Abstract
X-linked retinitis pigmentosa (XLRP) is a clinically and genetically heterogeneous degenerative disease of the retina. At least five loci have been mapped for XLRP; of these, RP2 and RP3 account for 10%–20% and 70%–90% of genetically identifiable disease, respectively. However, mutations in the respective genes, RP2 and RPGR, were detected in only 10% and 20% of families with XLRP. Mutations in an alternatively spliced RPGR exon, ORF15, have recently been shown to account for 60% of XLRP in a European cohort of 47 families. We have performed, in a North American cohort of 234 families with RP, a comprehensive screen of the RP2 and RPGR (including ORF15) genes and their 5′ upstream regions. Of these families, 91 (39%) show definitive X-linked inheritance, an additional 88 (38%) reveal a pattern consistent with X-linked disease, and the remaining 55 (23%) are simplex male patients with RP who had an early onset and/or severe disease. In agreement with the previous studies, we show that mutations in the RP2 gene and in the original 19 RPGR exons are detected in <10% and ∼20% of XLRP probands, respectively. Our studies have revealed RPGR-ORF15 mutations in an additional 30% of 91 well-documented families with X-linked recessive inheritance and in 22% of the total 234 probands analyzed. We suggest that mutations in an as-yet-uncharacterized RPGR exon(s), intronic changes, or another gene in the region might be responsible for the disease in the remainder of this North American cohort. We also discuss the implications of our studies for genetic diagnosis, genotype-phenotype correlations, and gene-based therapy.
Introduction
Retinitis pigmentosa (RP) is a clinically and genetically heterogeneous group of retinal degenerative diseases, characterized by night blindness, progressive restriction of the visual field, and pigmentary retinopathy. At least 28 different genetic loci have been mapped for autosomal dominant, autosomal recessive, and X-linked forms of RP (see RetNet Web site). The X-linked RP subtype (XLRP [MIM 268000]) is relatively severe, with an early age at onset and rapid progression, accounting for 10%–20% of families with RP (Bird 1975; Fishman 1978). XLRP is genetically heterogeneous, with five mapped loci: RP2 (MIM 312600) (Bhattacharya et al. 1984), RP3 (MIM 312610) (Musarella et al. 1988), RP6 (MIM312612) (Breuer et al. 2001), RP23 (Hardcastle et al. 2000) and RP24 (MIM 300155) (Gieser et al. 1998). On the basis of linkage analysis, RP2 is predicted to account for 10%–20% and RP3 for 70%–90% of XLRP (Musarella et al. 1990; Ott et al. 1990; Teague et al. 1994; Fujita et al. 1997). The RP2 and RPGR (for RP3) genes for these two major loci have now been cloned (Meindl et al. 1996; Roepman et al. 1996; Schwahn et al. 1998).
The RP2 gene is composed of five exons, encoding a ubiquitously expressed polypeptide of 350 amino acids (Schwahn et al. 1998). The N-terminus of the protein is homologous to cofactor C, which is involved in β-tubulin folding (Tian et al. 1996). A majority of the mutations in RP2 are localized in the cofactor C homologous domain and are predicted to generate a truncated protein (Schwahn et al. 1998; Mears et al. 1999). Further sequence analysis and secondary structure predictions show that residues 207–244 of the RP2 protein are homologous to a pig microtubule-associated protein, the γ-subunit of T-complex, and residues 250–317 show homology to NM23, a member of the nucleoside diphosphate kinase family (Miano et al. 2001). In transiently transfected cultured cells, the RP2 protein is localized to the plasma membrane, and mutations in the first five amino acids disrupt this localization (Chapple et al. 2000; Schwahn et al. 2001). These studies suggest that RP2 is a membrane- and/or tubulin-associated chaperone or signaling protein.
RPGR, the RP3 gene, spans 70 kb and was originally reported to include 19 exons encoding a protein of 815 amino acids. The N-terminal region of RPGR contains tandem repeats similar to RCC1 (called RCC1-like domain or RLD), which is a guanine nucleotide exchange factor (GEF) for Ran-GTPase (Ohtsubo et al. 1987; Drivas et al. 1990). On the basis of RCC1 crystal structure (Renault et al. 1998), the RPGR protein is predicted to have a seven-blade β-propeller structure and to function as a GEF for a small GTP-binding protein. Since the original reports (Meindl et al. 1996; Roepman et al. 1996), several groups have identified new exons and multiple RPGR transcripts (at least 14) with extensive alternative splicing (Yan et al. 1998; Kirschner et al. 1999; Vervoort et al. 2000; Kirschner et al. 2001). One of these new exons is ORF15, which is contiguous with exon 15 (located in the original intron 15). It spans 1.6 kb, is highly repetitive, and encodes a putative glutamic acid– and glycine-rich domain. ORF15 was also shown to be a mutational hot spot, with presumed mutations in 28 of 47 patients with XLRP (Vervoort et al. 2000). Inherent complexity generated by extensive alternative splicing has produced inconsistent results of RPGR intracellular localization and made the functional studies difficult. A mouse model generated by an in-frame deletion of Rpgr exons 4–6 showed a phenotype of slow cone and rod degeneration (Hong et al. 2000). Two proteins, PDE-δ and RPGR-interacting protein (RPGRIP), have been identified by the yeast two-hybrid method (Linari et al. 1999; Boylan and Wright 2000; Roepman et al. 2000b). The RPGR and RPGRIP proteins have been localized to the connecting cilia and rod outer segments (Hong et al. 2001; Roepman et al. 2000a).
Consistent with genetic studies, mutations in RP2 are observed in 7%–18% of XLRP (Hardcastle et al. 1999; Mears et al. 1999). However, the reported mutation screens have shown a remarkably low frequency of disease-causing sequence changes (11%–26%) in RPGR (Buraczynska et al. 1997; Sharon et al. 2000). RPGR-ORF15 was recently reported to account for 60% of missing mutations in a European XLRP cohort (Vervoort et al. 2000). Recently, RPGR-ORF15 mutations have also been described in families with X-linked cone degeneration (Mears et al. 2000; Demirci et al. 2002; Yang et al. 2002). These previous investigations, however, undertook limited mutation screenings of relatively small patient cohorts. To determine the spectrum of mutations in a large XLRP cohort from North America, we performed a comprehensive analysis of the RP2 and RPGR genes, including all reported exons and their 5′ upstream regions. The results demonstrate putative disease-causing mutations in 56 (62%) of 91 well-documented North American families with XLRP and in 100 (43%) of the total of 234 probands analyzed.
Subjects, Material, and Methods
Patients and Families
We have performed mutation analysis in a cohort of 234 putative XLRP families of predominantly (>95%) white ancestry from North America. Although many families are believed to have mixed ethnicity, we can be certain only of five Hispanic and two African American families in our cohort. Of the 234 families, 160 were referred by collaborating clinicians, and 74 were obtained by referral from the Foundation Fighting Blindness as part of the Retinitis Pigmentosa Research Center (RPRC). The diagnosis of XLRP in the clinician-referred families was made by ocular examination, visual field testing, and/or electroretinograms (ERGs), as well as by pedigree analysis (Jacobson et al. 1989; Sieving 1995). For individuals referred by the RPRC, the clinical information was less complete and the diagnosis was based on ophthalmic examination and pedigree data. Families with XLRP were subdivided into three categories that reflected the strength of the clinical documentation and the family history of disease (see table 5). Of the clinician-referred patients, 39% had a family history with at least two generations of affected males that were related through an unaffected or carrier female (total of 91; class A). An additional 38% were referred either by clinicians or through the RPRC and reported a family history of two or more affected male subjects, in a single or multiple generations (total of 88; class B). The remaining probands were the only affected males in the family with RP (simplex males), but, in most instances, were given the tentative diagnosis of XLRP on the basis of clinical presentation (e.g., early onset and severity of disease) (total of 55; class C).
Table 5.
Distribution of Mutations in the North American XLRP Cohort
|
No. (%) of Mutations Found in |
|||||
| Category | No. ofFamilies | RP2 | RPGR | ORF15 | Total |
| A | 91 | 6 (7) | 23 (25) | 27 (30) | 56 (62) |
| B | 88 | 6 (7) | 6 (7) | 16 (18) | 26 (32) |
| C | 55 |
3 (5) |
5 (9) |
8 (15) |
16 (29) |
| Total | 234 | 15 (6) | 34 (15) | 51 (22) | 100 (43) |
| Clinician-referred | 160 | 12 (8) | 26 (16) | 42 (26) | 80 (50) |
| RPRC-referred | 74 | 3 (4) | 8 (11) | 9 (12) | 20 (27) |
Informed consent was obtained from patients and healthy subjects after the nature of the procedures had been fully explained. The research protocols were approved by the institutional review boards of the University of Michigan and all other collaborating institutions and were in accordance with the Declaration of Helsinki. Referring clinicians provided control DNA samples from unaffected individuals with no family history of retinal disease. DNA was extracted from lymphocytes by use of standard procedures.
Mutation Analysis
Of the 234 families, 49 were previously screened for RPGR (Buraczynska et al. 1997) and 63 for RP2 (Mears et al. 1999). In the present study, we performed the mutation screening of 185 probands for RPGR and 171 for RP2 by directly sequencing the RPGR (including ORF15) and RP2 genes, including their 5′ upstream regions. The new mutations identified in this study were combined with the previously reported mutations for a comprehensive cohort analysis. Eighteen primer sets for RPGR-promoter exon 19 (Meindl et al. 1996), two primer sets for ORF15 (Vervoort et al. 2000), and five primer sets for RP2 exons (Schwahn et al. 1998) were synthesized at the Biomedical Research Core Facility of the University of Michigan and/or by Invitrogen. The primer sets amplify each exon and at least 25 bp of intronic sequence on either end. The primer pairs for the 5′ upstream regions of RP2 and RPGR were designed from the genomic sequences (GenBank accession numbers NT_011572 [RP2] and AJ318463 [RPGR]) and are as follows: RP2-1F: 5′-TTGCTCGAGAGGCTTTGATT-3′; RP2-1R: 5′-GACTTCAGCTATCCGCGTTC-3′; RPGR-1F: 5′-GACTAGCTGCCCACATGCTTAAAC-3′; and RPGR-1R: 5′-GGATGGGGGAGAACGGGAAA-3′. Amplified products were purified either with Qiaquick columns (Qiagen) or with the MultiScreen PCR system (Millipore). Products were sequenced by the University of Michigan DNA sequencing core using reagents (BigDye version 1) and automated sequencers (models 373, 377, and 3700) from Applied Biosystems, according to the manufacturer’s protocols. Sequence variations were confirmed using the ThermoSequenase Cycle Sequencing Kit (Amersham Pharmacia Biotech). Sequences were aligned using Sequencher (Gene Codes). Mutations were confirmed by analyzing available family members. Sequence variations were deemed putative mutations if they were predicted to result in a truncated protein, had not been previously reported as polymorphisms (Sharon et al. 2000; Vervoort et al. 2000), or were not identified in a screen of 200 normal chromosomes.
Results
Mutations and Sequence Variations in RP2
Direct sequencing of the PCR products spanning all exons, exon-intron boundaries, and the 5′ upstream region of the RP2 gene revealed nine different mutations in 10 of the 171 patients examined; of these, eight are novel mutations (table 1 and fig. 1). These mutations cosegregated with the disease in all available family members.
Table 1.
Mutations in the RP2 Gene in Affected Hemizygotes with XLRP
| Mutationa | Protein Changeb | Exon | Patient(s) |
| Deletion: | |||
| 409-411del | Ile37del | 2 | 1694 |
| 350-351del | Phe117fsTer155 | 2 | 471 |
| Insertion: | |||
| 515insG | Ser172fsTer173 | 2 | 790 |
| 670insC | Arg225fsTer234 | 2 | 1358 |
| Splice site: | |||
| IVS1+3A→G | Int1 | 115 | |
| Nonsense: | |||
| 82C→G | Tyr27Ter | 1 | 1645 |
| Missense: | |||
| 200G→A | Cys67Tyr | 2 | 1699 |
| 353G→A | Arg118His | 2 | 1324 and 1737 |
| 565T→C | Leu188Pro | 2 | 1063 |
Nucleotide positions are based on GenBank sequence (accession number NM_006915), with the first nucleotide of the initiation codon ATG designated as base 1.
Frameshift mutations are designated according to the following example: Phe117fsTer155 refers to a frameshift mutation, in which Phe117 is the first amino acid altered, with termination of the open reading frame at residue 155.
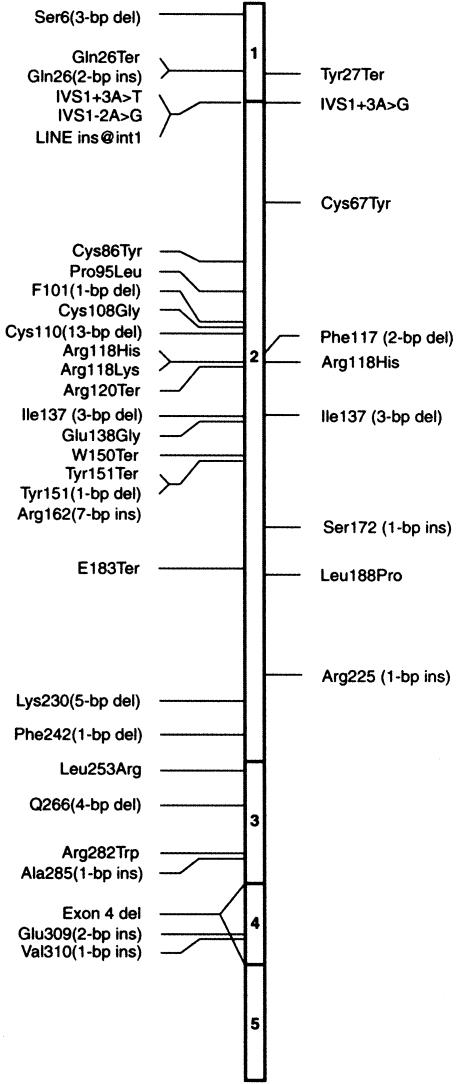
Figure 1.
Location of all known mutations in RP2. The right side designates mutations identified in this screen; all previously reported mutations are shown on the left.
Frameshift, nonsense, and splice-site mutations
Four frameshift mutations—small deletions or insertions—were detected in exon 2 in four patients. Tyr27Ter, a novel nonsense mutation, was found in one of the probands. IVS1+3A→G is a novel mutation and is predicted to affect the acceptor site of intron 1. The same nucleotide was altered in a previously reported mutation as a transversion (Sharon et al. 2000). Six of the seven mutations in this group are predicted to generate a truncated protein before or within the cofactor C homology domain. The seventh mutation has been previously reported and would result in the deletion of Ile137, a conserved residue in the cofactor C homology domain.
Missense mutations
Three missense mutations, two of which are novel, were found in four of the probands. These missense mutations were not observed in >200 chromosomes from healthy individuals. The Cys67Tyr mutation is predicted to change a potential disulfide bond–forming conserved residue to a larger neutral amino acid in the cofactor C homology domain. Arg118His and Leu188Pro are mutations in residues that are conserved between RP2 and the human cofactor C protein.
Sequence variations and polymorphisms
One polymorphic change, 844C→T, was observed in two patients in the mutation screen (table 2). This sequence variation has been reported elsewhere (Sharon et al. 2000) and was detected in both affected and unaffected individuals.
Table 2.
Sequence Variations and Polymorphisms in the RP2 and RPGR Genes
|
% in Population |
|||
| Sequence Change | Location | Patient | Unaffecteda |
| RP2: | |||
| 844C→Tb | Exon 3 | 1.6 | ND |
| RPGR: | |||
| 8C→A | Promoter | 2.2 | 2.5 |
| IVS1-15A→Gb | Intron 1 | 3.8 | ND |
| 212C→T | Exon 3 | .5 | ND |
| 1146T→A | Exon 10 | 0 | 1.4 |
| 1223G→Tb | Exon 10 | 13.0 | 4.9 |
| IVS10+16A→Gb | Intron 10 | 1.1 | ND |
| 1333G→Ab | Exon 11 | 2.2 | 9.2 |
| 1350A→Gb | Exon 11 | 1.1 | 6.3 |
| 1354A→G | Exon 11 | 0 | .5 |
| 1426A→G | Exon 11 | 1.6 | 1.9 |
| IVS12−101T→Ab | Intron 12 | 2.7 | ND |
| IVS12−100 1-bp insb | Intron 12 | 2.2 | ND |
| IVS12−97T→Cb | Intron 12 | 1.6 | ND |
| IVS12−93 2-bp insb | Intron 12 | 13.5 | ND |
| IVS13+11A→Gb | Intron 13 | 1.6 | ND |
| 1635-1637del | Exon 14 | 1.1 | ND |
| 1657C→T | Exon 14 | 1.1 | 1.4 |
| 1746G→A | Exon 14 | 2.7 | 7.2 |
| IVS16−137T→A | Intron 16 | 2.7 | ND |
| IVS17+46C→Tb | Intron 17 | 2.7 | ND |
| IVS18+11T→Cb | Intron 18 | 23.2 | ND |
| RPGR-ORF15:c | |||
| 863ins, 21 bp | .8 | ND | |
| 915-917del | .8 | ND | |
| 965-967del | 7.4 | ND | |
| 1052-1072dup | 1.7 | ND | |
| 1221-1235del | .8 | ND | |
| 1299-1310del | 9.1 | ND | |
| 1309-1320del | .8 | ND | |
Mutations and Sequence Variations in RPGR
Analysis of RPGR exons 1–19 revealed eighteen different mutations in 19 of the 185 independent XLRP patients that were examined (table 3 and fig. 2). All mutations cosegregated with the disease in available family members.
Table 3.
Mutations in the RPGR Gene in Affected Hemizygotes with XLRP
| Mutationa | Protein Changeb | Exon | Patient(s) |
| Deletion: | |||
| 541-542del | Phe162fsTer165 | 6 | 913 |
| 688-698del | Glu210fsTer214 | 7 | 310 |
| 830-832del | Leu258del | 7 | 652 |
| 1431-1432del | Cys458fsTer461 | 11 | 774 |
| 1744-1745del | His562fsTer581 | 14 | 688 |
| Insertion: | |||
| 1335ins 2bp | Ile407fsTer452 | 10 | 770 and 413 |
| Splice site: | |||
| IVS4+1 G→C | Int4 | 163 | |
| IVS10+2 T→C | Int10 | 1689 | |
| Nonsense: | |||
| 474G→T | Glu139Ter | 5 | 333 |
| 551G→A | Trp164Ter | 6 | 102 |
| 1293C→T | Arg412Ter | 10 | 725 |
| 1404C→T | Arg449Ter | 11 | 902 |
| Missense: | |||
| 438A→G | Arg127Gly | 5 | 1745 |
| 576G→C | Gly173Arg | 6 | 1734 |
| 808G→A | Cys250Tyr | 7 | 555 |
| 859G→A | Gly266Arg | 8 | 1708 |
| 924A→G | Glu285Gly | 8 | 1661 |
| 1366A→G | Gly436Asp | 11 | 767 |
Nucleotide positions and nomenclature are based on the work of Meindl et al. (1996) and Buraczynska et al. (1997).
Frameshift mutations are designated according to the following example: Phe162fsTer165 refers to a frameshift mutation, in which Phe162 is the first amino acid altered, with termination of the open reading frame at residue 165.
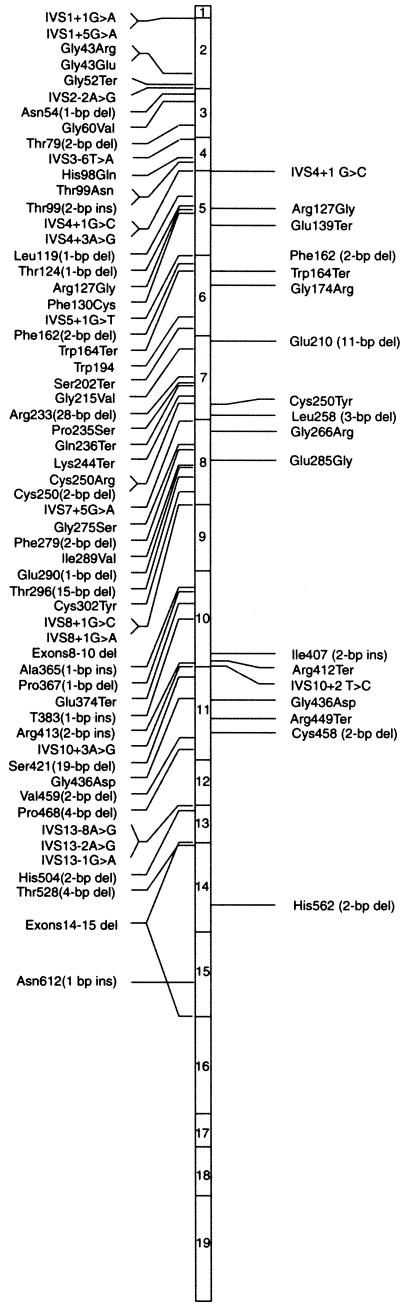
Figure 2.
Location of all known mutations in RPGR. The right side designates mutations identified in this screen; all previously reported mutations are shown on the left.
Frameshift, nonsense, and splice-site mutations
Of the 18 mutations, 11 (five frameshift, two splice-site, and four nonsense mutations) are predicted to result in a truncated protein. Two of the frameshift mutations have been reported previously, whereas four are novel. Nine of the mutations are predicted to truncate a part of the RLD. Two other mutations are predicted to truncate the protein after exon 11 or 14, thereby leaving the RLD intact but deleting the entire ORF15 region. An additional previously reported mutation is predicted to delete the Leu258 codon.
Two mutations, one of which is novel, were detected in conserved residues at intron/exon splice sites. One of these is in the donor site of intron 4, and the other is in the donor site of intron 10. Four nonsense mutations, three of which are novel, were identified in four independent patients. Each of these is predicted to result in a truncation of the RPGR protein prior to exon 12 and delete a part of RLD.
Missense mutations
In this mutation screen, we identified six missense mutations that were not detected in at least 200 normal chromosomes. One of these, Arg137Gly, has been previously reported, whereas the remaining five are novel. Gly173Arg affects a residue conserved between RPGR and RCC1, whereas Cys250Tyr, Gly266Arg, Glu285Gly, and Gly436Asp mutations alter residues in the RLD, supporting the importance of this domain in RPGR function.
Sequence variations and polymorphisms
Twenty-one sequence variations, nine of which are novel, were identified in this study. Table 2 lists the variations and their frequency in our normal and patient cohort.
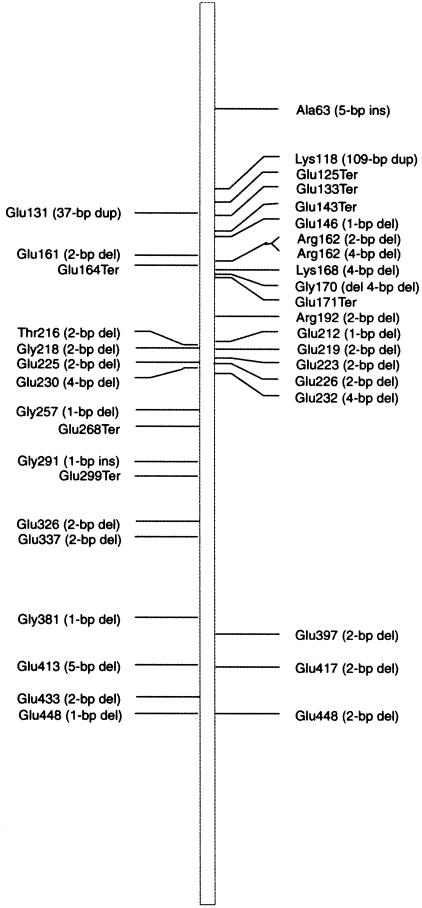
Mutations in RPGR-ORF15
The RPGR exon ORF15 was analyzed for mutations by direct sequencing of PCR products with multiple internal primers. Our analysis revealed 21 different mutations in 41 of the 195 patients examined and includes 17 frameshift and four nonsense mutations (table 4 and fig. 3). All of the frameshift mutations alter the predicted protein sequence for at least 19 amino acids prior to termination. One of these is predicted to truncate the RPGR protein after 86 altered amino acids.
Table 4.
Mutations in RPGR-ORF15 in Affected Hemizygotes with XLRP
| Mutationa | Protein Changeb | No. ofPatientsAffected |
| Deletion: | ||
| 432del | Glu146fsTer232 | 1 |
| 481-2del | Arg162fsTer184 | 9 |
| 481-4del | Arg162fsTer231 | 1 |
| 500-3del | Lys168fsTer231 | 1 |
| 504-7del | Gly170fsTer231 | 1 |
| 571-2del | Arg192fsTer250 | 1 |
| 632del | Glu212fsTer231 | 1 |
| 651-2del | Glu219fsTer250 | 6 |
| 653-4del | Glu219fsTer250 | 1 |
| 659-670del | Glu223fsTer250 | 1 |
| 673-4del | Glu226fsTer250 | 2 |
| 690-3del | Glu232fsTer235 | 3 |
| 1183-4del | Glu397fsTer495 | 1 |
| 1243-4del | Glu417fsTer495 | 1 |
| 1338-9del | Glu448fsTer495 | 1 |
| Insertion: | ||
| 186ins, 5bp | Ala63fsTer113 | 1 |
| 353-461dup | 1 | |
| Nonsense: | ||
| 370G→T | Glu125Ter | 1 |
| 394G→T | Glu133Ter | 1 |
| 424G→A | Glu143Ter | 3 |
| 508G→T | Glu171Ter | 3 |
Nucleotide positions and nomenclature are based on the work of Vervoort et al. (2000).
Frameshift mutations are designated according to the following example: Glu146fsTer232 refers to a frameshift mutation, in which Glu146 is the first amino acid altered, with termination of the open reading frame at residue 232.
Figure 3.
Location of all known mutations in RPGR-ORF15. The right side designates mutations identified in this screen; all mutations reported by Vervoort et al. (2000) are shown on the left.
Discussion
This is the first comprehensive report of mutation screening in a large cohort affected with XLRP in which the RP2 and RPGR (including ORF15) genes and their 5′ upstream regions were directly sequenced in affected male probands. Our results establish a molecular definition of XLRP in a North American population and provide essential information for the development of genotype-phenotype correlations for this heterogeneous disease. Together with our previous studies (Buraczynska et al. 1997; Fujita et al. 1997; Mears et al. 1999), we have performed a systematic analysis of 234 independent probands with a confirmed diagnosis of RP and probable X-linked recessive inheritance and have identified potential disease-causing mutations in approximately half of the families. Several of the reported mutations in the two genes are novel (see figs. 1, 2, and 3). Almost all of the newly discovered mutations are localized in the cofactor C homology domain of RP2 or in the RLD or exon ORF15 of RPGR, illustrating their functional significance.
In our studies, the observed mutation frequency in RP2 and original RPGR exons 1–19 is 6% and 15%, respectively, and thus is in concordance with previous mutation screens (Buraczynska et al. 1997; Fujita et al. 1997; Hardcastle et al. 1999; Mears et al. 1999; Sharon et al. 2000). Surprisingly, however, we could identify RPGR-ORF15 mutations in only 22% of our cohort, which is considerably lower than the published report of 60% (Vervoort et al. 2000). Interestingly, no causative mutations were detected in the 5′ upstream regions of the two genes. Although all the probands in our study cohort carry a diagnosis of typical RP, the nature of several family structures, variations in disease manifestation in carrier females, and clinical phenotypes might support an autosomal pattern of inheritance in some of the families. Consequently we divided our cohort into categories defined by the evidence supporting X-linked inheritance (table 5). In category A, where the evidence for X-linked inheritance was the strongest, causative mutations were identified in 62% of patients (7% in RP2, 25% in RPGR, and 30% in RPGR-ORF15). The disease locus in several category A families has been tightly linked to RP3; however, this screen failed to identify causative RPGR mutations (Fujita et al. 1997; D. K. Breuer, B. M. Yashar, and A. Swaroop, unpublished data). In class B, only 32% of the patients had mutations (7% in RP2, 7% in RPGR, and 18% in RPGR-ORF15). This frequency is similar to class C (single affected males) where 29% of patients have mutations (7% in RP2, 7% in RPGR, and 18% in RPGR-ORF15). The latter finding is particularly significant since it suggests that mutations in RP2 and RPGR may account for disease in a considerable proportion of simplex cases that account for almost 50% of RP. We estimate that RPGR mutations would therefore be responsible for 15%–20% of all cases of RP, higher than any other single genetic locus.
Several possibilities can be put forward to explain why no mutation has been detected in almost 40% of even class A XLRP patients.
-
1.
We examined >300 bp of the 5′ upstream regions of the RP2 and RPGR genes. Sequence changes in further upstream promoter or enhancer elements may affect gene expression.
-
2.
Disease-associated mutations present within intronic sequences may alter the splicing pattern. Mutations at the branch-point are known to result in defective splicing in the FBN2 gene (Maslen et al. 1997).
-
3.
RPGR is a large and highly complex gene in which new exons continue to be identified (Kirschner et al. 1999; Vervoort et al. 2000). We had also screened another reported exon 15a (Kirschner et al. 1999) in 43 families, but we did not detect any sequence variation. An additional exon, ORF14, was identified in the same analysis that yielded ORF15 (Vervoort et al. 2000); however, it was not screened in this study, since a previous study found no evidence for disease mutations. Another mutational hotspot in an as-yet-unidentified RPGR exon remains a possibility.
-
4.
Another gene tightly linked to the region offers an alternative scenario. RP6, a locus for which the gene has yet to be cloned, is located <5 cM from RP3 (Breuer et al. 2001) and is a valid candidate for future investigations.
The findings in this study may reflect a population difference, with the disease in our North American cohort attributable to a mutational hotspot in another exon of RPGR or another gene.
Since the vast majority of RPGR or RP2 mutations are predicted to result in a truncated protein product, our study strongly argues that the XLRP disease state is caused by the loss of function of RP2 or RPGR. This finding has significant implications for genetic diagnosis and gene-based therapy. The utility of a genetic test is dependent on specificity (the frequency with which the test yields a negative result when the disease is absent) and sensitivity (the frequency with which the test yields a positive result when the disease is present) (Gelehrter et al. 1998). Because of extensive alternative splicing and a lack of identifiable RPGR mutations even in genetically-linked RP3 families, molecular testing by direct sequencing will have low specificity; nevertheless, it might be possible to greatly enhance the informativeness of testing by analyzing protein products in lymphocytes of at-risk males and carrier females. Protein-truncation tests have been applied successfully in studies of the BRCA1 gene for breast cancer, the APC gene for colon cancer, and the CHM gene mutated in choroideremia (MacDonald et al. 1998; Balhausen 2000; Moore et al. 2000). With suitable RPGR- or RP2-specific antibodies, it may also be possible to identify mutations in the remainder of our cohort, if such changes alter the protein structure. We also suggest that a gene-based therapy that delivers a wild-type RPGR or RP2 gene to the retina might be sufficient to correct the gene defect. Early applications of gene therapy to the treatment of retinopathies have provided strong proof of principle (Bennett and Maguire 2000; Hauswirth and Beaufrere 2000; Bessant et al. 2001). A successful movement of this approach from research to clinical application will also depend on understanding the kinetics of the retinal degenerative process and administering the therapy early in the disease progression, while there are still viable photoreceptors. XLRP, like most genetic diseases, displays inter- and intrafamilial variability in the average age at onset of disease (Bird 1975; Fishman et al. 1988). Early molecular diagnosis of at-risk males should allow identification of the best patient candidates for therapeutic intervention. In summary, our study offers an important molecular definition of the XLRP population in North America and lays the groundwork for the future development of diagnostic testing and gene-based therapies for XLRP.
Acknowledgments
The authors sincerely appreciate the participation of the patients and the families. We wish to acknowledge a number of clinicians, researchers, and genetic counselors who assisted in various aspects of this study; these include Drs. Kirk Alek, Sten Andreasson, Jean Bennett, Monika Buraczynska, Ricardo Fujita, Jacquie Greenberg, Albert Maguire, Michael Marmur, Helen Mintz-Hittner, Rajkumar Ramesar, and Marcela Pena; Ms. Janice Edwards; Mr. Eric Krivchenia; Ms. Kathleen O’Brien; Ms. Gina Osland; and Ms. Diana Wheaton. Special thanks are due to The Foundation Fighting Blindness, of Owings Mills, MD, for supporting these investigations. This research was also supported, in part, by the National Institutes of Health (grants EY07961, EY07003, EY05627, and EY05235), the Food and Drug Administration (grant FDR-001232), Research to Prevent Blindness, the Grant Healthcare Foundation, the Le Bonheur Children’s Medical Center Research Program, the British Retinitis Pigmentosa Society, and the Scientific and Technical Research Council of the Turkish Republic (TUBITAK) and the State Planning Organization of Turkey (DPT) (grant 00K120660).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- National Center for Biotechnology Information, http://www4.ncbi.nlm.nih.gov/ (for descriptions of genes and sequences)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RP2 [MIM 312600], RP3 [MIM 312610], RP6 [MIM312612], RP24 [MIM 300155], and XLRP [MIM 26800])
- RetNet, http://www.sph.uth.tmc.edu/Retnet/ (for map locations of retinal disease loci and genes)
References
- Balhausen WG (2000) Genetic testing for familial adenomatous polyposis. Ann NY Acad Sci 910:36–47 [DOI] [PubMed] [Google Scholar]
- Bennett J, Maguire AM (2000) Gene therapy for ocular disease. Mol Ther 1:501–505 [DOI] [PubMed] [Google Scholar]
- Bessant DA, Ali RR, Bhattacharya SS (2001) Molecular genetics and prospects for therapy of the inherited retinal dystrophies. Curr Opin Genet Dev 11:307–316 [DOI] [PubMed] [Google Scholar]
- Bhattacharya SS, Wright AF, Clayton JF, Price WH, Phillips CI, McKeown CM, Jay M, Bird AC, Pearson PL, Southern EM (1984) Close genetic linkage between X-linked retinitis pigmentosa and a restriction fragment length polymorphism identified by recombinant DNA probe L1.28. Nature 309:253–255 [DOI] [PubMed] [Google Scholar]
- Bird AC (1975) X-linked retinitis pigmentosa. Br J Ophthalmol 59:177–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JP, Wright AF (2000) Identification of a novel protein interacting with RPGR. Hum Mol Genet 9:2085–2093 [DOI] [PubMed] [Google Scholar]
- Breuer DK, Affer M, Andreasson S, Birch DG, Fishman GA, Heckenlively JR, Hiriyanna S, Hoffman DR, Jacobson SG, Mears AJ, Musarella MA, Redolfi E, Sieving PA, Wright AF, Yashar BM, Zucchi I, Swaroop A (2001) X-linked retinitis pigmentosa: current status. In: Anderson RE, LaVail MM, Hollyfield JG (ed) New insights into retinal degenerative diseases. Kluwer Academic/Plenum Publishers, New York, pp 11–22 [Google Scholar]
- Buraczynska M, Wu W, Fujita R, Buraczynska K, Phelps E, Andreasson S, Bennett J, Birch DG, Fishman GA, Hoffman DR, Inana G, Jacobson SG, Musarella MA, Sieving PA, Swaroop A (1997) Spectrum of mutations in the RPGR gene that are identified in 20% of families with X-linked retinitis pigmentosa. Am J Hum Genet 61:1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple JP, Hardcastle AJ, Grayson C, Spackman LA, Willison KR, Cheetham ME (2000) Mutations in the N-terminus of the X-linked retinitis pigmentosa protein RP2 interfere with the normal targeting of the protein to the plasma membrane. Hum Mol Genet 9:1919–1926 [DOI] [PubMed] [Google Scholar]
- Demirci FY, Rigatti BW, Wen G, Radak AL, Mah TS, Baic CL, Traboulsi EI, Alitalo T, Ramser J, Gorin MB (2002) X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am J Hum Genet 70:1049–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drivas GT, Shih A, Coutavas E, Rush MG, D'Eustachio P (1990) Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol Cell Biol 10:1793–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman GA (1978) Retinitis pigmentosa: genetic percentages. Arch Ophthalmol 96:822–826 [DOI] [PubMed] [Google Scholar]
- Fishman GA, Farber MD, Derlacki DJ (1988) X-linked retinitis pigmentosa: profile of clinical findings. Arch Ophthalmol 106:369–375 [DOI] [PubMed] [Google Scholar]
- Fujita R, Buraczynska M, Gieser L, Wu W, Forsythe P, Abrahamson M, Jacobson SG, Sieving PA, Andreasson S, Swaroop A (1997) Analysis of the RPGR gene in 11 pedigrees with the retinitis pigmentosa type 3 genotype: paucity of mutations in the coding region but splice defects in two families. Am J Hum Genet 61:571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelehrter T, Collins F, Ginsburg D (1998) Principles of medical genetics. Williams & Wilkins, Baltimore, MD [Google Scholar]
- Gieser L, Fujita R, Goring HH, Ott J, Hoffman DR, Cideciyan AV, Birch DG, Jacobson SG, Swaroop A (1998) A novel locus (RP24) for X-linked retinitis pigmentosa maps to Xq26-27. Am J Hum Genet 63:1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle AJ, Thiselton DL, Van Maldergem L, Saha BK, Jay M, Plant C, Taylor R, Bird AC, Bhattacharya S (1999) Mutations in the RP2 gene cause disease in 10% of families with familial X-linked retinitis pigmentosa assessed in this study. Am J Hum Genet 64:1210–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle AJ, Thiselton DL, Zito I, Ebenezer N, Mah TS, Gorin MB, Bhattacharya SS (2000) Evidence for a new locus for X-linked retinitis pigmentosa (RP23). Invest Ophth Vis Sci 41:2080–2086 [PubMed] [Google Scholar]
- Hauswirth WW, Beaufrere, L. (2000) Ocular gene therapy: quo vadis? Invest Ophth Vis Sci 41:2821–2826 [PubMed] [Google Scholar]
- Hong DH, Pawlyk BS, Shang J, Sandberg MA, Berson EL, Li T (2000) A retinitis pigmentosa GTPase regulator (RPGR)–deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc Natl Acad Sci USA 97:3649–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DH, Yue G, Adamian M, Li T (2001) A retinitis pigmentosa GTPase regulator (RPGR)–interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem 276:12091–12099 [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Yagasaki K, Feuer WJ, Roman AJ (1989) Interocular asymmetry of visual function in heterozygotes of X-linked retinitis pigmentosa. Exp Eye Res 48:679–691 [DOI] [PubMed] [Google Scholar]
- Kirschner R, Erturk D, Zeitz C, Sahin S, Ramser J, Cremers FP, Ropers HH, Berger W (2001) DNA sequence comparison of human and mouse retinitis pigmentosa GTPase regulator (RPGR) identifies tissue-specific exons and putative regulatory elements. Hum Genet 109:271–278 [DOI] [PubMed] [Google Scholar]
- Kirschner R, Rosenberg T, Schultz-Heienbrok R, Lenzner S, Feil S, Roepman R, Cremers FP, Ropers HH, Berger W (1999) RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum Mol Genet 8:1571–1578 [DOI] [PubMed] [Google Scholar]
- Linari M, Ueffing M, Manson F, Wright A, Meitinger T, Becker J (1999) The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proc Natl Acad Sci USA 96:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald IM, Mah DY, Ho YK, Lewis RA, Seabra MC (1998) A practical diagnostic test for choroideremia. Ophthalmology 105:1637–1640 [DOI] [PubMed] [Google Scholar]
- Maslen C, Babcock D, Raghunath M, Steinmann B (1997) A rare branch-point mutation is associated with missplicing of fibrillin-2 in a large family with congenital contractural arachnodactyly. Am J Hum Genet 60:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Gieser L, Yan D, Chen C, Fahrner S, Hiriyanna S, Fujita R, Jacobson SG, Sieving PA, Swaroop A (1999) Protein-truncation mutations in the RP2 gene in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet 64:897–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Hiriyanna S, Vervoort R, Yashar B, Gieser L, Fahrner S, Daiger SP, Heckenlively JR, Sieving PA, Wright AF, Swaroop A (2000) Remapping of the RP15 locus for X-linked cone-rod degeneration to Xp11.4 to p21.1, and identification of a de novo insertion in the RPGR exon ORF15. Am J Hum Genet 67:1000–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A, Carvalho MR, Achatz H, Hellebrand H, Lennon A, Migliaccio C, Porter K, Zrenner E, Bird A, Jay M, Lorenz B, Wittwer B, D'Urso M, Meitinger T, Wright A (1996) A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet 13:35–42 [DOI] [PubMed] [Google Scholar]
- Miano MG, Testa F, Filippini F, Trujillo M, Conte I, Lanzara C, Millan JM, De Bernardo C, Grammatico B, Mangino M, Torrente I, Carrozzo R, Simonelli F, Rinaldi E, Ventruto V, D'Urso M, Ayuso C, Ciccodicola A (2001) Identification of novel RP2 mutations in a subset of X-linked retinitis pigmentosa families and prediction of new domains. Hum Mutat 18:109–119 [DOI] [PubMed] [Google Scholar]
- Moore W, Bogdarina I, Patel UA, Perry M, Crane-Robinson C (2000) Mutation detection in the breast cancer gene BRCA1 using the protein truncation test. Mol Biotechnol 14:89–97 [DOI] [PubMed] [Google Scholar]
- Musarella MA, Anson-Cartwright L, Leal SM, Gilbert LD, Worton RG, Fishman GA, Ott J (1990) Multipoint linkage analysis and heterogeneity testing in 20 X-linked retinitis pigmentosa families. Genomics 8:286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarella MA, Burghes A, Anson-Cartwright L, Mahtani MM, Argonza R, Tsui LC, Worton R (1988) Localization of the gene for X-linked recessive type of retinitis pigmentosa (XLRP) to Xp21 by linkage analysis. Am J Hum Genet 43:484–494 [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Kai R, Furuno N, Sekiguchi T, Sekiguchi M, Hayashida H, Kuma K, Miyata T, Fukushige S, Murotsu T (1987) Isolation and characterization of the active cDNA of the human cell cycle gene (RCC1) involved in the regulation of onset of chromosome condensation. Genes Dev 1:585–593 [DOI] [PubMed] [Google Scholar]
- Ott J, Bhattacharya S, Chen JD, Denton MJ, Donald J, Dubay C, Farrar GJ, Fishman GA, Frey D, Gal A (1990) Localizing multiple X chromosome-linked retinitis pigmentosa loci using multilocus homogeneity tests. Proc Natl Acad Sci USA 87:701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A (1998) The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392:97–101 [DOI] [PubMed] [Google Scholar]
- Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, Ropers HH, Cremers FP, Ferreira PA (2000a) The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet 9:2095–2105 [DOI] [PubMed] [Google Scholar]
- Roepman R, Schick D, Ferreira PA (2000b) Isolation of retinal proteins that interact with retinitis pigmentosa GTPase regulator by interaction trap screen in yeast. Methods Enzymol 316:688–704 [DOI] [PubMed] [Google Scholar]
- Roepman R, van Duijnhoven G, Rosenberg T, Pinckers AJ, Bleeker-Wagemakers LM, Bergen AA, Post J, Beck A, Reinhardt R, Ropers HH, Cremers FP, Berger W (1996) Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet 5:1035–1041 [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19:327–332 [DOI] [PubMed] [Google Scholar]
- Schwahn U, Paland N, Techritz S, Lenzner S, Berger W (2001) Mutations in the X-linked RP2 gene cause intracellular misrouting and loss of the protein. Hum Mol Genet 10:1177–1183 [DOI] [PubMed] [Google Scholar]
- Sharon D, Bruns GA, McGee TL, Sandberg MA, Berson EL, Dryja TP (2000) X-linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest Ophth Vis Sci 41:2712–2721 [PubMed] [Google Scholar]
- Sieving PA (1995) Diagnostic issues with inherited retinal and macular dystrophies. Semin Ophthalmol 10:279–294 [DOI] [PubMed] [Google Scholar]
- Teague PW, Aldred MA, Jay M, Dempster M, Harrison C, Carothers AD, Hardwick LJ, Evans HJ, Strain L, Brock DJH, Bundey S, Jay B, Bird AC, Bhattacharya SS, Wright AF (1994) Heterogeneity analysis in 40 X-linked retinitis pigmentosa families. Am J Hum Genet 55:105–111 [PMC free article] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ (1996) Pathway leading to correctly folded beta-tubulin. Cell 86:287–296 [DOI] [PubMed] [Google Scholar]
- Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG, Meindl A, Meitinger T, Ciccodicola A, Wright AF (2000) Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet 25:462–466 [DOI] [PubMed] [Google Scholar]
- Yan D, Swain PK, Breuer D, Tucker RM, Wu W, Fujita R, Rehemtulla A, Burke D, Swaroop A (1998) Biochemical characterization and subcellular localization of the mouse retinitis pigmentosa GTPase regulator (mRpgr). J Biol Chem 273:19656–19663 [DOI] [PubMed] [Google Scholar]
- Yang Z, Peachey NS, Moshfeghi DM, Thirumalaichary S, Chorich L, Shugart YY, Fan K, Zhang K (2002) Mutations in the RPGR gene cause X-linked cone dystrophy. Hum Mol Genet 11:605–611 [DOI] [PubMed] [Google Scholar]