Abstract
Phosphatidylinositol 4,5 bisphosphate (PIP2) is a key lipid messenger for regulation of cell migration. PIP2 modulates many effectors, but the specificity of PIP2 signalling can be defined by interactions of PIP2-generating enzymes with PIP2 effectors. Here, we show that type Iγ phosphatidylinositol 4-phosphate 5-kinase (PIPKIγ) interacts with the cytoskeleton regulator, IQGAP1, and modulates IQGAP1 function in migration. We reveal that PIPKIγ is required for IQGAP1 recruitment to the leading edge membrane in response to integrin or growth factor receptor activation. Moreover, IQGAP1 is a PIP2 effector that directly binds PIP2 through a polybasic motif and PIP2 binding activates IQGAP1, facilitating actin polymerization. IQGAP1 mutants that lack PIPKIγ or PIP2 binding lose the ability to control directional cell migration. Collectively, these data reveal a synergy between PIPKIγ and IQGAP1 in the control of cell migration.
Keywords: cell migration, IQGAP1, PIP2, PIPKIγ, polybasic motif
Introduction
Cell migration is a highly orchestrated, multistep process requiring the establishment of polarity, the regulation of cytoskeleton dynamics and spatiotemporal signalling (Ridley et al, 2003; Parsons et al, 2010). Cell migration is initiated in response to extracellular stimuli, such as cytokines and signals from the extracellular matrix (ECM). These extracellular signals activate intracellular signalling cascades that promote changes in the cytoskeleton. A diverse array of proteins are implicated in these processes, but scaffold proteins that integrate signals from multiple structural and signalling molecules play pivotal roles in transmitting cellular information (Rodriguez et al, 2003; Good et al, 2011). Previous work has focussed on how scaffold proteins coordinate different signals. However, the exact mechanism of how scaffold proteins themselves are targeted and activated remains largely unknown.
IQ motif containing GTPase activating protein 1 (IQGAP1) is a multidomain protein that regulates cytoskeletal dynamics, proliferation, adherens junction integrity and vesicular trafficking, by serving as a scaffold for key signals (Brown and Sacks, 2006; Brandt and Grosse, 2007; Osman, 2010). IQGAP1 targets to the leading edge, where it promotes actin polymerization through Rac1 and Cdc42 and their effectors, such as N-WASP and Dia1 (Ho et al, 1999; Li et al, 1999; Brown and Sacks, 2006; Brandt et al, 2007; Le Clainche et al, 2007). IQGAP1 also controls microtubule (MT) behaviour. IQGAP1 interacts with MT plus end regulators, CLIP-170 and adenomatous polyposis coli (APC), and recruits MTs to the leading edge membrane (Fukata et al, 2002; Watanabe et al, 2004). By targeting MTs to the leading edge, IQGAP1 is believed to facilitate the polarized trafficking of protein to the migrating front (Watanabe et al, 2005; Osman, 2010). Yet, how IQGAP1 interacts with the leading edge membrane is largely undefined. A recent study has shown that phosphatidylinositol 4,5 bisphosphate (PIP2)-dependent microdomains are required for the recruitment of MTs to the plasma membrane (PM), and Cdc42, N-WASP and IQGAP1 are also required in this process (Golub and Caroni, 2005). However, the exact role for PIP2 in IQGAP1 regulation of the cytoskeleton at the PM is unknown.
At a molecular level, IQGAP1 is kept inactive through an autoinhibitory interaction between the GRD domain and RGCT domain (Brandt and Grosse, 2007). This autoinhibition can be relieved by RhoGTPase binding to the GRD domain or phosphorylation on Ser1443 to activate IQGAP1 (Grohmanova et al, 2004; Li et al, 2005). In agreement with this model of activation, a mutant IQGAP1, defective in RhoGTPase binding on the GRD domain, induces multiple leading edges (Fukata et al, 2002) and a phosphomimetic variant of IQGAP1 on Ser1443 stimulates neurite outgrowth (Li et al, 2005).
PIP2 comprises ∼1% of membrane phospholipids and is the most abundant phosphoinositide species at the PM. Besides serving as a precursor for other lipid messengers, PIP2 exerts direct signalling roles by interacting with protein targets (Anderson et al, 1999; Heck et al, 2007). Though PIP2 binding is often achieved by defined modules on proteins, including C2, pleckstrin homology (PH) and band 4.1/ezrin/radixin/moesin (FERM) domains, many PIP2-interacting proteins lack canonical modules and instead contain clusters of basic amino acids, known as polybasic motifs (PBMs) that bind PIP2 (McLaughlin et al, 2002). The interaction of PBMs with phosphoinositides is largely mediated by the positively charged residues in the PBM that interact with the phosphate head group. Therefore, these interactions in some cases can be promiscuous for phosphoinositides (McLaughlin and Murray, 2005). Recent advances in proteomic analyses have identified hundreds of putative PIP2 binding proteins, but most of them do not contain canonical modules (Catimel et al, 2008; Dixon et al, 2011), and thus many PBMs or atypical phosphoinositide binding motifs remain to be characterized.
PIP2 modulates the activity and targeting of cytoskeleton regulatory proteins, controlling cytoskeletal dynamics and, ultimately, migration (Yin and Janmey, 2003; Zhang et al, 2012). Although the roles for PIP2 in cytoskeleton regulation are extensively studied, the roles for PIP2-generating enzymes in this process are still emerging (Ling et al, 2006; Zhang et al, 2012). In mammalian cells, PIP2 is primarily generated by type I PIP kinases (PIPKIs), and three isoforms, α, β and γ, are expressed in humans with multiple isoforms (van den Bout and Divecha, 2009). For example, four different isoforms of PIPKIγ are expressed in humans and each displays unique cellular distribution. PIPKIγi1 is the most abundant isoform in most cell types and largely locates to the PM (Mao and Yin, 2007). PIPKIγi2 is found at focal adhesions and cell–cell contacts (Ling et al, 2002, 2007). PIPKIγi4 is found largely in the nucleus, while PIPKIγi5 localizes to cell–cell contacts and intracellular compartments (Schill and Anderson, 2009). Often, protein–protein interactions recruit PIPKI isoforms to specific cellular regions, and many of these targeting proteins are themselves PIP2 effectors (Anderson et al, 1999; Heck et al, 2007). For example, talin recruits PIPKIγi2 to focal adhesions, while the site-specific generation of PIP2 by PIPKIγi2 strengthens talin binding to β1-integrin (Ling et al, 2006).
PIPKIγ and IQGAP1 are implicated in cancer progression and metastasis (Johnson et al, 2009; Sun et al, 2010). Overexpression of PIPKIγ in breast cancer was found to correlate with poor prognosis (Sun et al, 2010). Loss of the PIPKIγi2 isoform from metastatic breast cancer cell lines reduces cell motility (Thapa et al, 2012), but the role of other PIPKIγ isoforms and molecular mechanisms remain elusive. Similarly, loss of IQGAP1 from malignant breast epithelial cells reduces cell motility (Mataraza et al, 2003) and cell growth (Jadeski et al, 2008). IQGAP1 overexpression is reported in cancers originating from many different tissues (White et al, 2009). IQGAP1 is shown to regulate the function of many oncoproteins. Notably, IQGAP1 is found at the invasive front of aggressive cancers (Johnson et al, 2009) without knowing the underlying mechanism.
Here, we report IQGAP1 as a novel PIP2 effector that is tightly regulated by PIP2-generating enzyme PIPKIγ. PIPKIγ and IQGAP1 interact and function together in regulation of directional cell migration. Mechanistically, IQGAP1 requires PIPKIγ for targeting to the leading edge membrane of migrating cells. Also, IQGAP1 is activated specifically by PIP2, disrupting IQGAP1 autoinhibition to induce actin polymerization. Directional cell migration is dramatically attenuated in cells expressing IQGAP1 mutants that lack PIPKIγ or PIP2 interaction. Given that expression of both proteins is deregulated in cancers, this study identifies the PIPKIγ/IQGAP1 signalling nexus as a putative therapeutic target in the early steps of cancer progression.
Results
IQGAP1 and PIPKIγ interact
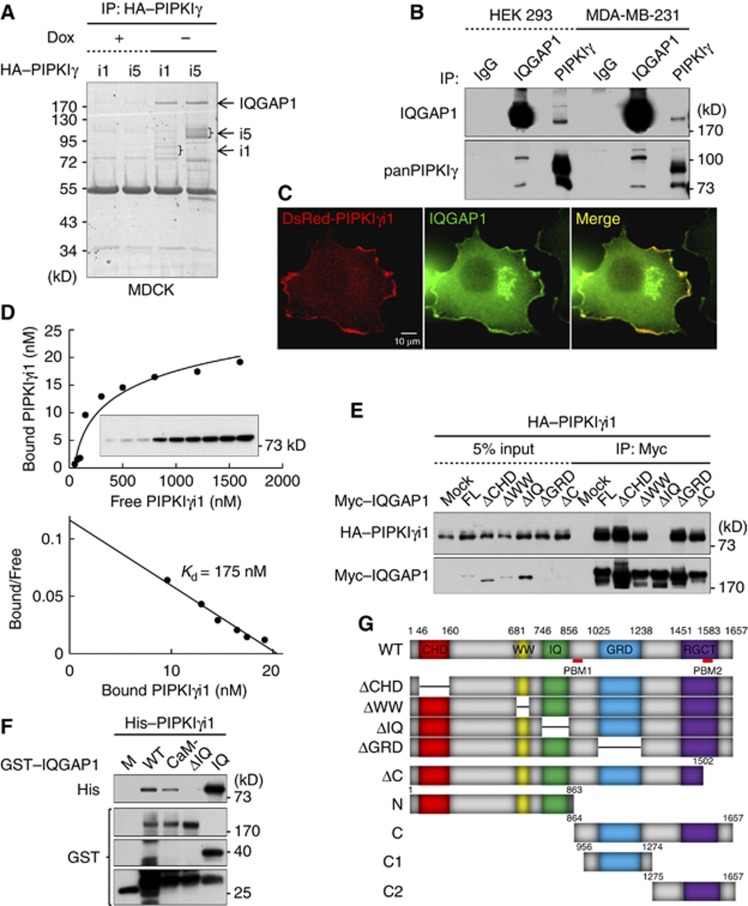
Interacting proteins often determine the function and intracellular targeting of PIPKIs (Heck et al, 2007). To identify interacting proteins for PIPKIγ, i1 and i5 isoforms were inducibly expressed and immunoprecipitated (IP’ed) from MDCK cell lysates. The isolated complexes were separated by SDS–PAGE and the gels visualized by Coomassie staining. Then, protein bands were analysed by mass spectrometry. IQGAP1 was identified to interact with the PIPKIγi1 and i5 complexes (Figure 1A).
Figure 1.
PIPKIγ interacts with the IQ motif of IQGAP1. (A) HA–PIPKIγi1 and i5 were expressed in tet-off MDCK cells, and an anti-HA antibody used to IP i1- and i5-containing complexes. Samples were resolved by SDS–PAGE and protein bands visualized by Coomassie staining. Dox, doxycycline. (B) PIPKIγ and IQGAP1 were separately IP’ed and association of the other protein examined by immunoblotting. IgG, isotype immunoglobulin control. (C) DsRed-PIPKIγi1 was transiently expressed in MCF-7 cells and endogenous IQGAP1 was immunostained. Cells were photographed under × 600 magnification. (D) GST-IQGAP1 (50 pM) was incubated with 5 to 1600, nM His–PIPKIγi1. Binding was detected by immunoblotting with an anti-His antibody (top). Kd was determined by standard Scatchard analysis (bottom). (E) Myc–IQGAP1 proteins were coexpressed with HA–PIPKIγi1 in HEK293 cells and proteins were IP’ed with an anti-Myc antibody. Associated PIPKIγi1 was analysed by immunoblotting with an anti-HA antibody. (F) Purified GST–IQGAP1 proteins were incubated with His–PIPKIγi1. The associated protein complex was examined by immunoblotting with the indicated antibodies. Some degraded products of GST–IQGAP1 proteins were detected by immunoblotting with an anti-GST antibody. Data above are representative of at least four independent experiments. (G) Schematic representation of IQGAP1 domains and IQGAP1 constructs used for this study.
Source data for this figure is available on the online supplementary information page.
The interaction between PIPKIγ and IQGAP1 was confirmed in human cell lines. Endogenous proteins were IP’ed and association was examined by immunoblotting. IQGAP1 co-IP’ed with PIPKIγ, and vice versa, from HEK 293 and MDA-MB-231 cell lysates (Figure 1B). The cellular location of the proteins was examined via immunostaining. DsRed-PIPKIγi1 colocalized with endogenous IQGAP1 at the periphery of MCF7 cells and to a lesser extent at a perinuclear compartment (Figure 1C). To characterize binding, His-PIPKIγi1 and GST-IQGAP1 were expressed in E. coli, purified and in vitro binding was assessed. As shown in Figure 1D, the binding was saturable and Scatchard analysis revealed that the dissociation constant (Kd) for the interaction is ∼175 nM, demonstrating that in vitro PIPKIγ directly interacts with IQGAP1 with a moderate affinity.
PIPKIγ interacts with the IQ domain
IQGAP1 integrates many signalling pathways by forming interactions through its calponin homology (CHD), WW, IQ, GAP-related (GRD) and RasGAP C-terminal (RGCT) domains (Brown and Sacks, 2006). To identify the PIPKIγ binding site on IQGAP1, we coexpressed Myc-IQGAP1 wild type (WT) or deletion mutants of each domain with HA-PIPKIγi1 in HEK 293 cells and performed an IP. Deletion of the IQ domain (ΔIQ) abrogated IQGAP1 co-IP with PIPKIγ (Figure 1E), and in vitro the ΔIQ mutant also failed to interact with PIPKIγ (Figure 1F). Furthermore, the IQ domain alone was capable of interacting with IQGAP1 (Figure 1F and Supplementary Figure S2A). These data indicate that the IQ domain is both necessary and sufficient to interact with PIPKIγ.
The IQ domain is composed of four tandem IQ motifs. The CaM− mutant, which contains point mutations in the IQ motifs and abrogates calmodulin binding (Li and Sacks, 2003), bound PIPKIγ to a lesser extent than WT (Figure 1F). Furthermore, deletion or mutation of individual motifs reduced binding to PIPKIγ, compared to WT, and the combined mutation of multiple IQ motifs further reduced binding (Figure 1F; and S Choi, unpublished observations). These data indicate that the intact IQ domain is required for the interaction with PIPKIγ. Further studies used the ΔIQ mutant to examine the functional importance of the PIPKIγ interaction.
Migration and lamellipodium formation require PIPKIγ
A role for PIPKIγi2 in migration is emerging (Sun et al, 2007; Thapa et al, 2012). To further define a role of other PIPKIγ isoforms in the regulation of migration, we stably knocked down PIPKIγ in MDA-MB-231 cells using two different shRNAs (Thapa et al, 2012). ShRNA 1 and 2 reduced total PIPKIγ (panPIPKIγ) expression by ∼75 and 90%, respectively. PIPKIγi2 expression was also slightly reduced (∼24 and 36%, respectively), whereas i4 and i5 expression were not changed (Supplementary Figure S1B), as reported previously (Wang et al, 2004). These data indicate that PIPKIγi1 is the predominant isoform in these cells (Mao and Yin, 2007). By bright field microscopy, PIPKIγ knockdown cells were less spread than control cells with fewer protrusions (Supplementary Figure S1A). Serum-induced migration using a Transwell assay was significantly attenuated by PIPKIγ knockdown (Supplementary Figure S1B). These data indicate that PIPKIγ is required for proper spreading and migration.
Knockdown of PIPKIγi2 has a defined migration defect (Sun et al, 2007; Thapa et al, 2012), but PIPKIγi1 could not be knocked down specifically as it is a splice variant with no unique coding sequence compared to the other isoforms. To explore the role of PIPKIγi1 and i2, we separately re-expressed these two isoforms to determine if they restore migration. The shRNA-resistant DsRed-PIPKIγ was stably re-expressed in PIPKIγ knockdown cells. Cells were then sorted to isolate cells with expression levels similar to endogenous PIPKIγ in control cells. Re-expression of PIPKIγi2 rescued migration (Supplementary Figure S1C), as reported previously (Thapa et al, 2012). Interestingly, PIPKIγi1 WT also rescued the migration whereas i1 kinase dead (KD) did not rescue, indicating that PIPKIγi1 or i2 are sufficient for serum-induced migration, and PIP2 synthesis is required for this process.
Migrating cells extend lamellipodia at the leading edge and persistent formation of lamellipodia is critical for directional migration (Ridley, 2011). To test how PIPKIγ regulates lamellipodium formation, a lamellipodial marker ARPC2 (Le Clainche et al, 2007) was immunostained following initiation of migration by scratch-wounding confluent cells. At 3 h after scratching, ARPC2 localized at the periphery of protrusions in the control cells (Supplementary Figure S1D). In PIPKIγ knockdown cells, formation of protrusions was retarded and ARPC2 no longer localized at the membrane extensions. PIPKIγi1 or i2 re-expression could recover lamellipodium formation, whereas PIPKIγi1 KD had no effect. Early protrusion formation was indistinguishable in different cells but persistent formation was diminished (Supplementary Figure S1E). This demonstrates that PIPKIγ, by generation of PIP2, regulates persistent lamellipodium formation that is required for migration.
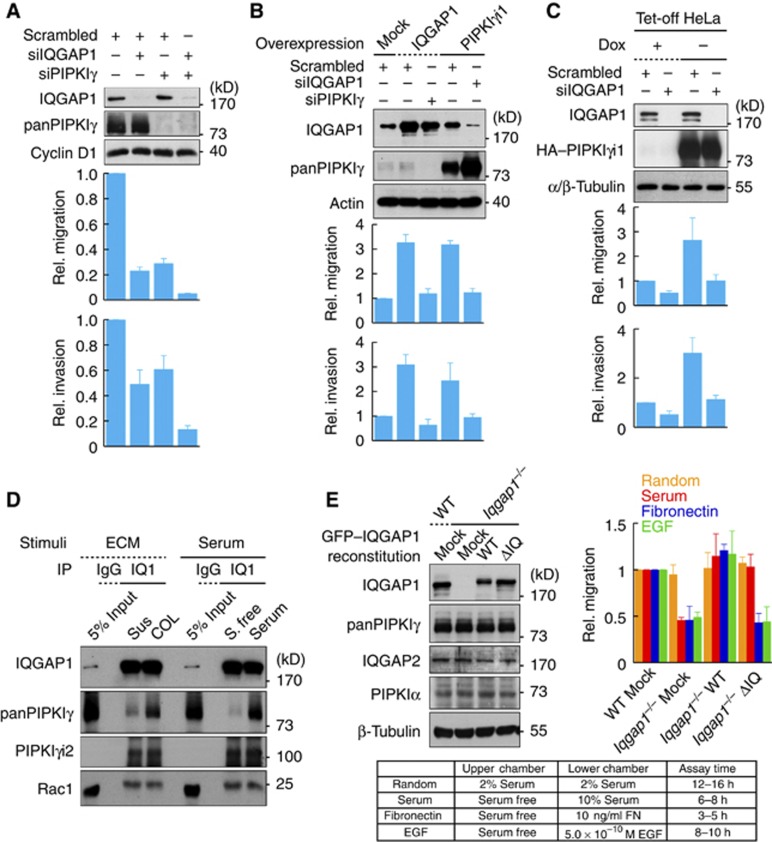
PIPKIγ and IQGAP1 interdependently control cell motility
Upon stimulation, IQGAP1 targets to the leading edge and recruits regulators of the cytoskeleton that control migration (Watanabe et al, 2005; Brown and Sacks, 2006). As described above, PIPKIγ also regulates migration (Thapa et al, 2012). Pip5k1c, a gene coding PIPKIγ in mice, knockout (KO) mice are embryonic lethal with migration defects of cardiovascular cell precursors (Wang et al, 2007), and cells from these mice have a defective association between the membrane and the cytoskeleton (Wang et al, 2008). To investigate how PIPKIγ and IQGAP1 control cell motility, serum-induced cell motility was measured using a Transwell system. Individual knockdown of PIPKIγ or IQGAP1 significantly reduced both migration and invasion (Figure 2A). Knockdown of both proteins dramatically reduced cell motility, indicating a synergistic role. To better define the relationship of the two proteins, we overexpressed IQGAP1 that is reported to enhance cell motility (Mataraza et al, 2003). Overexpression of IQGAP1 in MDA-MB-231 cells increased cell motility over three-fold, whereas knockdown of PIPKIγ in IQGAP1 overexpressing cells reduced cell motility to the basal level. Consistently overexpression of PIPKIγi1 increased cell motility and this increase was inhibited by knockdown of IQGAP1 (Figure 2B). Similar results were obtained in HeLa cells. Here, inducible expression of PIPKIγi1 increased cell motility and depletion of IQGAP1 under these conditions returned motility to the basal level (Figure 2C). Together these data indicate that PIPKIγ and IQGAP1 interdependently control cell motility.
Figure 2.
PIPKIγ and IQGAP1 cooperate to regulate migration. (A) MDA-MB-231 cells were transfected with the indicated siRNA for 48 h. Knockdown was confirmed by immunoblotting with the indicated antibodies (top). Using Transwell, 10% serum-induced migration (middle) and invasion through 2 mg/ml Matrigel (bottom) were measured. (B) Cells were transfected with the indicated DNA and siRNA combinations for 24 h. Expression level was analysed by immunoblotting with the indicated antibodies (top). Migration and invasion were measured as in (A) (bottom). (C) PIPKIγi1 was expressed in HeLa tet-off cells by removing doxycycline from media for 24 h. Protein expression and cell motility were measured as above. Data are shown as mean±s.d. for four independent experiments. (D) Cells maintained in suspension were either plated on 10 ng/ml collagen I or kept in suspension for 30 min. Serum-starved cells were treated with or without 10% serum for 15 min. Endogenous IQGAP1 was IP’ed and associated PIPKIγ was analysed by immunoblotting. COL, type I collagen; IQ1, IQGAP1; S, serum; Sus, suspension. (E) Iqgap1 KO MEFs were stably reconstituted with the indicated IQGAP1 proteins, and four different modes of migration were measured with a Transwell (top right). Protein expression was analysed by immunoblotting (top left). Conditions used for treating Transwells (bottom). Data are shown as mean±s.d. of four independent experiments. Data above are representative of at least four independent experiments.
Source data for this figure is available on the online supplementary information page.
The PIPKIγ–IQGAP1 interaction is required for migration
To investigate how PIPKIγ and IQGAP1 function together, we tested if their association is altered by stimuli that promote migration. Migration is initiated by a variety of extracellular stimuli, including chemokines or ECM (Ridley et al, 2003). To define the pathway in which PIPKIγ and IQGAP1 function, cells were stimulated with type I collagen (COL) or serum and changes in the association were examined by IP. In response to either stimulus there was an increase in the panPIPKIγ-IQGAP1 complex, whereas the Rac1 interaction with IQGAP1 remained unchanged (Figure 2D). This demonstrates that the PIPKIγ interaction with IQGAP1 is enhanced by factors that stimulate migration. Furthermore, phosphorylation of Ser1441 and Ser1443 residues of IQGAP1 (Grohmanova et al, 2004; Li et al, 2005) is required to enhance the interaction (Supplementary Figure S2C). Interestingly, the PIPKIγi2 interaction was unaffected, suggesting that migration enhances IQGAP1 interaction with the predominant isoform, PIPKIγi1 (Mao and Yin, 2007). This is consistent with results indicating that PIPKIγi2 modulates cell migration by a different mechanism (Sun et al, 2007; Thapa et al, 2012).
The IQGAP1 mutant that lacks interaction with PIPKIγ (ΔIQ) was examined to determine if this interaction is required for migration. For this, Iqgap1 KO mouse embryonic fibroblasts (MEFs) (Ren et al, 2007) were reconstituted with WT or ΔIQ IQGAP1 and migration was examined under various conditions (Keely, 2001). To avoid nonspecific effects from overexpression, we maintained IQGAP1 expression levels similar to the WT MEFs by the cell sorting method as above (Supplementary Figure S1). Iqgap1 KO MEFs showed >50% reduction in migration in response to serum, fibronectin or epidermal growth factor (EGF) stimuli. WT IQGAP1 fully rescued migration under all of these conditions, while the ΔIQ mutant showed no recovery of migration induced by fibronectin or EGF (Figure 2E). This indicates that the PIPKIγ–IQGAP1 interaction is necessary for integrin- and EGF receptor-mediated migration. Intriguingly, the ΔIQ mutant still rescued serum-induced migration. Serum contains a collection of factors that induce migration and the contribution of each factor in PIPKIγ-regulated migration varies by cell types (Sun et al, 2007). Collectively, the PIPKIγ–IQGAP1 interaction specifically regulates fibronectin- or EGF-induced migration in MEFs (Supplementary Figure S2D), indicating that the PIPKIγ–IQGAP1 nexus is regulated by these pathways.
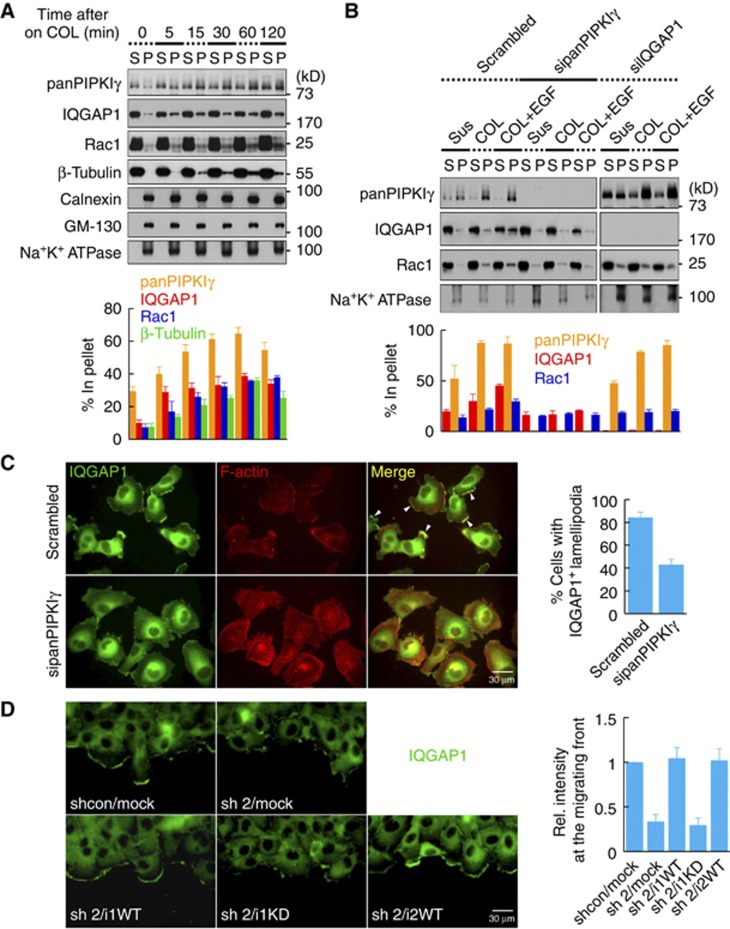
PIPKIγ controls IQGAP1 translocation to the leading edge membrane
At the onset of migration, many cytoskeleton regulatory proteins translocate to the leading edge membrane to mediate directional migration (Del Pozo et al, 2002; Ling et al, 2006; Ridley, 2011). To further define how PIPKIγ and IQGAP1 regulate migration, we examined their targeting to the membrane by cell fractionation. Cells were plated on COL, then lysed and fractionated into membrane and cytosolic components (Chao et al, 2010). In response to integrin activation, both PIPKIγ and IQGAP1 increased in the membrane fraction (Figure 3A). Rac1 also increased in the membrane fraction, as reported previously (Del Pozo et al, 2002). However, membrane proteins, such as calnexin, GM-130 and Na+K+ channel, remained unchanged (Figure 3A).
Figure 3.
PIPKIγ regulates IQGAP1 targeting to the leading edge membrane. (A) MDA-MB-231 cells maintained in suspension were plated on 10 ng/ml COL for the indicated times. Cells were lysed with a hypotonic buffer and the membrane fraction was separated from the cytosolic fraction by centrifugation. Then, 10 μg of each protein was resolved by SDS–PAGE and analysed by immunoblotting with the indicated antibodies (top). The percentage of protein bound in the pellet relative to total (S+P) was calculated by quantifying the immunoblots (bottom). S, supernatant. P, pellet. (B) After transient knockdown with the indicated siRNA, cells were treated as in (A) in the presence or absence of 50 ng/ml EGF for 30 min. Cells were fractionated and analysed as above. (C) Serum-starved control or PIPKIγ knockdown cells were treated with 20 ng/ml EGF for 1 h. Cells were fixed and stained for IQGAP1 and F-actin. Cells were photographed at × 400 magnification. For quantification, at least 300 cells were counted. White arrowheads indicate IQGAP1-positive lamellipodia. Data are shown as mean±s.d. of three independent experiments. (D) Cells grown to confluence were wounded and fixed 3 h later, followed by immunostaining for IQGAP1. Cells were photographed at × 400 magnification. Intensity of fluorescent signal at the migrating front was measured from at least 10 different images of each condition and quantified using ImageJ software. Data are shown as mean±s.d. of three independent experiments. All the experiments described above were performed independently at least three times.
Source data for this figure is available on the online supplementary information page.
In response to receptor activation, IQGAP1 translocates to the leading edge membrane (Brandt and Grosse, 2007; White et al, 2012). Yet, the mechanism for IQGAP1 interaction with the membrane is largely unknown. To examine if PIPKIγ regulates IQGAP1 membrane targeting, PIPKIγ was knocked down using RNAi and cells were fractionated. Knockdown of PIPKIγ significantly reduced IQGAP1 in the membrane fraction upon COL and/or EGF stimulation (Figure 3B). The knockdown of PIPKIγ also reduced the membrane content of Rac1, supporting reports that PIPKI and Rac1 interdependently control PM targeting (Chao et al, 2010; Halstead et al, 2010). To test the contribution of PIPKIγ in IQGAP1 targeting, we utilized a Rac1 binding defective mutant PIPKIγ (E111L) (Halstead et al, 2010). The mutant co-immunoprecipitated with IQGAP1 similar to WT PIPKIγ (Supplementary Figure S2E), indicating that Rac1 binding to PIPKIγ is not required for PIPKIγ interaction with IQGAP1. Notably, the E111L mutant enhanced IQGAP1 association with the membrane fraction similar to WT PIPKIγ (Supplementary Figure S2F). These data suggest that the IQGAP1 recruitment to the leading edge is largely regulated by PIPKIγ independent of Rac1. Knockdown of IQGAP1 reduced Rac1 in the membrane fraction, but had no effect on PIPKIγ accumulation in the membrane fraction.
To assess targeting in vivo, serum-starved cells were treated with EGF to induce lamellipodia formation (Baumgartner et al, 2006) and IQGAP1 localization was observed by immunostaining. As shown in Figure 3C, the number of PIPKIγ knockdown cells with IQGAP1-positive protrusions was reduced by >50% compared to the control cells. To assess PIPKIγ regulation of IQGAP1 localization in migrating cells, endogenous IQGAP1 was immunostained in cells migrating into the scratch wound. IQGAP1 nicely localized at the leading edge in the control cells, but in PIPKIγ knockdown cells the IQGAP1 staining at the cell periphery was significantly reduced (Figure 3D). Reconstitution with either PIPKIγi1 or i2 WT, but not i1 KD, rescued IQGAP1 localization at the leading edge. The difference between WT and KD is not due to an improper interaction with IQGAP1 because the amount of PIPKIγi1 KD that co-IP’ed with IQGAP1 was indistinguishable from that of WT (Supplementary Figure S2B). Taken together, these results demonstrate that PIPKIγ and generation of PIP2 are required for IQGAP1 targeting to the leading edge membrane in response to migratory signals.
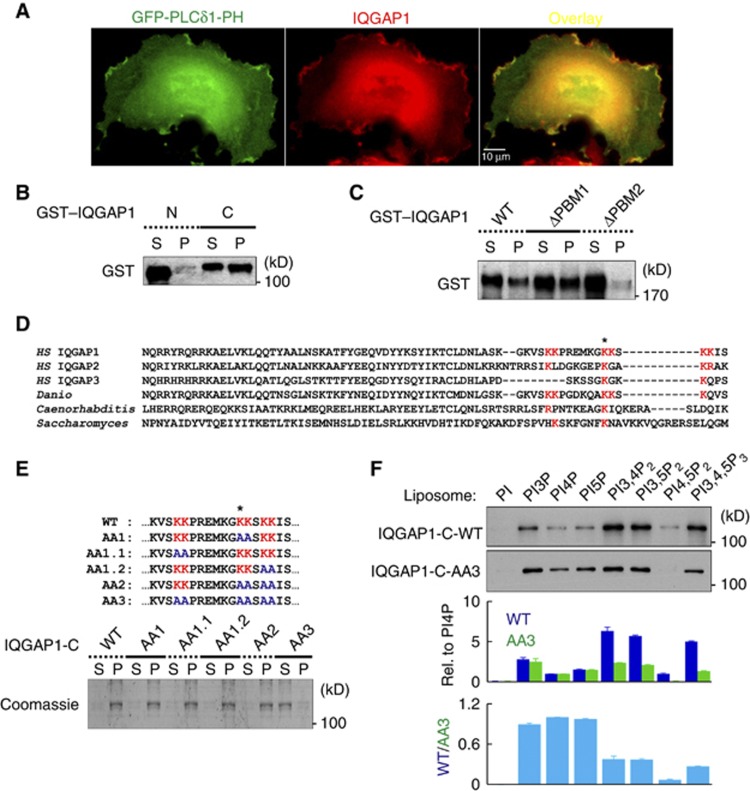
IQGAP1 interacts with PIP2 through a polybasic motif
Signalling specificity of PIP2 can be defined by interaction of PIPKIs with PIP2 effectors (Anderson et al, 1999; Heck et al, 2007). There is emerging evidence that PIPKIγ controls the cytoskeleton by interacting with cytoskeleton regulatory proteins, which are PIP2 effectors, such as talin (Ling et al, 2002) and trafficking components (Bairstow et al, 2006; Thapa et al, 2012). Because PIPKIγ associated with IQGAP1 physically (Figure 1) and functionally (Figures 2 and 3), we hypothesized that IQGAP1 could be a PIP2 effector. Consistent with this hypothesis, two independent proteomic analyses suggest that IQGAP1 interacts with PIP2 (Catimel et al, 2008; Dixon et al, 2011). To understand how IQGAP1 interacts with PIP2, their cellular distributions were examined by immunostaining. PH domain from phospholipase Cδ1 (PLCδ1) has been extensively used to probe cellular PIP2 (Czech, 2000; Raucher et al, 2000; Di Paolo and De Camilli, 2006) but excessive expression prevents targeting of PIP2 binding proteins to the plasma membrane (Raucher et al, 2000). Thus, we titrated the GFP-PLCδ1-PH expression and analysed endogenous IQGAP1 localization (Supplementary Figure S4B). In the optimal amount of expression, endogenous IQGAP1 partially colocalized with GFP-PLCδ1-PH (Figure 4A and Supplementary Figure S4B), indicating that both IQGAP1 and PIP2 are present at regions of the PM containing PIP2. To define PIP2 binding, liposomes were synthesized containing membrane lipids (57.5% of phosphatidylcholine, 20% of phosphatidylethanolamine and 20% of phosphatylserine in molar ratio) and 2.5% PI4,5P2. A co-sedimentation assay was used to define the PIP2 binding site on IQGAP1. IQGAP1-N or -C (Figure 1G) were examined and only IQGAP1-C co-sedimented with PIP2 liposomes, indicating that PIP2 binds to the C-terminal half (Figure 4B).
Figure 4.
IQGAP1 interacts with the phosphoinositides through a polybasic motif. (A) GFP-PLCδ1-PH was transiently expressed in MDA-MB-231 cells and endogenous IQGAP1 was immunostained. Cells were photographed at × 600 magnification. (B) PIP2 liposomes (2.5%) were incubated with 0.5 μM GST-IQGAP1-N or -C for 10 min. Liposome-bound IQGAP1 was pelleted by centrifugation. Equal volume of the supernatant and the pellet were resolved by SDS–PAGE and IQGAP1 in each fraction was analysed by immunoblotting with an anti-GST antibody. (C) GST–tagged WT or deletion mutants were used for a sedimentation assay with 2.5% PIP2 liposomes. (D) Amino acid sequence alignment of the PBM2 region among IQGAPs from the indicated species. (E) Selected lysine residues were mutated to alanines to generate a series of AA mutants (top). Binding of WT and the AA mutants to 5% PIP2 liposomes were tested (bottom). (F) Binding of GST–tagged WT and the AA3 mutant to 5 μM of 5% phosphoinositide liposomes were tested. Samples were analysed as above and liposome-bound proteins were detected by immunoblotting with anti-GST antibody. Immunoblots were quantified and the graph is shown as mean±s.d. of three independent experiments. All the experiments described above were performed independently at least four times.
Source data for this figure is available on the online supplementary information page.
A lysine cluster mediates IQGAP1 interaction with PIP2
IQGAP1 does not contain known PIP2 binding modules, but we found at least two potential PBMs within AA 921-970 and 1491-1560, named PBM1 and PBM2, respectively (Figure 1G). Deletion of PBM2 dramatically reduced IQGAP1 interaction with PIP2 liposomes, whereas deletion of PBM1 had little effect, indicating that IQGAP1 interacts with PIP2 through PBM2 (Figure 4C). To define a putative PIP2 binding site on PBM2, human IQGAP1, 2 and 3 sequences were aligned with IQGAP sequences from multiple species. As shown in Figure 4D, the sequence alignment identified a lysine residue, marked by an asterisk, which is conserved in PBM2. Around this lysine, there are other conserved basic residues, highlighted in red. We mutated these residues to alanines as illustrated in Figure 4E and tested the impact on PIP2 liposome binding. Mutating two or four lysine residues had little effect, whereas mutating all six residues (termed AA3) eliminated IQGAP1 binding to the PIP2 liposomes.
To examine phospholipid binding specificity, a lipid overlay assay was performed. IQGAP1 WT and -C bound to multiple phosphoinositides but not other phospholipids (Supplementary Figure S3A). To better define IQGAP1 phosphoinositide binding, the IQGAP1-C fragment was used in liposome sedimentation assays, with liposomes containing 5% phosphoinositide (Papayannopoulos et al, 2005). In this assay, PI3,4P2, PI3,5P2 and PI3,4,5P3 bound with a higher affinity than PI3P, PI4P, PI5P and PI4,5P2 (Figure 4F). Although the apparent affinity for other bis- and tris-phosphate species is up to seven-fold higher than PI4,5P2, PI4,5P2 is estimated to be present in the PM at a concentration 20- to 100-fold higher than other phosphoinositide species (Papayannopoulos et al, 2005), indicating that PI4,5P2 is the major in vivo ligand for IQGAP1. The AA3 mutation reduced binding to PI3,4P2, PI3,5P2 and PI3,4,5P3 but not monophosphate species (Figure 4F). Strikingly, the AA3 mutant lost binding to PI4,5P2. The combined data indicate that IQGAP1-C has multiple distinct phosphoinositide binding sites (Dixon et al, 2012) and the lysine cluster mutated in AA3 defines a specific PI4,5P2 binding site.
The IQGAP1 PIP2 binding mutant exhibited multiple leading edges and loss of migration
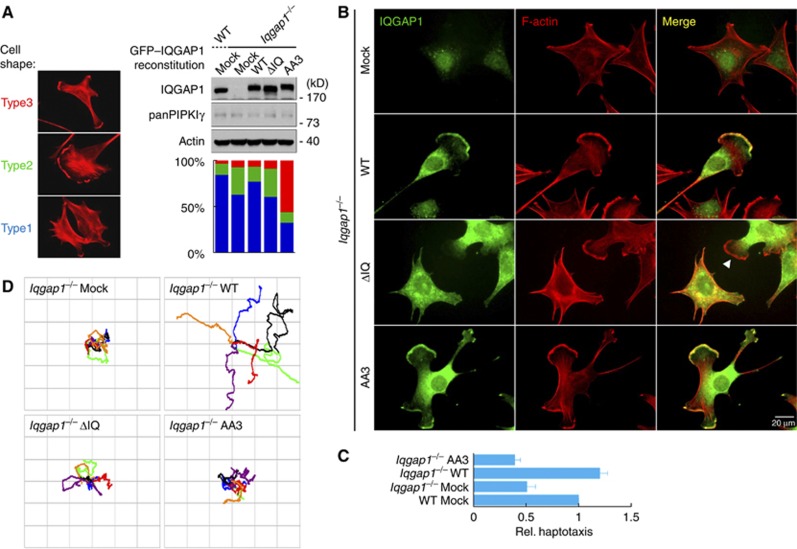
To determine how PIP2 binding modulates IQGAP1 function, the AA3 mutant was expressed in Iqgap1 KO MEFs and the cell morphology was examined. When plated on a stiff substratum (glass or plastic) coated with COL, fibronectin or gelatin, all types of cells indistinguishably highly spread and formed massive stress fibres (S Choi, unpublished observations). Cytoskeleton organization and cell shape are greatly influenced by substrate stiffness (Solon et al, 2007), and therefore cells were plated on pliant gelatin gel and cell morphology was observed by staining F-actin. Three distinct cell morphologies were observed compared to the star-shaped cells (type 1 morphology) that were predominant in WT MEFs (Figure 5A). Iqgap1 KO resulted in an increase in the number of cells with a single leading edge (type 2). Reconstitution of IQGAP1 WT partially recovered shapes of WT MEFs, whereas the ΔIQ mutant had a limited effect. Interestingly, the number of cells with multiple leading edges (type 3) was increased in the AA3-reconstituted cells (Figure 5A). To closely examine localization of the reconstituted proteins, IQGAP1 was immunostained. WT IQGAP1 localized at the leading edge where active actin polymerization occurs. The ΔIQ mutant was largely cytoplasmic and failed to localize at the leading edge (Figure 5B, arrowhead), supporting the results in Figure 3 indicating that the interaction with PIPKIγ controls IQGAP1 targeting.
Figure 5.
PIP2 binding of IQGAP1 is important for cell morphology and migration. For both (A) and (B), Iqgap1 KO MEFs, reconstituted with the indicated proteins, were plated on 0.2% gelatin gel for 3 h. Fixed cells were stained for IQGAP1 and F-actin. Cells were photographed at × 600 magnification. (A) At least 300 cells were counted for each condition and categorized based on cell morphology (left). The graph is shown as mean of three independent experiments (right bottom). Expression levels of the proteins were analysed by immunoblotting with antibodies against the indicated molecules (right top). (B) IQGAP1 and F-actin staining. Arrowhead indicates the lamellipodium that is deficient of the ΔIQ mutant. (C) With the reconstituted MEFs, fibronectin-induced haptotaxis was measured as described in Figure 2E. (D) Reconstituted MEFs were plated on gelatin gel for 3 h before recording using time-lapse microscopy. Images were collected every min for 6 h at × 100 magnification and combined into a time-lapse movie. The migration path of six individual cells was then traced and plotted on a grid, with the origin of each cell placed in the centre of the grid. All the experiments described above were performed independently at least three times.
Source data for this figure is available on the online supplementary information page.
The AA3-reconstituted cells formed multiple leading edges and the AA3 mutant localized at these sites (Figure 5B). Consistent with this morphological phenotype, the AA3-reconstituted cells did not rescue haptotactic migration (Figure 5C). The functional defects of AA3 were not due to a change in interaction with PIPKIγ as co-IP of the AA3 mutant with PIPKIγ was indistinguishable from that of WT IQGAP1 (Supplementary Figure S3B and C). Rather, the defects result from the loss of directional persistence (Figure 5D, Supplementary Figure S4 and Supplementary Movies 1, 2, 3). This indicates that the IQGAP1 interaction with PIPKIγ is required for IQGAP1 targeting to the leading edge, but PIP2 binding is required for the role of IQGAP1 in normal membrane protrusions (lamellipodia formation) and migration.
IQGAP1–PIP2 interaction regulates actin polymerization
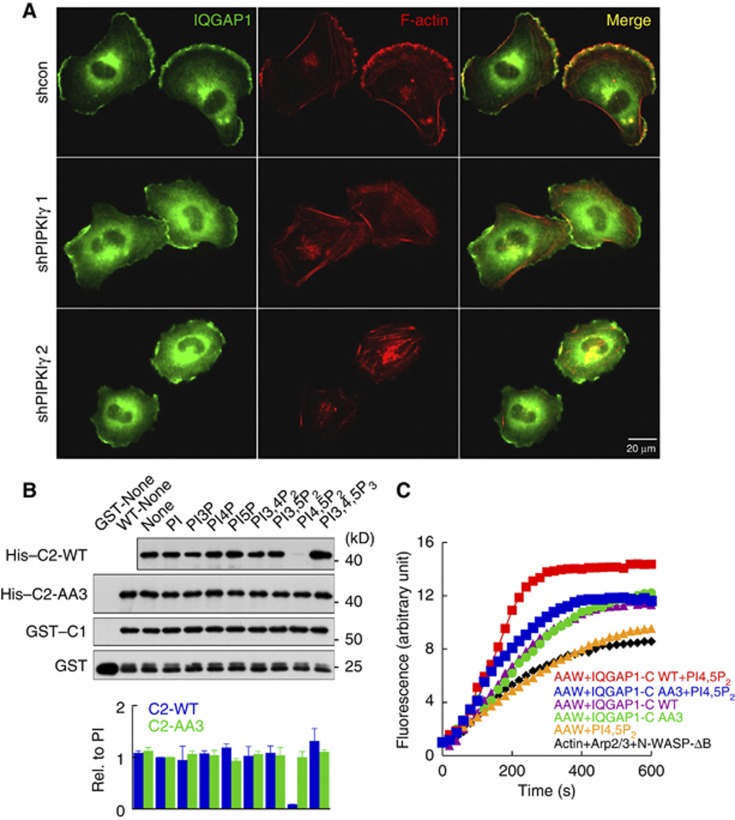
Knockdown of PIPKIγ reduced IQGAP1 targeting to the leading edge membrane. Also, in knockdown cells actin polymerization at the leading edge, indicated by strong F-actin staining, was lost and stress fibre formation was increased (Figures 3C and 6A), signifying that PIPKIγ controls actin polymerization at the leading edge by regulating IQGAP1 targeting. However, the AA3 mutant is capable of interacting with PIPKIγ and localizes at the leading edge membrane, but forms multiple leading edges (Figure 5B). These data suggest that PIPKIγ regulates activity of IQGAP1 required for persistent formation of a single leading edge.
Figure 6.
Phosphoinositide binding regulates IQGAP1 function in actin polymerization. (A) Control or PIPKIγ knockdown MDA-MB-231 cells were grown on cover glass for 24 h. Cells were fixed and endogenous IQGAP1 and F-actin were stained. Cells were photographed at × 600 magnification. (B) A total of 0.05 μM of His–C2 WT or AA3 mutant was incubated with 1 μM of GST–C1 immobilized on glutathione beads in the absence or presence of the indicated phosphoinositide liposomes (2 μM) for 10 min. Liposome-bound proteins were detected by immunoblotting with an anti-His antibody. Immunoblots were quantified and the graph is shown as mean±s.d. of three independent experiments. (C) Actin polymerization in the presence of the indicated combinations of GST–IQGAP1-C (50 nM) or 5% PI4,5P2 liposomes (2 μM). The experiments described above were performed independently at least four times.
Source data for this figure is available on the online supplementary information page.
IQGAP1 folds into an inactive conformation through an intramolecular interaction between the GRD and the RGCT domains (Brandt and Grosse, 2007). RhoGTPase binding to the GRD or phosphorylation of Ser1443 disrupts autoinhibition and activates IQGAP1 (Grohmanova et al, 2004). We identified a PIP2 binding PBM within the RGCT domain close to Ser1443, suggesting that PIP2 binding to this PBM may open the inactive conformation (Brandt et al, 2007; Le Clainche et al, 2007). To test this hypothesis, we examined how phosphoinositides affect binding between the GRD and the RGCT domains. For this analysis, His-C2 was incubated with immobilized GST-C1 (Figure 1G) in the presence or absence of phosphoinositide liposomes. In the absence of liposomes, C1 bound to C2 as reported previously (Grohmanova et al, 2004). Intriguingly, the binding was dramatically decreased in the presence of PI4,5P2 liposomes, while other phosphoinositides or phosphatidylinositol had no significant effect. Introduction of the AA3 mutation in the C2 fragment eliminated the effect of PI4,5P2 on the C1–C2 binding (Figure 6B and Supplementary Figure S5C). Although the AA3 IQGAP1-C interacts with other phosphoinositide species, it lacks PI4,5P2 binding (Figure 4F). This indicates that there are multiple phosphoinositide binding sites in IQGAP1-C (Dixon et al, 2012), but only PI4,5P2 binding to the PBM modulates the activation of IQGAP1 as indicated by a loss of the C1–C2 interaction.
The C-terminal fragment of IQGAP1 (AA 746–1657) enhances actin polymerization by activating N-WASP (Le Clainche et al, 2007). Using this system, the influence of phosphoinositides in IQGAP1-mediated actin polymerization was assessed. Since the actin polymerization activity of N-WASP is also regulated by PI4,5P2, a N-WASP-ΔB mutant, which lacks the PI4,5P2-responsive element (Rohatgi et al, 2000), was used for this assay. Addition of PI4,5P2 liposomes had no effect while addition of IQGAP1-C enhanced actin polymerization as shown previously (Le Clainche et al, 2007). Introduction of PI4,5P2 liposomes in combination with WT IQGAP1-C significantly enhanced actin polymerization activity, whereas PI4,5P2 had a limited effect on actin polymerization by the AA3 mutant (Figure 6C). Strikingly, stimulation of actin polymerization was highly specific for PI4,5P2 (Supplementary Figure S5A–D).
Discussion
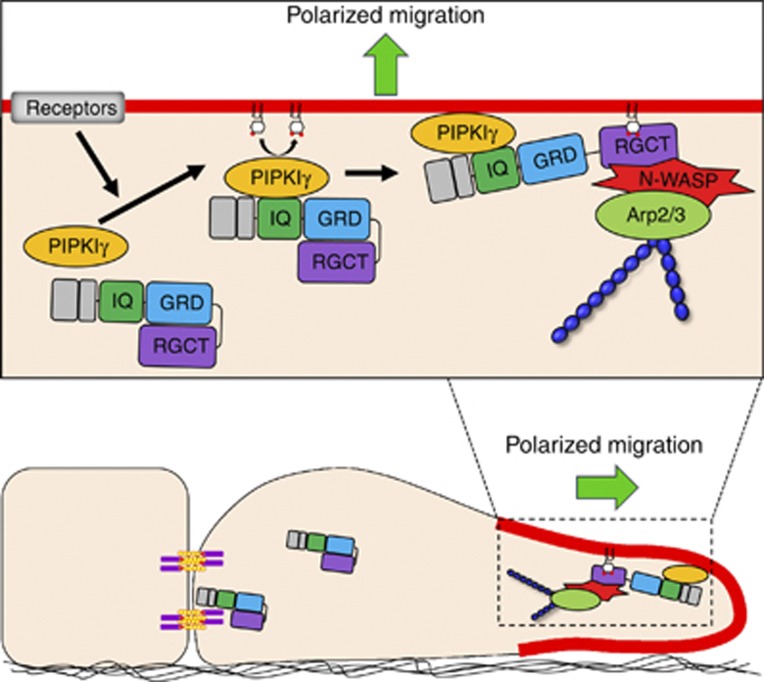
Here, we define a novel mechanism of how PIPKIγ and IQGAP1 function together as a signalling nexus to regulate migration (Figure 7). In polarized epithelial cells, IQGAP1 is largely localized to cell–cell contacts (Li et al, 1999; Fukata et al, 2001; Watanabe et al, 2004; Noritake et al, 2005). In directionally migrating cells, IQGAP1 translocates to the leading edge (Mataraza et al, 2003) and facilitates actin polymerization. In response to receptor signalling, PIPKIγ associates with IQGAP1 and recruits IQGAP1 to the leading edge membrane. There, generation of PIP2 by PIPKIγ activates IQGAP1, as PIP2 binding to a PBM relieves autoinhibition between the RGD and RGCT domains. This allows the RGCT domain to recruit N-WASP and the Arp2/3 complex to facilitate actin polymerization (Supplementary Figure S3E) (Brandt and Grosse, 2007). Overall, extracellular stimuli control the spatiotemporal activation of the PIPKIγ/IQGAP1 nexus to regulate actin polymerization required for persistent formation of lamellipodia and migration.
Figure 7.
Model of PIP2-mediated IQGAP1 activation. In response to receptor activation, PIPKIγ recruits IQGAP1 to the leading edge membrane of migrating cells. Then, PIP2 generated by PIPKIγ interacts with a PBM of IQGAP1 to block the autoinhibitory interaction between the GRD and RGCT domains. The relieved RGCT domain mediates actin polymerization by recruiting N-WASP and the Arp2/3 complex.
All PIPKIγ isoforms have the potential to interact with IQGAP1 (Figure 1A) and this suggests that IQGAP1 may mediate isoform-specific functions at different compartments. For example, IQGAP1 is found in the nucleus and ectopic expression of IQGAP1 enhances transcriptional activity of β-catenin (Briggs et al, 2002). Similarly, PIPKIγ also modulates β-catenin-mediated transcriptional co-activation (Schramp et al, 2011). IQGAP1 associates with the exocyst complex and regulates cancer cell invasion, a function also regulated by PIPKIγi2 (Sakurai-Yageta et al, 2008). Here, we demonstrate that receptor signalling stimulates the recruitment of IQGAP1 to the leading edge through an interaction with PIPKIγ, likely the PIPKIγi1 isoform (Figure 2D). PIPKIγi2 isoform plays an analogous role by interaction with talin, linking the trafficking of integrin-containing vesicles to talin-rich adhesions (Thapa et al, 2012).
The C-terminal half of IQGAP1 (IQGAP1-C) binds to different phosphoinositide species with a varying binding affinity (Figure 4F). A recent study shows that the distal portion of the C-terminus of IQGAP1 (AA 1559–1657) forms a pseudo C2 domain fold and binds to class I phosphoinositide 3-kinase products, PI3,4P2 and PI3,4,5P3 (Dixon et al, 2012). According to the solved structure, Lys1562 and Lys1604 are important for ligand recognition. Here we define a distinct PI4,5P2-binding site at Lys1546, Lys1547, Lys1554, Lys1555, Lys1557 and Lys1558 (Figure 4). These data indicate that there could be multiple phosphoinositide binding sites on IQGAP1-C. Consistent with this possibility, the IQGAP1-C1 interaction with IQGAP1-C2 is specifically inhibited by PI4,5P2, while mutating the six lysine residues blocks the inhibition (Figure 6B). Further work is necessary to define other phosphoinositide binding sites on IQGAP1. These studies will give us mechanistic insight into how IQGAP1 is found at the intracellular compartments where different phosphoinositide species are predominant (Di Paolo and De Camilli, 2006; Osman, 2010).
PIPKIγ regulates IQGAP1 targeting to the leading edge and this event requires PIP2 generation (Figure 3). IQGAP1 is widely believed to target to the PM by association with Rac1 and Cdc42 (Fukata et al, 2002; Watanabe et al, 2004; Brandt and Grosse, 2007). Rac1 and Cdc42 contain PBMs near the C-termini and these PBMs contribute to membrane targeting (Del Pozo et al, 2002; Heo et al, 2006). This raises the possibility that PIP2 controls IQGAP1 targeting to the PM by indirectly regulating Rac1 targeting. Consistently, sequestration of cellular PIP2 by either neomycin treatment (Gabev et al, 1989) or PLCδ1-PH expression (Raucher et al, 2000) blocks both Rac1 and IQGAP1 translocation to membrane in response to integrin activation (Supplementary Figure S3D). To examine the sole contribution of PIP2 binding for IQGAP1 targeting, we generated and expressed a PIP2-binding-defective mutant of IQGAP1 in Iqgap1−/− MEFs. The PIP2-binding-defective mutant still localizes to the PM, while the PIPKIγ-binding-defective (ΔIQ) mutant is largely cytosolic (Figure 5B). These data indicate that the physical interaction between the two proteins is more important than PIP2 binding for targeting IQGAP1 to the PM.
Cells expressing the PIP2 binding IQGAP1 mutant (AA3) form multiple leading edges, suggesting that PIP2 regulation of IQGAP1 is important for maintaining polarity and leading edge integrity (Figure 5B). These cells exhibit perpetual formation and retraction of leading edges but display little movement (Figure 5D and Supplementary Movie 3). Consistent with this observation, IQGAP1 is suggested to maintain polarity of migrating cells through local capture of MTs at the leading edge by interaction with MT regulators (Watanabe et al, 2005). The interaction sites for these proteins are within the RGCT domain, which also contains the PIP2 binding site (Brown and Sacks, 2006). We envision that the autoinhibitory interaction between the GRD and RGCT domains may also block MT recruitment, and PIP2 binding may relieve this (Figure 3A). In this model, the AA3 mutant may remain inactive at the leading edge and fail to recruit MTs, which would result in loss of cell polarity. Alternatively, multiple leading edges could be induced by perturbation of actin dynamics. In support of this possibility, cells display multiple leading edges after manipulation of certain actin regulatory proteins. For example, multiple leading edges also form in Cdc42 KO dendritic cells (Lammermann et al, 2009) and in Vero cells after expression of an IQGAP1 mutant that is defective in Rac1/Cdc42 binding (Fukata et al, 2002).
Finally, multiple reports suggest roles for both PIPKIγ and IQGAP1 in cancer metastasis (Johnson et al, 2009; White et al, 2009; Sun et al, 2010). The current findings define a molecular mechanism of how these two proteins interact and function together in migration and invasion, and potentially other processes required for cancer progression.
Materials and methods
Cell culture and constructs
MDA-MB-231, HEK 293, MCF-7 and MEF cells were maintained in DMEM supplemented with 10% fetal bovine serum (Gibco). MDCK and HeLa tet-off cells were cultured as previously described (Ling et al, 2007) and induction of transgene was achieved by removing of doxycycline from media for 24 h. The constructs used for this work have been described previously (Sokol et al, 2001; Li et al, 2005; Papayannopoulos et al, 2005; Le Clainche et al, 2007; Ren et al, 2007).
Stable cell line generation
To generate stable MDA-MB-231 cell lines, cells were transfected with vectors expressing DsRed-PIPKIγ isoforms using Lipofectamine 2000 (Invitrogen) and selected with 1.2 mg/ml Geneticin (Gibco) for 15 days, and further selected for DsRed expression using cell sorter. Cells expressing the transgene at a level similar to the endogenous level of PIPKIγ were used for experiments. For generation of stable cell lines in MEFs, cells were infected with retrovirus for 24 h. Then, cells expressing GFP-IQGAP1 were first selected for GFP expression, and then further sorted by expression level.
Antibodies and siRNAs
Monoclonal antibodies against IQGAP1, β-tubulin, Myc-tag, Na+K+ ATPase, GST–tag, His–tag (Millipore), α/β-tubulin, cyclin D1 (Cell Signaling Technology), Rac1, calnexin, GM-130 (BD Biosciences), HA–tag (Covence Biotechnology), actin (MP Biomedicals) and polyclonal antibody against IQGAP2 (Santa Cruz Biotechnology) were used for this study. Polyclonal and monoclonal antibodies against total and specific isoforms of PIPKIγ were produced as described previously (Schill and Anderson, 2009). Pooled siRNAs against PIPKIγ were obtained from Dharmacon and IQGAP1 from Santa Cruz Biotechnology.
IP and immunoblotting
Cells were lysed in a buffer containing 1% Brij58, 150 mM NaCl, 20 mM HEPES, pH 7.4, 2 mM MgCl2, 2 mM CaCl2, 1 mM Na3VO4, 1 mM Na2MoO4 and protease inhibitors. Protein concentration of lysates was measured by the BCA method (Pierce) and equal amounts of protein were used for further analysis. For IP, 0.5 to 1 mg of proteins were incubated with 1 μg of antibodies at 4°C for 8 h and then incubated with a 50% slurry of Protein G Sepharose (GE Life Sciences) for another 2 h. After washing 5 × with lysis buffer, the protein complex was eluted with SDS sample buffer. For immunoblotting, 10 to 20 μg of proteins were loaded. After developing immunoblots, the film was scanned using a transmitted light scanner (resolution=600 d.p.i.). Protein bands were quantified using ImageJ, and statistical analysis of the data was performed with Microsoft Excel. The statistical analysis was performed using data from at least three independent experiments.
In vitro binding assay
Recombinant proteins were expressed in BL21 E. coli strain. GST–tagged proteins were then purified with GST Sepharose 4B (GE Life Sciences) and His–tagged proteins were purified with His-Bind Resin (Novagen). GST–tagged proteins were incubated with glutathione beads before binding assays. The binding assay was performed in the lysis buffer used for IP by adding 10 nM to 5 μM of His–tagged proteins and 20 μl of GST–tagged protein bound to glutathione beads. After incubation for 1 h at 25°C, unbound proteins were washed out and the protein complex was analysed by immunoblotting. For the binding assay with liposomes, analysis was performed for 10 m at 25°C without detergent (150 mM KCl, 50 mM HEPES, pH 7.4, 2 mM MgCl2, 2 mM CaCl2 and protease inhibitors) to maintain the integrity of liposomes.
Transwell motility assay
Motility assays were performed with a Transwell (Corning) as described before (Keely, 2001). Briefly, equal numbers of cells were loaded on the upper chamber and cells that migrated towards attractants were fixed with 4% paraformaldehyde followed by staining with 0.5% crystal violet. Cells were counted in photographs taken from at least five random fields with a Nikon Eclipse TE2000U at × 200 resolution. Statistical analysis was performed with Microsoft Excel, using data from at least three independent experiments. A Transwell with 3.0 μm pores was used for migration assay and 8.0 μm pores for invasion assay.
Subcellular fractionation assay
Cells were lysed in a hypotonic lysis buffer (Del Pozo et al, 2002) for 10 min. Then cell lysates were homogenized with 15 strokes of a Dounce homogenizer. Homogenates were centrifuged at 700 g for 3 min to pellet nuclei and intact cells. The supernatants were spun at 100 000 g for 30 min at 4°C to sediment particulates. The cytosol-containing supernatant was removed and the crude membrane pellet was gently washed with the lysis buffer. Protein concentration was determined in the membrane and cytosolic fractions. Equal amounts of protein were resolved by SDS–PAGE and further analysed by immunoblotting.
Fluorescence microscopy
Glass coverslips were coated with 10 ng/ml COL, fibronectin, gelatin or 10% serum before seeding cells. For Figure 5, coverslips were coated as described previously (Sakurai-Yageta et al, 2008). Cells were grown on coverslips placed inside six-well plates until experimental manipulation. Coverslips were washed twice in 37°C PBS, and then fixed with 4% paraformaldehyde, followed by permeabilization with 0.5% Triton X-100 in PBS. The cells were then blocked for 1 h at 25°C in 3% BSA. Primary antibody incubation was performed at 4°C for 12 h, while incubation with fluorophore-conjugated secondary antibodies occurred at 37°C for 45 min. Fluorescence microscopy was performed using a × 60 plan-fluor objective on a Nikon Eclipse TE2000U equipped with a Photometrics CoolSNAP ES CCD camera. Images were captured using MetaMorph v6.3 (Molecular Devices). Images were exported to Photoshop CS2 (Adobe) for final processing and assembly.
Liposome sedimentation assay
Liposomes were prepared as previously described (Papayannopoulos et al, 2005). Dried lipids were resuspended with a buffer containing 150 mM KCl, 50 mM HEPES, pH 7.4, 2 mM MgCl2, 2 mM CaCl2 and 300 mM sucrose. After bath sonication for 20 min, the rehydrated lipids were subjected to at least five cycles of freezing and thawing and extruded through a 0.1 μm filter with a lipid extruder (Avanti). Liposome co-sedimentation assay was performed by mixing 0.5 μM of proteins with 2.5 μM of liposomes in the buffer without sucrose. After 10 min of incubation at 25°C, samples were centrifuged at 100 000 g for 30 min at 4°C. Pellets were gently washed and resuspended in SDS sample buffer for a final volume equal to the supernatant. Samples were resolved by SDS–PAGE and proteins were detected by either Coomassie staining or immunoblotting.
Live cell imaging
Delta TPG dish (Fisher Scientific) were coated with a gelatin gel as described previously (Sakurai-Yageta et al, 2008). Cells were seeded at a density of 1.0 × 104 cells/dish in L15 culture medium and placed in a temperature-controlled chamber of a Nikon Eclipse TE2000U. Time-lapse recording started 3 h after cell plating. Images were collected every 30 or 60 s for over 5 h with a Photometrics CoolSNAP ES CCD camera (Roper Scientific) operated by Metamorph image analysis software (Molecular Devices). Analyses of collected images including tracking the migration path of individual cells and generation of movies were performed with Metamorph.
Actin polymerization assay
Actin polymerization assay was performed as described before (Le Clainche et al, 2007). Pyrene-conjugated G-actin (Cytoskeleton) was prepared according to the manufacturer’s instructions. Then, 12.5 nM of Arp2/3 complex and 40 nM of N-WASP-ΔB in the presence of GST-IQGAP1-C (50 nM) and/or 5% phosphoinositide-liposomes (2 μM) were incubated for 5 min before the addition of 1.5 μM of pyrene-conjugated G-actin stock. Fluorescence was read immediately after the addition of actin using a PC1 photon counting spectrofluorometer (ISS) set on kinetic mode to read every 20 s for the duration of the assay. PC1 setting was as follows: excitation, 365 nm; emission, 407 mm. Obtained fluorescence density was converted to arbitrary units.
Supplementary Material
Acknowledgments
We are grateful to Ruth Kroschewski (ETH Zurich) for providing the IQGAP1-C1 and 2 constructs, to Wendell Lim (University of California, San Francisco) for the N-WASP constructs. We also thank Kurt Amann (University of Wisconsin-Madison) for technical advice for an actin polymerization assay. This work was supported by NIH grants to RAA, the Intramural Research Program of the National Institutes of Health to DBS and American Heart Association fellowships to SC, ACH and NT.
Author contributions: SC and RAA designed experiments; SC, NT and ZL performed experiments; SC, ACH, DBS and RAA wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson RA, Boronenkov IV, Doughman SD, Kunz J, Loijens JC (1999) Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J Biol Chem 274: 9907–9910 [DOI] [PubMed] [Google Scholar]
- Bairstow SF, Ling K, Su X, Firestone AJ, Carbonara C, Anderson RA (2006) Type Igamma661 phosphatidylinositol phosphate kinase directly interacts with AP2 and regulates endocytosis. J Biol Chem 281: 20632–20642 [DOI] [PubMed] [Google Scholar]
- Baumgartner M, Sillman AL, Blackwood EM, Srivastava J, Madson N, Schilling JW, Wright JH, Barber DL (2006) The Nck-interacting kinase phosphorylates ERM proteins for formation of lamellipodium by growth factors. Proc Natl Acad Sci USA 103: 13391–13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Grosse R (2007) Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep 8: 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R (2007) Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol 178: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Li Z, Sacks DB (2002) IQGAP1-mediated stimulation of transcriptional co-activation by beta-catenin is modulated by calmodulin. J Biol Chem 277: 7453–7465 [DOI] [PubMed] [Google Scholar]
- Brown MD, Sacks DB (2006) IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol 16: 242–249 [DOI] [PubMed] [Google Scholar]
- Catimel B, Schieber C, Condron M, Patsiouras H, Connolly L, Catimel J, Nice EC, Burgess AW, Holmes AB (2008) The PI(3,5)P2 and PI(4,5)P2 interactomes. J Proteome Res 7: 5295–5313 [DOI] [PubMed] [Google Scholar]
- Chao WT, Daquinag AC, Ashcroft F, Kunz J (2010) Type I PIPK-alpha regulates directed cell migration by modulating Rac1 plasma membrane targeting and activation. J Cell Biol 190: 247–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech MP (2000) PIP2 and PIP3: complex roles at the cell surface. Cell 100: 603–606 [DOI] [PubMed] [Google Scholar]
- Del Pozo MA, Kiosses WB, Alderson NB, Meller N, Hahn KM, Schwartz MA (2002) Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat Cell Biol 4: 232–239 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Gray A, Boisvert FM, Agacan M, Morrice NA, Gourlay R, Leslie NR, Downes CP, Batty IH (2011) A screen for novel phosphoinositide 3-kinase effector proteins. Mol Cell Proteomics 10: M110 003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Gray A, Schenning M, Agacan M, Tempel W, Tong Y, Nedyalkova L, Park HW, Leslie NR, van Aalten DM, Downes CP, Batty IH (2012) IQGAP proteins reveal an atypical phosphoinositide (aPI) binding domain with a pseudo C2 domain fold. J Biol Chem 287: 22483–22496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Nakagawa M, Itoh N, Kawajiri A, Yamaga M, Kuroda S, Kaibuchi K (2001) Involvement of IQGAP1, an effector of Rac1 and Cdc42 GTPases, in cell-cell dissociation during cell scattering. Mol Cell Biol 21: 2165–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K (2002) Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109: 873–885 [DOI] [PubMed] [Google Scholar]
- Gabev E, Kasianowicz J, Abbott T, McLaughlin S (1989) Binding of neomycin to phosphatidylinositol 4,5-bisphosphate (PIP2). Biochim Biophys Acta 979: 105–112 [DOI] [PubMed] [Google Scholar]
- Golub T, Caroni P (2005) PI(4,5)P2-dependent microdomain assemblies capture microtubules to promote and control leading edge motility. J Cell Biol 169: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332: 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmanova K, Schlaepfer D, Hess D, Gutierrez P, Beck M, Kroschewski R (2004) Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of rho-GTPase regulator. J Biol Chem 279: 48495–48504 [DOI] [PubMed] [Google Scholar]
- Halstead JR, Savaskan NE, van den Bout I, Van Horck F, Hajdo-Milasinovic A, Snell M, Keune WJ, Ten Klooster JP, Hordijk PL, Divecha N (2010) Rac controls PIP5K localisation and PtdIns(4,5)P synthesis, which modulates vinculin localisation and neurite dynamics. J Cell Sci 123(Pt 20): 3535–3546 [DOI] [PubMed] [Google Scholar]
- Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, Anderson RA (2007) A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol 42: 15–39 [DOI] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T (2006) PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314: 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YD, Joyal JL, Li Z, Sacks DB (1999) IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem 274: 464–470 [DOI] [PubMed] [Google Scholar]
- Jadeski L, Mataraza JM, Jeong HW, Li Z, Sacks DB (2008) IQGAP1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. J Biol Chem 283: 1008–1017 [DOI] [PubMed] [Google Scholar]
- Johnson M, Sharma M, Henderson BR (2009) IQGAP1 regulation and roles in cancer. Cell Signal 21: 1471–1478 [DOI] [PubMed] [Google Scholar]
- Keely PJ (2001) Ras and Rho protein induction of motility and invasion in T47D breast adenocarcinoma cells. Methods Enzymol 333: 256–266 [DOI] [PubMed] [Google Scholar]
- Lammermann T, Renkawitz J, Wu X, Hirsch K, Brakebusch C, Sixt M (2009) Cdc42-dependent leading edge coordination is essential for interstitial dendritic cell migration. Blood 113: 5703–5710 [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, Kroschewski R (2007) IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem 282: 426–435 [DOI] [PubMed] [Google Scholar]
- Li R, Debreceni B, Jia B, Gao Y, Tigyi G, Zheng Y (1999) Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of Cdc42. J Biol Chem 274: 29648–29654 [DOI] [PubMed] [Google Scholar]
- Li Z, McNulty DE, Marler KJ, Lim L, Hall C, Annan RS, Sacks DB (2005) IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J Biol Chem 280: 13871–13878 [DOI] [PubMed] [Google Scholar]
- Li Z, Sacks DB (2003) Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J Biol Chem 278: 4347–4352 [DOI] [PubMed] [Google Scholar]
- Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA (2007) Type I gamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol 176: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420: 89–93 [DOI] [PubMed] [Google Scholar]
- Ling K, Schill NJ, Wagoner MP, Sun Y, Anderson RA (2006) Movin' on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol 16: 276–284 [DOI] [PubMed] [Google Scholar]
- Mao YS, Yin HL (2007) Regulation of the actin cytoskeleton by phosphatidylinositol 4-phosphate 5 kinases. Pflugers Arch 455: 5–18 [DOI] [PubMed] [Google Scholar]
- Mataraza JM, Briggs MW, Li Z, Entwistle A, Ridley AJ, Sacks DB (2003) IQGAP1 promotes cell motility and invasion. J Biol Chem 278: 41237–41245 [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438: 605–611 [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D (2002) PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct 31: 151–175 [DOI] [PubMed] [Google Scholar]
- Noritake J, Watanabe T, Sato K, Wang S, Kaibuchi K (2005) IQGAP1: a key regulator of adhesion and migration. J Cell Sci 118(Pt 10): 2085–2092 [DOI] [PubMed] [Google Scholar]
- Osman M (2010) An emerging role for IQGAP1 in regulating protein traffic. ScientificWorldJournal 10: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA (2005) A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell 17: 181–191 [DOI] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 11: 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T (2000) Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 100: 221–228 [DOI] [PubMed] [Google Scholar]
- Ren JG, Li Z, Sacks DB (2007) IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci USA 104: 10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ (2011) Life at the leading edge. Cell 145: 1012–1022 [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR (2003) Cell migration: integrating signals from front to back. Science 302: 1704–1709 [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM (2003) Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol 5: 599–609 [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW (2000) Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol 150: 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, D'Souza-Schorey C, Chavrier P (2008) The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol 181: 985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill NJ, Anderson RA (2009) Two novel phosphatidylinositol-4-phosphate 5-kinase type Igamma splice variants expressed in human cells display distinctive cellular targeting. Biochem J 422: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramp M, Thapa N, Heck J, Anderson R (2011) PIPKIgamma regulates beta-catenin transcriptional activity downstream of growth factor receptor signaling. Cancer Res 71: 1282–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol SY, Li Z, Sacks DB (2001) The effect of IQGAP1 on Xenopus embryonic ectoderm requires Cdc42. J Biol Chem 276: 48425–48430 [DOI] [PubMed] [Google Scholar]
- Solon J, Levental I, Sengupta K, Georges PC, Janmey PA (2007) Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J 93: 4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Ling K, Wagoner MP, Anderson RA (2007) Type I gamma phosphatidylinositol phosphate kinase is required for EGF-stimulated directional cell migration. J Cell Biol 178: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Turbin DA, Ling K, Thapa N, Leung S, Huntsman DG, Anderson RA (2010) Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast Cancer Res 12: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Sun Y, Schramp M, Choi S, Ling K, Anderson RA (2012) Phosphoinositide signaling regulates the exocyst complex and polarized integrin trafficking in directionally migrating cells. Dev Cell 22: 116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bout I, Divecha N (2009) PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci 122(Pt 21): 3837–3850 [DOI] [PubMed] [Google Scholar]
- Wang Y, Lian L, Golden JA, Morrisey EE, Abrams CS (2007) PIP5KI gamma is required for cardiovascular and neuronal development. Proc Natl Acad Sci USA 104: 11748–11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Litvinov RI, Chen X, Bach TL, Lian L, Petrich BG, Monkley SJ, Kanaho Y, Critchley DR, Sasaki T, Birnbaum MJ, Weisel JW, Hartwig J, Abrams CS (2008) Loss of PIP5KIgamma, unlike other PIP5KI isoforms, impairs the integrity of the membrane cytoskeleton in murine megakaryocytes. J Clin Invest 118: 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Li WH, Wang J, Xu K, Dong P, Luo X, Yin HL (2004) Critical role of PIP5KI{gamma}87 in InsP3-mediated Ca(2+) signaling. J Cell Biol 167: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kaibuchi K (2005) Regulation of microtubules in cell migration. Trends Cell Biol 15: 76–83 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K (2004) Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell 7: 871–883 [DOI] [PubMed] [Google Scholar]
- White CD, Brown MD, Sacks DB (2009) IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett 583: 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CD, Erdemir HH, Sacks DB (2012) IQGAP1 and its binding proteins control diverse biological functions. Cell Signal 24: 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HL, Janmey PA (2003) Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65: 761–789 [DOI] [PubMed] [Google Scholar]
- Zhang L, Mao YS, Janmey PA, Yin HL (2012) Phosphatidylinositol 4, 5 bisphosphate and the actin cytoskeleton. Subcell Biochem 59: 177–215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.