Thus, the relative time spent in the male and female germlines may be a major determinant of the overall accumulation of nucleotide substitutions. However, since the extent of divergence differs significantly among autosomes, additional unknown factors must also influence the accumulation of substitutions in the human genome.
Introduction
Chimpanzees and humans are estimated to have shared a common ancestor only 4.6–6.2 million years ago (Chen and Li 2001). Thus, for a study of the accumulation of nucleotide substitutions in the human genome, chimpanzees offer the most relevant nonhuman comparison, for at least two reasons. First, the close relationship between the sequences compared minimizes the risk that multiple substitutions at the same sites will obscure the results. Second, because of the short divergence time, processes that may influence the accumulation of DNA sequence changes—for example, regional differences in recombination and mutation rates—can be assumed not to have changed drastically since the two species shared a common ancestor.
Early comparative studies of the human and chimpanzee genomes (King and Wilson 1975; Sibley and Ahlquist 1984; Goodman et al. 1990; Bailey et al. 1991) established that the extent of DNA sequence difference is on the order of 1.6%. Since that time, little additional knowledge about the pattern of divergence has accumulated. Only recently, a study of 53 intergenic autosomal regions in the chimpanzee genome (Chen and Li 2001) indicated that the extent of divergence is only 1.24%. However, different regions of the human genome differ in base composition (Bernardi 1995) and in extent of divergence from the chimpanzee (Dorit et al. 1995; Glusman et al. 2000). Therefore, sampling of a large number of DNA sequences is required to gain an overview of the extent and pattern of divergence between the chimpanzee and human genomes.
To better understand how DNA sequences have changed during recent human evolution, we have determined ∼3 Mb from >10,000 regions in the chimpanzee genome. About two thirds could be unambiguously aligned to DNA sequences in humans. The results reveal complex patterns of accumulation of DNA sequence differences that are distinct with regard both to various classes of substitutions and to different chromosomes.
Material and Methods
DNA Sequence Generation
Total genomic DNA from an individual male chimpanzee was mechanically fragmented using a nebulizer. Fragments were separated on a 1% agarose gel, and the size fraction of 300–600 bp was extracted from the gel with the QIAquick Gel Extraction Kit (Qiagen). Endfilling was performed for 30 min at 20°C with T4-Polymerase (3 μl, 4 U/μl) and Klenow-Enzyme (6 μl, 2 U/μl) in a buffer containing 5 mM MgSO4; 50 mM Tris/HCl, pH 7.5; 0.1 mM DTT; and 0.03 mM dNTPs. Fragments were ligated into dephosphorylated, SmaI-cleaved pUC18 (Pharmacia Biotech).
Escherichia coli (XL1blue; Stratagene) were transformed using electroporation, and colonies were picked. Recombinant plasmids were isolated from 1.2 ml Luria broth overnight cultures through use of the QIAprep 96 Turbo BioRobot Kit (Qiagen) on a Biorobot 9600 (Qiagen). Plasmid DNA (100–500 ng) was used as template in sequencing reactions with 10 pmol of the sequencing primer M13 for (5′-GTA AAA CGA CGG CCA GT-3′) and M13 rev (5′-CAG GAA ACA GCT ATG AC-3′), using the ABI Prism BigDye Cycle Sequencing Ready Reaction Kit (Applied Biosystems). After an initial denaturation at 96°C for 2.5 min, cycle sequencing was performed under the following conditions: 35 cycles of 96°C for 20 s, 55°C (M13 for) or 52°C (M13 rev) for 30 s, and 60°C for 4 min, on a PTC 200/PTC 225 Thermocycler (MJ Research). Sequencing reactions were precipitated with isopropanol, were dissolved in 25 μl H2O (Merck), and were analyzed on an ABI 3700 DNA sequencer (Applied Biosystems).
Overlapping end sequences from individual clones were assembled with the Phred/Phrap package (Genome Software Development Page) and were viewed with Consed (Gordon et al. 1998). Low-quality positions (i.e., Phred value <30) were masked and excluded from further analysis. To achieve an overall high sequence quality of the analyzed DNA fragments, stretches of four or more consecutive masked positions were removed from the sequence, and the resulting sequence parts were analyzed independently. Sequences shorter than 50 bp were discarded. The remaining sequences had a GC content of 47%, ∼6% above the genome average, because of the fragmentation method used. Repetitive elements were identified in the sequences through use of the program RepeatMasker, version 05/05/99 (Repeat Masker Server Home Page).
DNA Sequence Comparisons
Human counterparts of the chimpanzee sequences were identified by comparing the sequences to the University of California, Santa Cruz (UCSC), draft version of the human genome (freeze August 6, 2001), through use of the program Blat (Human Genome Working Draft Web site). Since this program tends to produce artificially prolonged alignments, blocks of contiguous aligned bases without gaps in either of the species were individually used as alignment starts. The score of the resulting alignments was calculated using the following parameters: match = +1; mismatch = −2; mismatch to masked position = 0; gap opening = −4; and gap extension = −3. If any of the shorter alignments had a higher alignment score than the original alignment, it was used for subsequent analyses.
Analysis was restricted to alignments with ⩾60 compared bases. Chimpanzee clones carrying ⩾60 consecutive unaligned bases in addition to an aligned segment were judged to be potentially chimeric. To identify the human counterpart of the unaligned sequence part, lower-ranking alignments were analyzed. In cases where alignments of two sequences of a potentially chimeric clone shared >10 identical bases, the entire clone was excluded from further analysis. To further avoid sequences that display multiple matches to the human genome, the alignment scores were compared with the scores of the second-best alignment in each case. Only when the difference between those scores was ⩾10 were the alignments used for further analysis.
To minimize the influence of potentially undetected nonorthologous comparisons in the sequence comparisons, we excluded the 2.5% of alignments with the highest extent of sequence differences from the analysis. This corresponds to a threshold of 96% in the sequence similarity. When lowered to 95%, 1.7% of all alignments are excluded, and the mean chimpanzee-human sequence difference changes from 1.24% to 1.27%.
Alignment start and end points were compared with the start and end points of exons and introns from RefSeq genes and mRNAs mapped to the draft version of the human genome (UCSC Genome Browser Gateway Web site), in order to identify the relation of alignments to known genes.
Alignments between DNA sequences of humans and chimpanzees were analyzed for number and type of sequence differences, mean GC content, and CpG content, through use of the Perl script mutanalyzer.pm (script available upon request). Per-site frequencies (pn1↔n2) of the individual classes of DNA sequence differences were calculated as: pn1↔n2=Nn1↔n2/(N1+N2), where Nn1↔n2 is the number of observed differences between the two bases n1 and n2, and N1 and N2 are the numbers of positions in the sequence that are n1 or n2, respectively. For insertions/deletions, every contiguous stretch of insertions/deletions in the alignment was treated as a single event, and the per-site frequency was assessed by dividing the number of insertion/deletion events by the total number of compared bases.
Human Polymorphism Data
Information about human single nucleotide polymorphisms (SNPs) was retrieved from the dbSNP Home Page (Build 96). SNPs that were not assigned to a chromosome, as well as SNPs for which the type of sequence change could not be determined unambiguously, were excluded.
Statistical Analyses
As a measure of homogeneity of DNA sequence differences among chromosomes, we compared the variation within the chromosomes to the variation between the chromosomes by means of a permutation test. Since the X and the Y chromosome are suspected to each evolve in a unique manner, the sex chromosomes were excluded from this analysis. First, we computed the χ2 value for the matches and mismatches among the different autosomes. We then shuffled the sequences randomly, keeping the same number of sequences on each autosome, and computed the χ2 value for this rearrangement. This procedure was repeated 1,000 times, and the resulting χ2 values were compared to the value obtained from the original sequence arrangement.
Under the hypothesis that the ratio rYX between the average substitution rates on the Y and the X chromosomes can be explained by the ratio α between substitution rates in the male and the female germlines, we have α=2rYX/(3-rYX). We estimate rYX by RYX=[(Y/nY)-πY]/[(X/nX)-πX], where Y and X are the numbers of substitutions observed on the Y and X chromosomes, nY and nX are the total lengths of the sequences compared on the Y and X chromosomes, and πY and πX are the mean nucleotide diversity in the ancestral species common to humans and chimpanzees (Li 1977).
We apply the “delta method” (Rice 1995) to approximate the SD, SR, of R by
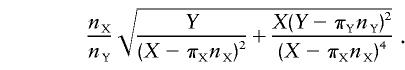 |
Using normal approximation for R, we get the 95% CI R±1.96SR for rYX. With α=2rYX/(3-rYX), we can use the estimate and the CI for rYX to calculate an estimate and a CI for α. We apply the same procedure with the ratio rYA of substitution rates on the Y chromosome and the autosomes and with the ratio rAX of substitution rates on the autosomes and the X chromosome, using α=rYA/(2-rYA) and α=(4rAX-3)/(3-2rAX), respectively.
Results
DNA Sequence Determination
Genomic DNA isolated from the blood of a male chimpanzee (Pan troglodytes) was sheared, and fragments 300–600 bp in length were isolated by agarose gel electrophoresis and were cloned into the plasmid vector pUC18. After transformation into E. coli, the inserts of randomly selected clones were sequenced from both ends. In total, we analyzed 10,549 DNA sequences, ranging in length from 60 bp to 950 bp. The total amount of DNA sequence analyzed was 3,000,286 bp.
Comparison to the Human Genome
The chimpanzee DNA sequences were compared to the human genome in the public domain through use of the program Blat. Twenty-eight percent of the total amount of sequence was excluded from the analysis, since the entire sequence, or parts of it, displayed more than one match in the human genome that was not due to known families of repeated sequences. For 7% of the chimpanzee sequences, no region with similarity could be detected in the human genome.
In total, 8,859 sequence pairs encompassing 1,944,162 nucleotides in the chimpanzee genome remained for analysis; 67% were located in intergenic regions, 31% in introns, and 2% in exons.
Table 1 shows the absolute and relative numbers of observed sequence differences between the two species. In total, 24,165 substitutional differences were seen, resulting in a genomewide average DNA sequence difference between humans and chimpanzees of 1.24%. Among these differences, a 2.4-fold excess of transitions over transversions is seen. Notably, transitions at CpG sites account for 28% of all substitutional differences, whereas CpG dinucleotides constitute only 3.5% of the analyzed sequences. Among the transversions, A↔C/G↔T transversions are the most abundant class. However, per positions where a given substitution can occur, G↔C transversions are found to be 1.2-fold more common than A↔C/G↔T transversions and 1.9-fold more common than A↔T transversions.
Table 1.
Differences between the Chimpanzee and Human DNA Sequences
| Difference Type | Observations | % ofTotal | FrequencyPer Position |
| Transitions: | |||
| Total | 16,990 | 70.3 | .0087 |
| At CpG sites | 6,770 | 28.0 | .1218 |
| Transversions: | |||
| G↔C | 2,185 | 9.0 | .0023 |
| A↔C/G↔T | 3,734 | 15.5 | .0019 |
| A↔T | 1,256 | 5.2 | .0012 |
| Insertions/deletions | 2,407 | … | .0012 |
In total, 2,407 insertions/deletions ranging in size from 1 bp to 65 bp were detected. Since 1,223 apparent deletions are observed among the chimpanzee DNA sequences, whereas 1,184 are observed among the human DNA sequences, insertions/deletions seem to occur at equal rates in the two species.
Analysis by Chromosome
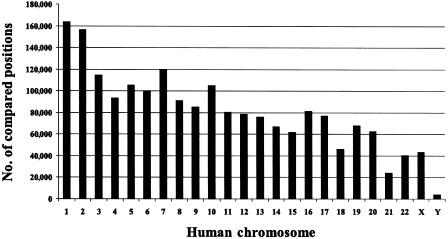
A total of 8,859 human-chimpanzee sequence pairs could be assigned to a human chromosome. The amount of sequence compared per chromosome ranges from 163 kb for human chromosome 1 to 3.9 kb for the nonrecombining portion of the human Y chromosome (fig. 1). The low representation of the Y chromosome is due to its small size, its hemizygous state in males, and its high repeat content.
Figure 1.
Histogram showing the number of DNA sequence positions compared per human chromosome
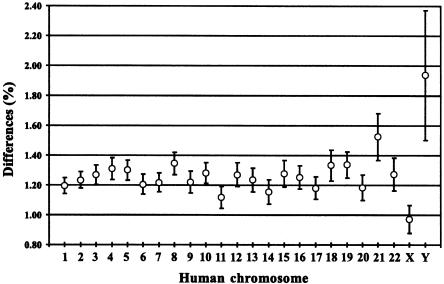
In figure 2, the mean DNA sequence difference between humans and chimpanzees is shown for the 24 human chromosomes. The X chromosome has accumulated the least amount of differences (1.0%), whereas the Y chromosome has diverged the most (1.9%). The divergence of the autosomes falls between those of the two sex chromosomes, with an average of 1.2%. Interestingly, the amount of differences that the individual autosomes have accumulated differs, ranging from a minimum of 1.1%, for human chromosome 11, to a maximum of 1.5%, for human chromosome 21. A permutation test shows that the autosomes differ significantly in the amount of differences accumulated (P<.001)—that is, the χ2 values from 1,000 random permutations are all smaller than the observed χ2 value. This is also seen when the analysis is restricted to sequences located in introns and in intergenic regions.
Figure 2.
Mean DNA sequence differences between humans and chimpanzees by human chromosome. Bars indicate 95% CIs.
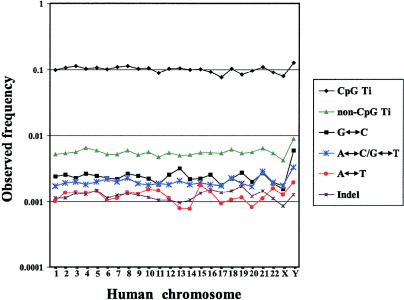
When the various types of DNA sequence differences are regarded separately for each chromosome (fig. 3), transitions at CpG sites dominate quantitatively, with a frequency of ∼12%. All other types of sequence differences have frequencies <1%. Among these, transitions at non-CpG sites are most abundant, followed by G↔C transversions and A↔C/G↔T transversions. A↔T transversions and insertions/deletions are the rarest differences, with mean frequencies of ∼0.12%. All types of substitutional differences, as well as insertions/deletions, have nonuniform distributions among the autosomes (P<.05 for each type).
Figure 3.
Frequency distribution of DNA sequence differences between chimpanzees and humans, for each human chromosome. Ti = transitions.
Discussion
Substitution Patterns
The comparison between 1.9 Mb of genomic DNA derived from >8,000 individual DNA sequences of the chimpanzee genome and the corresponding DNA sequences in the human genome reveals a mean sequence difference of 1.24%. This is in agreement with a recent study (Chen and Li 2001) in which the same sequence difference was seen when a total of 24 kb from 53 autosomal regions were compared.
Among the 24,165 substitutional differences seen, 28% were transitions at CpG dinucleotides, although such sites comprise only 3.5% of the entire sequence. Thus, a nucleotide has a 23-fold higher probability of carrying a transitional difference in the context of a CpG dinucleotide than in another sequence context (table 2). This is generally thought to be caused by the fact that many cytosines in CpG dinucleotides are methylated. When deaminated, methylated cytosines yield mismatched thymine residues that are often incorrectly repaired, resulting in a C→T transition (reviewed by Holliday and Grigg 1993). Thus, the rate at which two DNA sequences have diverged between humans and chimpanzees depends largely on the number of CpG dinucleotides in the sequence and, presumably, on their methylation status in the germline.
Table 2.
Nucleotide Differences per Position at CpG- and Non-CpG Sites
|
Frequency at |
|||
| NucleotideDifference | Non-CpGSites | CpGSites | Overrepresentation at CpG Sitesa |
| Transitions | .0054 | .1218 | 22.5 |
| G↔C | .0017 | .0133 | 7.8 |
| A↔C/G↔T | .0016 | .0130 | 6.6 |
Values are ratios of the frequencies at CpG and non-CpG sites.
Among the three types of transversional differences, G↔C transversions are the most frequent per position, whereas A↔T transversions are the least frequent (table 1). Interestingly, transversions are observed at CpG dinucleotides at a significantly higher frequency than they are at CpG-unrelated sites (χ2 test: P<10-10). This excess of transversions at CpG sites is 7.8-fold for G↔C transversions and 6.6-fold for A↔C/G↔T transversions (table 2), but they do not differ significantly from each other (P=.7). Similarly, when the frequencies of transversions at non-CpG sites are analyzed, no significant difference between G↔C and A↔C/G↔T transversions is seen (P=.4), although both of these transversions remain more frequent than A↔T transversions (P<10-10 in both cases) (table 2; fig. 3). The excess of transversions at CpG sites may be related to oxidative damage, since guanosine residues have been shown to be more susceptible to transversions than are other bases, when exposed to oxygen radicals in vitro, particularly in the CpG context (Agnez-Lima et al. 2001).
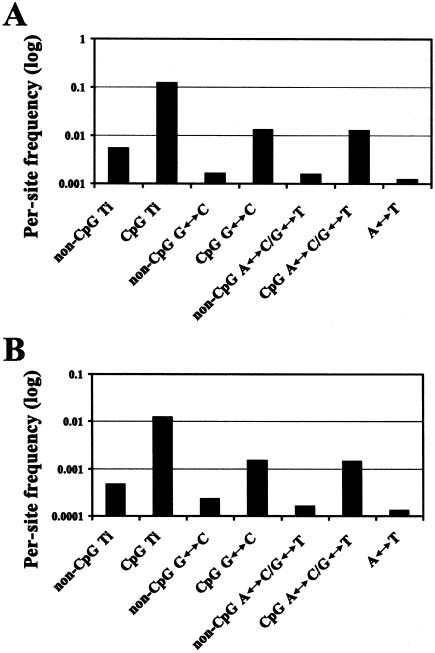
When the relative amounts of different substitutional types observed between the chimpanzee and the human are compared with the same substitutional types observed as polymorphisms among humans, they are found to be very similar (fig. 4). In general, each substitutional type is found among humans at approximately a tenth of the frequency found in the comparisons between humans and chimpanzees. This includes differences at CpG sites as well as non-CpG sites. Thus, the mutational and selective mechanisms that underlie the DNA sequence differences seen between humans and chimpanzees operate at approximately the same relative strengths within humans as well.
Figure 4.
Genomewide average frequencies for various nucleotide differences between chimpanzees and humans (A) and among humans (B). Ti = transitions.
Patterns of Sequence Differences among Chromosomes
The observation that the autosomes differ in their average extent of nucleotide divergence (fig. 2) is intriguing. Regional differences in divergence rates have been seen previously for expressed genes (Casane et al. 1997; Matassi et al. 1999). Furthermore, it was recently shown that when gene sequences are compared between humans and mice, as well as between mice and rats, genes located on the same human or rodent chromosome have similar divergence rates (Lercher et al. 2001). At present, the features of the genome that determine these differences have not been identified. However, it is interesting that the distribution of differences between the chimpanzee and human genomes (fig. 2) is similar (P<.01, excluding the sex chromosomes) to the distribution of differences at silent sites in coding sequences between human and mouse (Lercher et al. 2001). For example, among the five human autosomes that show the highest divergence from the chimpanzee (chromosomes 4, 8, 18, 19, and 21), three are among the five human autosomes (chromosomes 4, 13, 16, 19, and 21) that show the highest divergence at silent sites in human and mouse genes. Similarly, of the three chromosomes that have changed the least between humans and chimpanzees (chromosomes 11, 14, and 17), two are among the three chromosomes (14, 15, and 17) that show the lowest divergence at silent sites in human and mouse genes. Consequently, whatever factors determine the relative rates of divergence at the chromosomal scale are conserved over long time periods.
Higher Mutation Rate in Males
Two lines of evidence support the existence of a higher mutation rate in the male than in the female germline. First, the majority of point mutations causing certain diseases occur in the male germline and show a positive correlation with the age of the father (reviewed by Crow 2000). Second, DNA sequences located on chromosomes that spend more time in the male germline diverge faster between species than do sequences located on chromosomes spending less time in the male germline (Miyata et al. 1987; Ellegren and Fridolfsson 1997; Hurst and Ellegren 1998). Accordingly, it has been pointed out that the factor by which the male germline evolves faster than the female germline (termed “α”) can be inferred from the divergence rates of DNA sequences on the autosomes, the X chromosome, and the Y chromosome (Miyata et al. 1987). The current data set, supplemented with data from the literature (Anagnostopoulos et al. 1999; Kaessmann et al. 1999; Bohossian et al. 2000), allows us to calculate α from all three possible chimpanzee-human comparisons—that is, Y and X chromosomal comparisons (Y/X), Y chromosomal and autosomal comparisons (Y/A), and autosomal and X chromosomal comparisons (A/X). If differences between mutation rates in the male and female germlines are the dominant factor influencing the rate of DNA sequence evolution, we expect the estimates of α from these three comparisons to be similar.
Table 3 shows that the Y/X comparison results in an α value of 1.9, the Y/A comparison in an α value of 1.3, and the A/X comparison in an α value of 5.4. However, the average divergence between DNA sequences of two species is the sum of the divergence generated since the separation of the species and the amount of nucleotide diversity in the ancestral population at the time of speciation (Li 1977). Since autosomes and X and Y chromosomes differ substantially in their amount of nucleotide diversity (Sachidanandam et al. 2001) and since this is not correlated with the rate of divergence of these chromosomes between species, this might have substantial influence on the estimation of α values in closely related species. If the divergences used to calculate α values are corrected for the nucleotide diversity in the common ancestral species by subtraction of the nucleotide diversity observed for autosomes, the X chromosome, and the Y chromosome in contemporary humans (Sachidanandam et al. 2001), α values remain significantly different from each other (table 3). This would indicate that the rate of DNA sequence change would depend on factors in addition to the time spent in the male and female germlines. For example, it is possible that the X chromosome may have a specifically reduced mutation rate that may have been selected to compensate for its hemizygous state in males (McVean and Hurst 1997). However, it is not unreasonable to assume that the nucleotide diversity in the common ancestor of humans and chimpanzees was higher than in contemporary humans. For example, Chen and Li (2001) estimated the effective population size of the common ancestor of humans and chimpanzees to be five to nine times larger than in humans. Furthermore, the extent of polymorphism is higher in all the great ape species than in humans (Kaessmann et al. 2001). If the nucleotide diversity in the ancestral population is assumed to have been four times higher than in contemporary humans, α values for the Y/X, Y/A, and A/X comparisons are 2.8, 2.6, and 3.2, respectively. Since these values do not differ significantly from each other (table 3), this is compatible with the idea that the time DNA sequences spend in the male and female germlines determines their overall evolutionary rate. Interestingly, this could reflect a difference in numbers of genome replications coupled to cell divisions per generation in males and females. However, it is noteworthy that the differences in evolutionary rates among autosomes indicate that some hitherto-unknown large-scale factor or factors, in addition to sex-specific differences in substitution rates, influence the accumulation of substitutions in the human genome. The complete sequencing of the chimpanzee genome would be a major step towards unraveling these factors.
Table 3.
Estimation of the Male-to-Female Mutation Rate Ratio (α)
| ChromosomeComparisonand π Correctiona | R | α (95% CI) |
| Y/X | 1.48 ± .05 | 1.9 (1.7–2.2) |
| 1×π | 1.54 ± .06 | 2.1 (1.8–2.4) |
| 4×π | 1.76 ± .07 | 2.8 (2.3–3.4) |
| Y/A | 1.14 ± .03 | 1.3 (1.2–1.5) |
| 1×π | 1.20 ± .03 | 1.5 (1.3–1.7) |
| 4×π | 1.56 ± .05 | 2.6 (2.2–3.2) |
| A/X | 1.30 ± .03 | 5.4 (3.7–8.6) |
| 1×π | 1.27 ± .03 | 4.8 (3.3–7.6) |
| 4×π | 1.21 ± .04 | 3.2 (2.2–4.9) |
X-chromosome supplementary data are from Anagnostopoulos et al. (1999) (38,129 bp; 531 substitutions), Kaessmann et al. (1999) (13,960 bp; 97 substitutions), and the authors' unpublished data (87,710 bp from the region around the gene ZFX; 710 substitutions). Y-chromosome supplementary data are from Anagnostopoulos et al. (1999) (4,650 bp; 62 substitutions) and the authors' unpublished data (89,602 bp from the region around the gene ZFY; 1,262 substitutions). Sex-chromosome DNA sequences were determined from regions outside the pseudoautosomal regions. “1×π” and “4×π” represent multiples of the mean nucleotide diversity (π) of contemporary humans for autosomes and for X and Y chromosomes, used to correct for polymorphism present in the common ancestor of humans and chimpanzees.
Acknowledgments
We thank two anonymous reviewers for helpful comments on the manuscript (in particular, the suggestion to correct α values for ancestral diversity), Linda Vigilant for critical reading of the manuscript, Birgit Nickel and Michaela Winkler for technical assistance, Achim Radtke for database support, the Primate Foundation of Arizona for chimpanzee genomic DNA, and the Max Planck Society and the Bundesministerium für Bildung und Forschung for financial support.
Electronic-Database Information
The URLs for data in this article are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- Genome Software Development Page, http://www.phrap.org/
- Human Genome Working Draft, http://genome.ucsc.edu/
- Repeat Masker Server Home Page, http://repeatmasker.genome.washington.edu/
- UCSC Genome Browser Gateway, http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg10
References
- Agnez-Lima LF, Napolitano RL, Fuchs RP, Mascio PD, Muotri AR, Menck CF (2001) DNA repair and sequence context affect (1)O(2)-induced mutagenesis in bacteria. Nucleic Acids Res 29:2899–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos T, Green PM, Rowley G, Lewis CM, Giannelli F (1999) DNA variation in a 5-Mb region of the X chromosome and estimates of sex-specific/type-specific mutation rates. Am J Hum Genet 64:508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey WJ, Fitch DH, Tagle DA, Czelusniak J, Slightom JL, Goodman M (1991) Molecular evolution of the psi eta-globin gene locus: gibbon phylogeny and the hominoid slowdown. Mol Biol Evol 8:155–184 [DOI] [PubMed] [Google Scholar]
- Bernardi G (1995) The human genome: organization and evolutionary history. Annu Rev Genet 29:445–476 [DOI] [PubMed] [Google Scholar]
- Bohossian HB, Skaletsky H, Page DC (2000) Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406:622–625 [DOI] [PubMed] [Google Scholar]
- Casane D, Boissinot S, Chang BH, Shimmin LC, Li W (1997) Mutation pattern variation among regions of the primate genome. J Mol Evol 45:216–226 [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH (2001) Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 68:444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF (2000) The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet 1:40–47 [DOI] [PubMed] [Google Scholar]
- Dorit RL, Akashi H, Gilbert W (1995) Absence of polymorphism at the ZFY locus on the human Y chromosome. Science 268:1183–1185 [DOI] [PubMed] [Google Scholar]
- Ellegren H, Fridolfsson AK (1997) Male-driven evolution of DNA sequences in birds. Nat Genet 17:182–184 [DOI] [PubMed] [Google Scholar]
- Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, Demaille J, Lancet D (2000) Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics 63:227–245 [DOI] [PubMed] [Google Scholar]
- Goodman M, Tagle DA, Fitch DH, Bailey W, Czelusniak J, Koop BF, Benson P, Slightom JL (1990) Primate evolution at the DNA level and a classification of hominoids. J Mol Evol 30:260–266 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- Holliday R, Grigg GW (1993) DNA methylation and mutation. Mutat Res 285:61–67 [DOI] [PubMed] [Google Scholar]
- Hurst LD, Ellegren H (1998) Sex biases in the mutation rate. Trends Genet 14:446–452 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Pääbo S (1999) DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet 22:78–81 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S (2001) Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet 27:155–156 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188:107–116 [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Williams EJ, Hurst LD (2001) Local similarity in evolutionary rates extends over whole chromosomes in human-rodent and mouse-rat comparisons: implications for understanding the mechanistic basis of the male mutation bias. Mol Biol Evol 18:2032–2039 [DOI] [PubMed] [Google Scholar]
- Li WH (1977) Distribution of nucleotide differences between two randomly chosen cistrons in a finite population. Genetics 85:331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi G, Sharp PM, Gautier C (1999) Chromosomal location effects on gene sequence evolution in mammals. Curr Biol 9:786–791 [DOI] [PubMed] [Google Scholar]
- McVean GT, Hurst LD (1997) Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature 386:388–392 [DOI] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T (1987) Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol 52:863–867 [DOI] [PubMed] [Google Scholar]
- Rice JA (1995) Mathematical statistics and data analysis. Wadsworth Publishing, Belmont, CA [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, et al (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE (1984) The phylogeny of the hominoid primates, as indicated by DNA-DNA hybridization. J Mol Evol 20:2–15 [DOI] [PubMed] [Google Scholar]