Abstract
Mutations of CIAS1 have recently been shown to underlie familial cold urticaria (FCU) and Muckle-Wells syndrome (MWS), in three families and one family, respectively. These rare autosomal dominant diseases are both characterized by recurrent inflammatory crises that start in childhood and that are generally associated with fever, arthralgia, and urticaria. The presence of sensorineural deafness that occurs later in life is characteristic of MWS. Amyloidosis of the amyloidosis-associated type is the main complication of MWS and is sometimes associated with FCU. In FCU, cold exposure is the triggering factor of the inflammatory crisis. We identified CIAS1 mutations, all located in exon 3, in nine unrelated families with MWS and in three unrelated families with FCU, originating from France, England, and Algeria. Five mutations—namely, R260W, D303N, T348M, A439T, and G569R—were novel. The R260W mutation was identified in two families with MWS and in two families with FCU, of different ethnic origins, thereby demonstrating that a single CIAS1 mutation may cause both syndromes. This result indicates that modifier genes are involved in determining either a MWS or a FCU phenotype. The finding of the G569R mutation in an asymptomatic individual further emphasizes the importance of such modifier a gene (or genes) in determining the disease phenotype. Identification of this gene (or these genes) is likely to have significant therapeutic implications for these severe diseases.
Introduction
Muckle-Wells syndrome (MWS [MIM 191900]) and familial cold urticaria (FCU [MIM 120100]) belong to the group of hereditary fever syndromes that includes familial Mediterranean fever (FMF [MIM 249100]), hyperimmunoglobulinemia D with periodic fever syndrome (HIDS [MIM 260920]), and tumor necrosis factor receptor 1–associated periodic syndrome (TRAPS [MIM 142680 and MIM 134610]) (Delpech and Grateau 2001; Drenth and Van der Meer 2001). MWS and FCU are rare autosomal dominant disorders reported in the northern-European population. The first clinical signs occur during childhood and generally include fever, joint inflammation, myalgias, urticaria, and conjunctivitis (Kile and Rusk 1940; Muckle and Wells 1962). In MWS, a progressive sensorineural deafness occurs later in life, and a renal amyloidosis of the amyloidosis associated (AA) type can sometimes be observed (Messier et al. 1988). In contrast, FCU is defined by a highly specific clinical feature: the effect of cold on the skin, which triggers the general symptoms of inflammatory crises (Hoffman et al. 2001b). MWS and FCU have both been localized to chromosomal region 1q44 (Cuisset et al. 1999; Hoffman et al. 2000). Recently, Hoffman et al. (2001a) identified missense mutations in the CIAS1 gene, which cosegregated in three families with FCU and in one family with MWS. The gene is expressed in peripheral blood leukocytes and encodes a protein called “cryopyrin,” which contains several distinct motifs, including a pyrin domain (also found in the marenostrin/pyrin encoded by the MEFV gene involved in FMF [Masumoto et al. 2001]), a central nucleotide-binding site (NACHT subfamily; for explanation of the “NACHT” acronym, see the “Discussion” section) domain in exon 3, and a C-terminal domain containing seven leucin-rich repeats (LRR). Recently, it has been demonstrated that cryopyrin selectively interacts with apoptosis-associated specklike protein containing a CARD domain (ASC) and that ASC linked to cryopyrin activates nuclear factor κB (NF-κB) (Manji et al. 2002).
In this article, we present five new mutations, all located in exon 3 of CIAS1, in 12 families with MWS or FCU. Of these 12 families with MWS, 3 were previously studied for the localization of MWS to the 1q44 chromosomal region (Cuisset et al. 1999).
Patients, Material, and Methods
Patients
Mutations in the CIAS1 gene were searched in 12 unrelated families with MWS or FCU (table 1). Pedigrees are shown in figure 1. Each participant was examined and provided informed written consent. Studies of some members of these families have been published elsewhere. Patients presented characteristic clinical signs, such as urticaria, arthralgia, arthritis, fever, ocular signs, and AA amyloidosis. The presence of sensorineural deafness allowed us to ascertain MWS and to exclude FCU (Muckle 1979), whereas the triggering effect of cold was characteristic of FCU.
Table 1.
List of Families Affected with MWS or FCU
| Family | Syndrome | Nationality | Reference |
| 1 | MWS | French | Lagrue et al. 1972 |
| 2 | MWS | French | Prost et al. 1976; Barrière et al. 1977 |
| 3 | FCU | French | Present study |
| 4 | FCU | Algerian | Present study |
| 5 | MWS | English | Watts et al. 1994 |
| 6 | MWS | French | Hedon et al. 1983 |
| 7 | MWS | French | Present study |
| 8 | MWS | French | Present study |
| 9 | MWS | French | Present study |
| 10 | MWS | English | Present study |
| 11 | MWS | French | Present study |
| 12 | FCU | French | Bader-Meunier et al. 1996 |
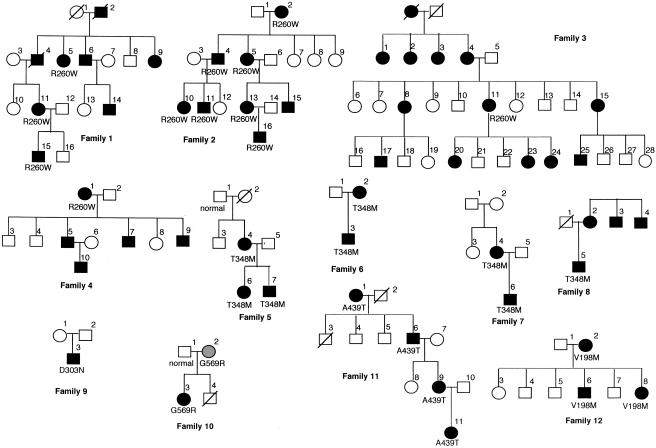
Figure 1.
Pedigree of the 12 families with MWS or FCU, with mutation analysis of CIAS1. Affected individuals are shown as blackened circles (females) or blackened squares (males). Deceased individuals are denoted by diagonal slashes. A gray circle represents an asymptomatic individual carrying an MWS mutation.
Mutation Detection
Peripheral blood was obtained from patients, and DNA was extracted as described elsewhere (Cuisset et al. 1999). A mutation search was performed on genomic DNA after PCR amplification of the nine exons of the CIAS1 gene, through use of oligonucleotides and experimental conditions described by Hoffman et al. (2001a). Free nucleotides and oligonucleotides were eliminated by exonuclease I (10 units) (USB) and shrimp alkaline phosphatase (1 unit) (USB) treatment at 37°C for 15 min, followed by incubation at 80°C for 15 min. Mutation detection was performed by fluorescent sequencing with dye-terminator chemistry (Perkin Elmer) on a 3100 automated sequencer (ABI Perkin Elmer).
The R260W mutation was analyzed in the control population by PCR amplification of genomic DNA, through use of the forward primer MWS-R260W-5′ (5′-GACCCCGATGATGAGCATTCT-3′) and the reverse modified primer MWS-R260W-3′ (5′-TCTCACAAGGCTCACCTCTC-3′). The PCR products were digested with the restriction enzyme TaqI, which revealed the presence of the R260W mutation. The other novel mutations (T348M, D303N, G569R, and A439T) either abolished or created a restriction site (table 2). This was used to test the frequency in control population.
Table 2.
CIAS1 Mutations Found in Families with MWS and FCU
| Family | NucleotideChange | AminoAcidChange | Enzyme-SiteChange | ControlFrequency |
| 1–4 | C778T | R260W | TaqI(+)a | 0/134 |
| 5–8 | C1043T | T348M | NlaIII(+) | 0/146 |
| 9 | G907A | D303N | TaqI(−) | 0/138 |
| 10 | G1705C | G569R | AciI(+) | 0/122 |
| 11 | G1315A | A439T | AciI(−) | 0/122 |
| 12 | G562A | V198M | …b | …b |
PCR was performed using modified oligonucleotides (see “Patients, Material, and Methods” section).
See Hoffmann et al. (2001a).
Microsatellite Analysis
Genotyping of the CIAS1 gene chromosomal region was performed with the following eight markers: AFMB358wg1, D1S423, AFMA274zc5, AFMB005wh9, D1S2836, D1S2215, AFM142wx1, and D1S2682. We performed PCR and automated fluorescent genotyping using standard procedures, as described elsewhere (Cuisset et al. 1999).
Single-Nucleotide Polymorphism (SNP) Analysis
The analysis of the sequences allowed us to identify four SNPs located in exon 3 of the CIAS1 gene. These SNPs are localized on the third base of codons 219 (C/T), 242 (A/G), 260 (A/G), and 434 (C/T), and they do not modify the encoded amino acid. The frequency of each allele in the control population is given in table 3.
Table 3.
CIAS1 SNPs Identified in the Present Study
| NucleotideChange | Codon | % (No.) of Alleles in theUnaffected Population |
| C/T | 219 | 92.5 (37) C, 7.5 (3) T |
| A/G | 242 | 45 (18) A, 55 (22) G |
| A/G | 260 | 10 (4) A, 90 (36) G |
| C/T | 434 | 5 (2) C, 95 (36) T |
Results
CIAS1 Mutation Analysis
A total of 12 families, including 9 with MWS and 3 with FCU, were studied (table 1). The nine unrelated families with MWS include three families previously reported to show linkage to the 1q44 region (Cuisset et al. 1999), as well as four French and two English families with a high suspicion of MWS. The three families with FCU included one Algerian and two French families. Genealogical trees are shown in figure 1. The main clinical signs of patients are summarized in table 4. Among these families, we identified a total of six mutations, all located in exon 3 of the CIAS1 gene; five of these are novel mutations and were not present in the control population (table 2). The R260W and T348M mutations are the main ones encountered in families 1–4 and in families 5–8, respectively (fig. 2). This R260W mutation was found in two families with MWS and in two families with FCU. In families 1–3, the disease segregated over four generations. Study of families 1, 2, and 5 had previously allowed us to localize MWS to the 1q44 chromosomal region (Cuisset et al. 1999). Patients in these families exhibited clinical features of the syndrome. AA amyloidosis was diagnosed by renal biopsy in family 1 (Messier et al. 1988). In families 3, 4, and 12, the patients had FCU with urticaria, arthralgia, and fever, since infancy. The inflammatory crisis in patient 11 of family 3 started after cold exposure. The triggering effect of cold was not observed in patients from families 4 and 12. Both parents of patient 1 of family 4 were from Algeria. In these three families, the disease was clearly dominantly inherited.
Table 4.
Main Clinical Signs of Patients with MWS and FCU[Note]
| Family andPatient (Sex) | Mutation | Age(years) | Age atOnseta(years) | Urticaria | Deafness | ArticularSigns | OcularSigns | Fever | Proteinuria | Renal AAAmyloidosis |
| 1: | ||||||||||
| 5 (F) | R260W | ? | ? | + | − | + | − | − | + | + |
| 11 (F) | R260W | 42 | B | + | + | + | + | − | ? | ? |
| 15 (M) | R260W | 14 | 3 | + | − | − | + | − | ? | ? |
| 2: | ||||||||||
| 2 (F) | R260W | 84 | ? | + | + | + | + | − | ? | ? |
| 4 (M) | R260W | 60 | I | + | + | + | + | − | ? | ? |
| 5 (F) | R260W | 60 | I | + | + | + | + | − | ? | ? |
| 10 (F) | R260W | 36 | 10 | + | + | + | + | − | ? | ? |
| 11 (M) | R260W | 34 | I | + | + | + | + | − | + | ? |
| 13 (F) | R260W | 39 | I | + | + | + | + | − | ? | ? |
| 16 (M) | R260W | 12 | 3 | + | − | + | + | − | ? | ? |
| 3: | ||||||||||
| 11 (F) | R260W | 53 | 1 | + | − | + | ? | + | ? | ? |
| 4: | ||||||||||
| 1 (F) | R260W | 53 | I | + | − | + | ? | + | ? | ? |
| 5: | ||||||||||
| 4 (F) | T348M | 57 | 2 | + | + | + | + | + | ? | ? |
| 6 (F) | T348M | 31 | B | + | + | + | + | + | ? | ? |
| 7 (M) | T348M | 28 | B | + | + | + | + | + | ? | ? |
| 6: | ||||||||||
| 2 (F) | T348M | 57 | ? | + | + | + | ? | − | ? | ? |
| 3 (M) | T348M | 30 | ? | + | + | + | + | + | ? | ? |
| 7: | ||||||||||
| 4 (F) | T348M | 47 | ? | + | + | + | + | + | ? | ? |
| 6 (M) | T348M | 9 | ? | + | + | + | + | + | ? | ? |
| 8: | ||||||||||
| 5 (M) | T348M | 41 | 3 | + | + | + | + | − | ? | ? |
| 9: | ||||||||||
| 3 (M) | D303N | ? | B | + | + | + | ? | − | ? | ? |
| 10: | ||||||||||
| 3 (F) | G569R | 31 | B | + | + | + | + | − | ? | ? |
| 11: | ||||||||||
| 6 (M) | A439T | ? | ? | + | + | + | + | − | ? | ? |
| 9 (F) | A439T | 36 | 3 | + | + | − | + | − | ? | ? |
| 11 (F) | A439T | 7 | 2 | + | ? | − | + | − | ? | ? |
| 12: | ||||||||||
| 2 (F) | V198M | 40 | 1 | + | − | + | − | + | ? | ? |
| 6 (M) | V198M | 13 | B | + | − | + | − | + | ? | ? |
| 12 (F) | V198M | 4 | B | + | + | − | − | − | ? | ? |
Note.— ? = unknown; + = present; − = absent.
B = at birth; I = in infancy.
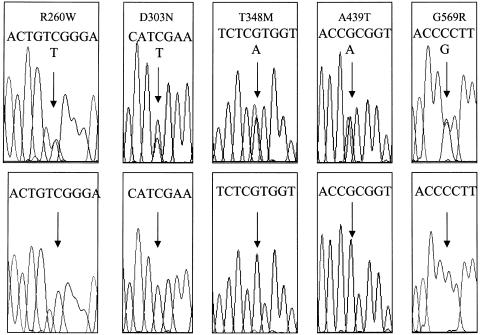
Figure 2.
DNA-sequence electrophoregrams for the five novel CIAS1 mutations identified in the present study. Upper electrophoregrams correspond to the patients, lower ones to the control. Arrows indicate the position of the mutation. Sequences showing mutations R260W and A439T are on the coding strand, and D303N, T348M, and G569R are on the opposite strand.
Families 5–8 with MWS displayed the same T348M mutation. In contrast to families with the R260W mutation, families with the T348M mutation are small. Inheritance of this mutation is found only in a two-generation family. No renal AA amyloidosis was associated with this mutation.
The D303N mutation was found in a single patient, who presented with all clinical criteria of MWS. Since the parents of this patient have no mutation, D303N can be considered to be a de novo mutation in this family.
In family 10, patient 3 is the only one affected by MWS. His mother, patient 2, shares the same mutation, but, so far, she has not developed clinical signs of MWS. The G569R mutation seems to be inherited with a low penetrance. In family 11, MWS is due to a new mutation, A439T, through four generations. Another mutation at the same codon, A439V, has been characterized previously in a family with FCU, which suggests the presence of a mutational hotspot in this region (Hoffman et al. 2001a). Patients from family 12 had FCU and carried the V198M mutation, which was previously described in a family with FCU (Hoffman et al. 2001a).
Haplotype Analysis
Do unrelated patients with the same R260W or T348M mutation share common haplotypes? Sequencing of the CIAS1 gene in patients with MWS or FCU allowed us to characterize several SNPs (table 3). We evaluated the frequency of these SNPs in the control population. We then used these intragenic SNPs and microsatellites to construct haplotypes in families 1–8. We did not find a common founder microsatellite haplotype for the R260W or T348M mutations, which suggests that these mutations appear on different alleles (fig. 3 and table 5).
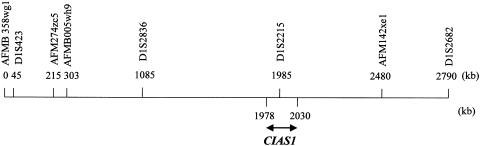
Figure 3.
Physical map of the region surrounding the CIAS1 gene, between microsatellite markers AFMB358wg1 and D1S2682 (UCSC Human Genome Project Working Draft).
Table 5.
Genotypes and Haplotypes of Patients with MWS and FCU Carrying the R260W and T348M Mutations
|
Genotype |
||||||||||||||
| Familyand Patient | AFMB358wg1 | D1S423 | AFM274zc5 | AFMB005wh9 | D1S2836 | D1S2215 | T219T (C/G) | A242A (A/G) | R260W | R260R (A/G) | T348M | S434S (C/T) | AFM142xe1 | D1S2682 |
| 1: | ||||||||||||||
| 5 | 2/2 | 1/2 | 3/5 | 2/2 | 1/4 | 1/2 | C/C | A/G | R260W | G/G | … | C/T | 3/3 | 4/9 |
| 11 | 2/3 | 1/1 | 3/7 | 2/2 | 1/1 | 2/3 | C/C | G/G | R260W | G/G | … | C/C | 3/3 | 4/4 |
| 15 | 2/2 | 1/2 | 3/4 | 2/2 | 1/1 | 2/3 | C/C | A/G | R260W | G/G | … | … | 2/3 | 1/4 |
| Haplotype | 2 | 1 | 3 | 2 | 1 | 2 | C | G | R260W | G | … | C | 3 | 4 |
| 2: | ||||||||||||||
| 2 | 1/2 | 2/2 | 1/5 | 1/2 | 2/3 | 2/3 | C/T | G/G | R260W | G/G | … | … | 1/4 | 1/4 |
| 4 | 1/2 | 2/2 | 1/7 | 1/1 | 2/6 | 2/3 | C/C | A/G | R260W | G/G | … | … | 1/3 | 4/4 |
| 5 | 1/2 | 2/2 | 1/7 | 1/1 | 2/6 | 2/3 | C/C | A/G | R260W | G/G | … | C/C | 1/3 | 4/4 |
| 10 | 1/2 | 1/2 | 1/6 | 1/2 | 1/2 | 2/3 | C/C | A/G | R260W | G/G | … | … | 1/1 | 1/4 |
| 11 | 1/2 | 1/2 | 1/6 | 1/2 | 1/2 | 2/3 | C/C | A/G | R260W | G/G | … | C/C | 1/1 | 1/4 |
| 13 | 1/2 | 2/2 | 1/8 | 1/2 | 1/2 | 2/3 | C/C | G/G | R260W | G/G | … | … | 1/1 | 2/4 |
| 16 | 1/2 | 2/2 | 1/2 | 1/3 | 2/4 | 2/3 | C/C | G/G | R260W | G/G | … | … | 1/3 | 4/4 |
| Haplotype | 1 | 2 | 1 | 1 | 2 | 2 | C | G | R260W | G | … | C | 1 | 4 |
| 5: | ||||||||||||||
| 4 | 2/2 | 1/2 | 2/3 | 1/2 | 1/6 | 3/? | C/C | A/A | … | G/G | T348M | C/C | 3/4 | 3/4 |
| 6 | 2/3 | 2/2 | 1/3 | 1/2 | 4/6 | 3/3 | C/C | A/A | … | G/G | T348M | C/C | 1/4 | 4/4 |
| 7 | 2/2 | 1/2 | 2/3 | 1/2 | 4/6 | 3/3 | C/C | A/A | … | G/G | T348M | … | 1/4 | 4/4 |
| Haplotype | 2 | 2 | 3 | 1 | 6 | 3 | C | A | … | G | T348M | C | 4 | 4 |
| 6: | ||||||||||||||
| 2 | 2/2 | 3/5 | 5/9 | 1/5 | 9/10 | 3/3 | … | … | … | … | T348M | … | 3/3 | 3/4 |
| 3 | 2/2 | 3/7 | 9/11 | 1/2 | 9/10 | 3/3 | C/C | G/G | … | G/G | T348M | C/C | 3/3 | 3/3 |
| Haplotype | 2 | 3 | 9 | 1 | 9 | 3 | C | G | G | T348M | C | 3 | 3 | |
| 3: | ||||||||||||||
| 11 | … | … | … | … | … | … | C/C | A/G | R260W | G/G | … | … | … | … |
| 4: | ||||||||||||||
| 1 | … | … | … | … | … | … | C/C | G/G | R260W | A/G | … | C/C | … | … |
| 7: | ||||||||||||||
| 6 | … | … | … | … | … | … | C/T | G/G | … | G/G | T348M | C/C | … | … |
| 8: | ||||||||||||||
| 5 | … | … | … | … | … | … | C/C | A/G | … | A/G | T348M | C | … | … |
Discussion
The role of CIAS1 in MWS and FCU was originally discerned on the basis of different missense mutations found in three families with FCU and in only one family with MWS (Hoffmann et al. 2001a). The present study presents the largest series of families and novel mutations reported since the discovery of the CIAS1 gene and confirms its involvement in MWS and FCU. This is the first demonstration that MWS and FCU can be due to the same mutation, R260W, implying that MWS and FCU are in fact the same syndrome, in which some clinical signs, such as deafness, AA amyloidosis, and cold sensitivity, are not always present. This strongly suggests that mutations in CIAS1 do not account for a unique genetic contribution and that unknown modifier genes can influence the phenotype. This observation has already been made in patients with FMF who carry the characteristic clinical signs and the same genotype but who do not always develop AA amyloidosis. The presence of AA amyloidosis is highly influenced by the presence of polymorphisms in the SAA1 gene (Cazeneuve et al. 2000). A similar observation in rheumatoid arthritis has shown that polymorphisms in the SAA1 promoter may influence the development of AA amyloidosis (Booth et al. 1998; Moriguchi et al. 1999, 2001). The clinical signs of MWS and FCU seem to be more pronounced than expected, since, in families 3 and 4, which had recurrent febrile urticaria, the triggering effect of cold and sensorineural deafness were absent. These two families are similar to an Indian family described by McDermott et al. (2000), which presented with incomplete clinical signs of MWS and FCU and showed linkage to the same region. The spectrum of diseases associated with CIAS1 mutations is thus not restricted to MWS and FCU and will probably include some forms of familial inflammatory urticaria that, so far, have no genetic explanation.
Several families in the present study displayed the same mutation—families 1–4 (R260W) and families 5–8 (T348M)—and therefore the question of a common origin can be raised. We studied microsatellites and biallelic polymorphisms in the patients carrying the same mutation. No common haplotypes based on microsatellites were found for these two mutations. The haplotype analysis covers a 2.9-Mb region, including the CIAS1 gene, which is probably too large to allow us to observe a common origin, especially if the mutations are ancient. On the basis of SNP analysis, we did not observe significant haplotypes for the R260W mutation. Unfortunately, it was not possible to obtain genomic DNA from different patients of families 3 and 4. The R260W mutation is carried in families who do not share a common ethnic origin. Until now, MWS and FCU had been reported only in northern-European populations; however, family 4 is from Algeria, which enlarges the ethnic spectrum. We therefore suspect that the R260W mutation appears on different chromosomes. Among families 5–8, which share the same common T348M mutation, preliminary results of SNP study seem to indicate that the T348M mutation appeared on different chromosomes.
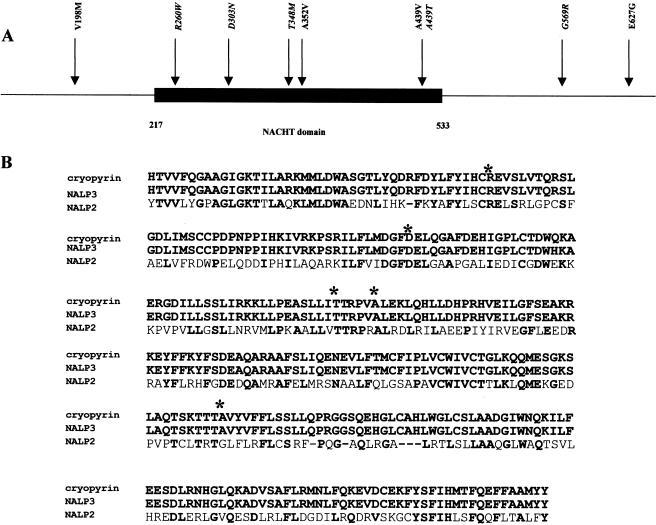
At this time, all mutations in both syndromes are missense mutations located in exon 3 of the CIAS1 gene and mainly involve the NACHT domain (fig. 4). The “NACHT” family acronym was recently used by Koonin and Aravind (2001) and is based on the following protein names: NAIP (neuronal apoptosis inhibitor protein), CIIA (MHC class II transcription activator CIITA), HET-E (bacterial nucleotide triphosphatase protein) and TP1 (telomerase-associated protein). The existence of this new family was assessed with the identification of a new proapoptotic protein—CARD4, which contains a nucleotide triphosphatase protein—that has a highly significant sequence similarity to CIITA and that activates NF-κB. The role of the NACHT domain is not actually known, but mutations in genes encoding a NACHT domain have recently been found to be involved in inflammatory diseases such as Blau syndrome and Crohn disease (Hugot et al. 2001; Miceli-Richard et al. 2001; Ogura et al. 2001). The highest degree of homology between NACHT domains has been found between cryopyrin, NALP2, and NALP3 (fig. 4). Mutations observed in cryopyrin are located on amino acids conserved among these three proteins. Of the 14 mutations responsible for MWS and FCU, 12 described so far consist of the replacement of either a cytosine by a thymine or a guanine by an adenine. This suggests that exon 3 of CIAS1 contains several mutational hotspots, and it supports the hypothesis that deamination of methylcytosines in CpG sites represents one major mechanism underlying the occurrence of MWS and FCU. This hypothesis could allow us to clarify several points: (1) two de novo mutations were found in four families (Hoffman et al. 2001a); (2) transmission of the disease phenotype was associated with the T348M mutation only over two generations; and (3) high mutability of two codons was observed—codon 439, where two different mutations, A439T and A439V, were found (GCG→ACG [present study] and GCG→GTG [Hoffman et al. 2001a], respectively), and codon 260, where a R260W mutation (CGG→TGG) and a frequent polymorphism (CGG→CGA) were observed.
Figure 4.
A, Exon 3 of CIAS1 (GenBank accession number AF427617). The blackened box represents the NACHT domain, and all mutations involved in MWS and FCU are shown. Mutations in italics are those described in the present study. B, Alignment of the NACHT-domain amino acid sequence from human cryopyrin, NALP2, and NALP3 (GenBank accession numbers AAG30289 and AF418985). Asterisks (*) represent positions of mutations in the NACHT domain. Amimo acid homologies are represented by boldface characters.
Discovery of involvement of the CIAS1 gene in MWS and in FCU confirms the genetic heterogeneity of dominantly inherited forms of recurrent fevers. The clinical spectrum of diseases associated with CIAS1 mutations is not limited to MWS and FCU and includes another form of familial urticaria, which does not meet the clinical criteria for MWS and FCU. The new diagnostic test will help in the clinical diagnosis and management of the conditions in patients affected with inflammatory recurrent syndromes, including those who belong to an ethnic group known to be at risk for FMF.
Acknowledgments
We are grateful to the family members for agreeing to participate in this study. We thank Chankannira Sân and Nicolas Lebrun, for technical assistance, and Luisa Dandolo and Jean-Pierre Hardelin, for critical reading of the manuscript. N.L.D. is a recipient of a fellowship from La Fondation de la Recherche Médicale. The French Hereditary Recurrent Inflammatory Disorder Study Group is supported by grants from the Institut National de la Santé et de la Recherche Médicale and from the Association Française Contre les Myopathies. The research on MWS was performed with financial support from Contrat de Recherche Clinique grant CRC 950162, 1995; from the Délégation à la Recherche Clinique; and from Assistance Publique-Hôpitaux de Paris.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CIAS1 [accession number AF427617], NALP2 [accession number AAG30289], and NALP3 [accession number AF418985])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MWS [MIM 191900], FCU [MIM 120100], FMF [MIM 249100], HIDS [MIM 260920], and TRAPS [MIM 142680 and MIM 134610])
- UCSC Human Genome Project Working Draft, http://genome.cse.ucsc.edu/
References
- Bader-Meunier B, Venencie PY, Vieillefond A, Le Touze P, Dommergues JP (1996) Hypergammaglobulinemia D and familial urticaria in children. Ann Dermatol Venereol 123:398–400 [PubMed] [Google Scholar]
- Barriere H, Prost A, Wallez B (1977) Syndrome de Muckle et Wells. Ann Dermatol Venereol 104:664–666 [PubMed] [Google Scholar]
- Booth DR, Booth SE, Gillmore JD, Hawkins PN, Pepys MB (1998) SAA1 alleles as risk factors in reactive systemic AA amyloidosis. Amyloid 5:262–265 [DOI] [PubMed] [Google Scholar]
- Cazeneuve C, Ajrapetyan H, Papin S, Roudot-Thoraval, Geneviéve D, Mndjoyan E, Papazian M, Sarkisian A, Babloyan A, Boissier B, Duquesnoy P, Kouyoumdjian J-C, Girodon-Boulandet E, Grateau G, Sarkisian T, Amselem S (2000) Identification of the MEFV-Independent modifying genetic factors for familial Mediterranean fever. Am J Hum Genet 67:1136–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuisset L, Drenth JPH, Berthelot JM, Meyrier A, Vaudour G, Watts R, Scott DGI, Nicholls A, Pavek S, Vasseur C, Beckmann JS, Delpech M, Grateau G (1999) Genetic linkage of the Muckle-Wells syndrome to chromosome 1q44. Am J Hum Genet 65:1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpech M, Grateau G (2001) Genetically determined recurrent fevers. Curr Opin Immunol 13:539–542 [DOI] [PubMed] [Google Scholar]
- Drenth JPH, Van der Meer JWM (2001) Hereditary periodic fever. N Engl J Med 345:1748–1756 [DOI] [PubMed] [Google Scholar]
- Hedon V, Kaplan C, Vaudour G, Muller JY (1983) The Muckle-Wells syndrome and the major histocompatibility complex. Tissue Antigens 21:318–319 [DOI] [PubMed] [Google Scholar]
- Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD (2001a) Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet 29:301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman HM, Wanderer AA, Broide DH (2001b) Familial cold inflammatory syndrome: phenotype and genotype of an autosomal dominant periodic fever. J Allergy Clin Immunol 108:615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman HM, Wright FA, Broide DH, Wanderer AA, Kolodner RD (2000) Identification of a locus on chromosome 1q44 for familial cold urticaria. Am J Hum Genet 66:1693–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot J-P, Chamaillard M, Zouali H, Lesage S, Cézard J-P, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig L, Gower-Rousseau C, Macry J, Colombel J-F, Sahbatou M, Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603 [DOI] [PubMed] [Google Scholar]
- Kile RL, Rusk HA (1940) A case of cold urticaria with unusual family history. JAMA 114:1067–1068 [Google Scholar]
- Koonin EV, Aravind L (2001) The NACHT family—a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem Sci 5:223–224 [DOI] [PubMed] [Google Scholar]
- Lagrue G, Vernant JP, Revuz J, Touraine R, Weil B (1972) Syndrome de Muckle et Wells. Cinquième observation familiale. Nouv Presse Med 1:2223–2226 [PubMed] [Google Scholar]
- Manji GA, Wang L, Geddes BJ, Brown M, Merriam S, Al-Garawi A, Mak S, Lora JM, Briskin M, Jurman M, Cao J, DiStefano PS, Bertin J (2002) PYPAF1: a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-κB. J Biol Chem 277:11570–11575 [DOI] [PubMed] [Google Scholar]
- Masumoto J, Taniguchi S, Sagara J (2001) Pyrin N-terminal homology domain-and caspase recruitment domain-dependent oligomerization of ASC. Biochem Biophys Res Commun 280:652–655 [DOI] [PubMed] [Google Scholar]
- McDermott MF, Aganna E, Hitman A, Ogunkolade BW, Booth DR, Hawkins PN (2000) An autosomal dominant periodic fever associated with AA amyloidosis in a north Indian family maps to distal chromosome 1q. Arthritis Rheum 43:2034–2040 [DOI] [PubMed] [Google Scholar]
- Messier G, Meyrier A, Rainfray M, Coste T, Callard P (1988) Overt or occult renal amyloidosis in the Muckle-Wells syndrome. Kidney Int 34:566 [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur A-M, Manouvrier-Hanu S, Häfner R, Chamaillard M, Zouali H, Thomas G, Hugot J-P (2001) CARD15 mutations in Blau syndrome. Nat Genet 29:19–20 [DOI] [PubMed] [Google Scholar]
- Moriguchi M, Terai C, Kaneko H, Koseki Y, Kajiyama H, Uesato M, Inada S (2001) A novel single-nucleotide polymorphism at the 5′-flanking region of the SAA1 associated with risk of type AA amyloidosis secondary to rheumatoid arthritis. Arthritis Rheum 44:1266–1272 [DOI] [PubMed] [Google Scholar]
- Moriguchi M, Terai C, Koseki Y, Uesato M, Nakajima A, Inada S, Nishinarita M, Uchida S, Nakajima A, Kim SY, Chen CL, Kamatani N (1999) Influence of genotypes at SAA1 and SAA2 loci on the development and length of latent period of secondary AA amyloidosis in patients with rheumatoid arthritis. Hum Genet 105:360–366 [DOI] [PubMed] [Google Scholar]
- Muckle TJ (1979) The Muckle-Wells syndrome. Br J Dermatol 100:87–92 [DOI] [PubMed] [Google Scholar]
- Muckle TJ, Wells M (1962) Urticaria deafness and amyloidosis: a new heredo-familial syndrome. Q J Med 31:235–248 [PubMed] [Google Scholar]
- OguraY, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411:603–606 [DOI] [PubMed] [Google Scholar]
- Prost A, Barriere H, Legent F, Cottin S, Wallez B (1976) Rhumatisme intermittent révélateur d'un syndrome familial arthrites-éruption urticarienne-surdité: syndrome de Muckle et Wells sans amylose rénale. Rev Rhum Mal Osteoartic 43:201–208 [PubMed] [Google Scholar]
- Watts RA, Nicholls A, Scott DG (1994) The arthropathy of the Muckle-Wells syndrome. Br J Rheumatol 33:1184–1187 [DOI] [PubMed] [Google Scholar]