Abstract
Noonan syndrome (NS) is a developmental disorder characterized by facial dysmorphia, short stature, cardiac defects, and skeletal malformations. We recently demonstrated that mutations in PTPN11, the gene encoding the non–receptor-type protein tyrosine phosphatase SHP-2 (src homology region 2-domain phosphatase–2), cause NS, accounting for ∼50% of cases of this genetically heterogeneous disorder in a small cohort. All mutations were missense changes and clustered at the interacting portions of the amino-terminal src-homology 2 (N-SH2) and protein tyrosine phosphatase (PTP) domains. A gain of function was postulated as a mechanism for the disease. Here, we report the spectrum and distribution of PTPN11 mutations in a large, well-characterized cohort with NS. Mutations were found in 54 of 119 (45%) unrelated individuals with sporadic or familial NS. There was a significantly higher prevalence of mutations among familial cases than among sporadic ones. All defects were missense, and several were recurrent. The vast majority of mutations altered amino acid residues located in or around the interacting surfaces of the N-SH2 and PTP domains, but defects also affected residues in the C-SH2 domain, as well as in the peptide linking the N-SH2 and C-SH2 domains. Genotype-phenotype analysis revealed that pulmonic stenosis was more prevalent among the group of subjects with NS who had PTPN11 mutations than it was in the group without them (70.6% vs. 46.2%; P<.01), whereas hypertrophic cardiomyopathy was less prevalent among those with PTPN11 mutations (5.9% vs. 26.2%; P<.005). The prevalence of other congenital heart malformations, short stature, pectus deformity, cryptorchidism, and developmental delay did not differ between the two groups. A PTPN11 mutation was identified in a family inheriting Noonan-like/multiple giant-cell lesion syndrome, extending the phenotypic range of disease associated with this gene.
Introduction
Noonan syndrome (NS [MIM 163950]), which was first recognized as distinct entity almost 40 years ago, is a clinically heterogeneous disorder defined by short stature, facial dysmorphia, and a wide spectrum of congenital heart defects (Noonan 1968; Allanson 1987). The distinctive facial features consist of a broad forehead, hypertelorism, down-slanting palpebral fissures, a high-arched palate, and low-set, posteriorly rotated ears. Cardiac involvement is present in up to 90% of affected individuals. Pulmonic stenosis and hypertrophic cardiomyopathy (HCM) are the most common forms of cardiac disease in NS, but a wide range of other lesions, including atrioventricular septal defects and aortic coarctation, are also observed (Marino et al. 1999; Bertola et al. 2000). Additional relatively frequent features of NS include multiple skeletal defects (chest and spine deformities), webbed neck, mental retardation, cryptorchidism, and bleeding diasthesis. Although precise epidemiological data are not available, the prevalence of NS is estimated to be between 1:1,000 and 1:2,500 live births (Nora et al. 1974).
From a genetic point of view, NS was a poorly understood condition until recently. Autosomal dominant inheritance is apparent for the majority of families with the disorder, although there is evidence for an autosomal recessive form of NS (van der Burgt and Brunner 2000). A locus at chromosomal band 12q24 (NS1) was established in a study of two large families inheriting NS (Jamieson et al. 1994; Brady et al. 1997; Legius et al. 1998). Genetic heterogeneity was also documented on the basis of linkage exclusion (Jamieson et al. 1994). Recently, PTPN11, which encodes the non–receptor-type protein tyrosine phosphatase SHP-2 (src homology region 2-domain phosphatase–2), was identified as the NS1 disease gene, through use of a positional candidacy approach (Tartaglia et al. 2001). Missense mutations in PTPN11 accounted for 50% of cases in a cohort of 22 unrelated subjects with familial or sporadic NS (Tartaglia et al. 2001).
SHP-2 (previously named “Syp,” “SH-PTP2,” “PTP1D,” or “PTP2C”) is a member of a small subfamily of cytosolic protein tyrosine phosphatases (PTPs) that includes SHP-1 (encoded by PTPN6) and the Drosophila SHP-2–homolog corkscrew (csw). These proteins exhibit a high degree of homology in amino acid sequence; share a common structure composed of two tandemly arranged amino-terminal src-homology 2 (SH2) domains (N-SH2 and C-SH2), a PTP, and a carboxy-terminal tail (Stein-Gerlach et al. 1998; Feng 1999); and are essential during development (Kozlowski et al. 1993; Tsui et al. 1993; Tang et al. 1995; Arrandale et al. 1996; Perkins et al. 1996; Saxton et al. 1997). In contrast to SHP-1, which is expressed primarily in the hematopoietic cell lineage, SHP-2 is ubiquitously expressed and is involved in mesodermal patterning (Tang et al. 1995), limb development (Saxton et al. 2000), hematopoietic cell differentiation (Qu and Feng 1998; Qu et al. 1998), and semilunar valvulogenesis (Chen et al. 2000). SHP-2 is a key molecule in the cellular response to growth factors, hormones, cytokines, and cell adhesion molecules. It is required for activation of the Ras/mitogen-activated protein (MAP) kinase cascade induced by epidermal, fibroblast, and hepatocyte growth factors (Maroun et al. 2000; Shi et al. 2000; Cunnick et al. 2002). Depending on its subcellular localization and binding partners, SHP-2 can also negatively modulate signaling (Milarski and Saltiel 1994; Stofega et al. 2000; Maile and Clemmons 2002).
Activation of SHP-2 results from binding of the SH2 domains to short amino acid motifs containing a phosphotyrosyl residue. Although both SH2 domains positively modulate the level of phosphatase activity of SHP-2 (Pluskey et al. 1995), the N-SH2 domain has the major role in the control of its activation. Crystallographic data on SHP-2 indicate that the N-SH2 domain interacts with the PTP domain in the inactive conformation, blocking the catalytic site (Hof et al. 1998). After the N-SH2 domain binds a phosphotyrosyl residue, a conformational change that reduces the intermolecular interaction between the N-SH2 and PTP domains makes the catalytic site available to substrate. Thus, the N-SH2 domain acts as a molecular switch, activating and inactivating SHP-2. The PTPN11 mutations that cause NS clustered in the interacting portions of the N-SH2 and PTP domains, and energetics-based structural analysis suggested that these mutations would stabilize SHP-2 in the active conformation (Tartaglia et al. 2001). Thus, it was proposed that the PTPN11 mutations in NS induce a gain of function.
For the present study, we screened for PTPN11 coding-region defects in a large, well-characterized cohort of individuals with sporadic or familial NS. Our results define more accurately the range of molecular defects causing NS and establish a correlation between genotype and the cardiac phenotypes, pulmonic stenosis, and hypertrophic cardiomyopathy. In addition, we show that a family with a NS-related disorder, Noonan-like/multiple giant-cell lesion syndrome (MIM 163955), inherited a PTPN11 mutation that segregated with the phenotype.
Material and Methods
Clinical Evaluation
Subjects were examined by clinicians (M.A.P. and I.vdB.) experienced with NS. Electrocardiograms, echocardiograms, and clinical photographs were obtained routinely for the probands, as well as for most of other affected family members in the kindreds segregating the disorder. NS was diagnosed on the basis of the presence of the following major characteristics: typical facial dysmorphia, pulmonic stenosis or HCM plus abnormal electrocardiogram pattern, pectus carinatum/excavatum, height >2 SD below the mean, and cryptorchidism in male subjects. To have a diagnosis of NS, individuals with typical facial dysmorphia had to have at least one additional major feature, whereas individuals with suggestive facial findings had to have at least two other major characteristics (van der Burgt et al. 1994). HCM was diagnosed when the left-ventricular maximal end diastolic wall thickness was >1.5 cm in adults (Shapiro and McKenna 1983) or >2 SD above the mean for a given age in children (Burch et al. 1993). The clinical description of a kindred with Noonan-like/multiple giant-cell lesion syndrome was reported elsewhere (Bertola et al. 2001). Informed consent was obtained from all subjects included in the study.
Mutational Analysis
Genomic DNAs were isolated from peripheral blood lymphocytes (Gentra). The entire PTPN11 coding region (exons 1–15) was screened for mutations. For exons 2–15, PCRs were performed in a 25-μl reaction volume containing 20–80 ng genomic DNA, 1 U AmpliTaq Gold (Roche), 20 pmol each primer, 1.5 mM MgCl2, 75 μM each dNTP, and 1× PCR Buffer II (Roche), through use of a GeneAmp PCR System 9700 (Applied Biosystems). Exon 1 amplifications were performed using the GC-rich PCR System (Roche), according to the manufacturer’s specifications. Cycling parameters were as follows: 94°C for 8 min (first denaturing step); 33 cycles of 94°C for 45 s, 54–60°C (see table 1) for 30 s, and 72°C for 45 s; and 72°C for 10 min (last extension step). Primer pairs were designed to amplify exons, exon/intron boundaries, and short intron flanking stretches. Primer sequences, annealing temperatures, and sizes of PCR products are listed in table 1.
Table 1.
Primer Pairs and Annealing Temperatures Used to Amplify the PTPN11 Coding Sequence and Sizes of PCR Products
|
Primer Sequence(5′→3′) |
||||
| Exon | Forward | Reverse | Annealing Temperature(°C) | Product Length(bp) |
| 1 | GCTGACGGGAAGCAGGAAGTGG | CTGGCACCCGTGGTTCCCTC | 60 | 589 |
| 2 | ACTGAATCCCAGGTCTCTACCAAG | CAGCAAGCTATCCAAGCATGGT | 60 | 405 |
| 3 | CGACGTGGAAGATGAGATCTGA | CAGTCACAAGCCTTTGGAGTCAG | 60 | 384 |
| 4 | AGGAGAGCTGACTGTATACAGTAG | CATCTGTAGGTGATAGAGCAAGA | 58 | 447 |
| 5 | CTGCAGTGAACATGAGAGTGCTTG | GTTGAAGCTGCAATGGGTACATG | 60 | 329 |
| 6 | TGCATTAACACCGTTTTCTGT | GTCAGTTTCAAGTCTCTCAGGTC | 54 | 282 |
| 7 | GAACATTTCCTAGGATGAATTCC | GGTACAGAGGTGCTAGGAATCA | 56 | 271 |
| 8 | GACATCAGGCAGTGTTCACGTTAC | CCTTAAAGTTACTTTCAGGACATG | 57 | 350 |
| 9 | GTAAGCTTTGCTTTTCACAGTG | CTAAACATGGCCAATCTGACATGTC | 56 | 357 |
| 10 | GCAAGACTTGAACATTTGTTTGTTGC | GACCCTGAATTCCTACACACCATC | 60 | 284 |
| 11 | CAAAAGGAGACGAGTTCTGGGAAC | GCAGTTGCTCTATGCCTCAAACAG | 60 | 453 |
| 12 | GCTCCAAAGAGTAGACATTGTTTC | GACTGTTTTCGTGAGCACTTTC | 56 | 250 |
| 13 | CAACACTGTAGCCATTGCAACA | CGTATCCAAGAGGCCTAGCAAG | 60 | 356 |
| 14a | ACCATTGTCCCTCACATGTGC | CAGTGAAAGGCATGTGCTACAAAC | 60 | 259 |
| 15 | CAGGTCCTAGGCACAGGAACTG | ACATTCCCAAATTGCTTGCCT | 60 | 321 |
GC clamps were added at the 5′ end for DHPLC analysis: forward primer, 5′-CCCGCCGCCCCCGCCG-3′; reverse primer, 5′-CCGCGCCCCCGCCCG-3′ (product length = 290 bp).
Mutational analysis of the amplimers was performed by means of denaturing high-performance liquid chromatography (DHPLC), through use of the Wave DNA Fragment Analysis System (Transgenomics) at column temperatures recommended by the WaveMaker version 4.1.31 software (Transgenomics). DHPLC buffers and run conditions were as follows: buffer A (0.1M triethylammonium acetate [TEAA], 0.025% acetonitrile [ACN]), buffer B (0.1M TEAA, 25% ACN); a flow rate of 0.9 ml/min; and a gradient duration of 3 min, with active clean (75% ACN). Buffer B gradients and temperatures are reported in table 2. Positive controls—that is, PCR products expected to result in variant elution profiles—were used in all DHPLC runs. Heterozygous templates with previously identified mutations or single-nucleotide polymorphisms (SNPs) were used as positive controls for exons 3, 4, 7, 8, and 13. For each of the remaining exons, a synthetic template containing a single nucleotide change was constructed using the overlap extension method in a two-step PCR procedure. Wild-type and mutated PCR products were denatured together at 94°C for 5 min and were slowly cooled at room temperature, to allow heteroduplex formation. Bidirectional direct sequencing of purified PCR products (Qiagen) was performed using the ABI BigDye Terminator Sequencing Kit (Perkin Elmer) and an ABI 3700 Capillary Array Sequencer (Perkin Elmer). Sequences were analyzed by the Sequencing Analysis v.3.6.1 and AutoAssembler v.1.4.0 software packages (Perkin Elmer). Cosegregation analysis and exclusion of the mutations in control samples (n=100) were performed by means of RFLP, DHPLC, or direct sequencing.
Table 2.
Percent Buffer B and Temperatures Used in DHPLC Analysis for PTPN11 Mutation Detection
|
% Buffer Ba |
||||
| Exon | Loading | Initial | Final | Temperature(s)(°C) |
| 1 | 56 | 61 | 67 | 67 |
| 2 | 55 | 60 | 66 | 56, 57 |
| 3 | 54 | 59 | 65 | 57, 58 |
| 4 | 53 | 58 | 64 | 56, 57 |
| 5 | 51 | 56 | 62 | 56, 58 |
| 6 | 50 | 55 | 61 | 56, 57 |
| 7 | 50 | 55 | 61 | 56, 57 |
| 8 | 51 | 56 | 62 | 57, 58 |
| 9 | 52 | 57 | 63 | 56, 57 |
| 10 | 50 | 55 | 61 | 57, 58 |
| 11 | 54 | 59 | 65 | 59 |
| 49 | 54 | 60 | 64 | |
| 12 | 48 | 53 | 59 | 58, 59 |
| 13 | 51 | 56 | 62 | 59 |
| 50 | 55 | 61 | 60 | |
| 14 | 52 | 57 | 63 | 57 |
| 49 | 54 | 60 | 60 | |
| 15 | 51 | 56 | 62 | 56, 57 |
% bufferA=100-% buffer B.
Genotype-phenotype correlations were performed using 2×2 contingency-table analysis. The significance threshold was set at P<.05.
Results
Spectrum of PTPN11 Mutations
The study population comprised 119 propositi with NS, including 70 with sporadic NS and 49 apparently unrelated families. Of the familial cases, the phenotype was linked to the NS1 locus in 11. Linkage exclusion for the NS1 locus was documented in four families. The small size of the remaining 34 families (typically, an affected parent and child) did not allow for linkage analysis. All subjects were of northern-European descent, except for two, one of Afro-Caribbean and one of Indian origin.
DNAs from the 119 propositi with NS were screened for PTPN11 coding-region mutations. Exons 1–15 and flanking intron sequences were PCR amplified and were analyzed by means of DHPLC, and products with variant elution profiles were sequenced bidirectionally. PTPN11 mutations were identified in 54 subjects, comprising 22 different nucleotide changes (table 3). All mutations were missense exonic changes, with the majority clustering in exons 3 and 8. The mutations cosegregated completely with the disease in all familial cases. None of the 22 different mutations was observed in at least 100 control Europeans or Americans of European descent. Parental DNAs were available for 43 of 52 (83%) individuals with sporadic NS who had PTPN11 mutations. Only one parent was identified as carrying a mutation—the 925A→G transition, predicting an Ile309Val substitution. The variant profile for this mutation was not observed in >400 control chromosomes.
Table 3.
PTPN11 Mutations in NS
| NucleotideSubstitution | No. ofCases | Amino Acid Substitution | Domain |
| Exon 2: | |||
| 124A→G | 2 | Thr42Ala | N-SH2 |
| Exon 3: | |||
| 179G→C | 2 | Gly60Ala | N-SH2 |
| 181G→A | 1 | Asp61Asn | N-SH2 |
| 182A→G | 2 | Asp61Gly | N-SH2 |
| 184T→G | 2 | Tyr62Asp | N-SH2 |
| 188A→G | 4 | Tyr63Cys | N-SH2 |
| 215C→G | 1 | Ala72Gly | N-SH2 |
| 218C→T | 1 | Thr73Ile | N-SH2 |
| 228G→C | 1 | Glu76Asp | N-SH2 |
| 236A→G | 5 | Gln79Arg | N-SH2 |
| 317A→C | 3 | Asp106Ala | N-SH2/C-SH2 linker |
| Exon 4: | |||
| 417G→C | 1 | Glu139Asp | C-SH2 |
| 417G→T | 1 | Glu139Asp | C-SH2 |
| Exon 7: | |||
| 836A→G | 1 | Tyr279Cys | PTP |
| 844A→G | 1 | Ile282Val | PTP |
| 853T→C | 1 | Phe285Leu | PTP |
| Exon 8: | |||
| 854T→C | 1 | Phe285Ser | PTP |
| 922A→G | 17 | Asn308Asp | PTP |
| 923A→Ga | 2 | Asn308Ser | PTP |
| 925A→G | 1 | Ile309Val | PTP |
| Exon 13: | |||
| 1502G→A | 1 | Arg501Lys | PTP |
| 1510A→G | 3 | Met504Val | PTP |
Affected members of one family segregating the 923A→G change (Asn308Ser) exhibited the Noonan-like/multiple giant-cell lesion condition.
The A→G transition at position 922 in exon 8, predicting the Asn308Asp substitution within the PTP domain, was the most common mutation, accounting for one-third of the total. Of note, the Asn308Asp mutation was identified in the large three-generation family that was used to originally establish linkage to the NS1 locus (Jamieson et al. 1994; van der Burgt et al. 1994) (fig. 1A). The two occurrences of the adjacent 923A→G mutation, predicting an Asn308Ser change, indicate that codon 308 represents a hotspot for NS.
Figure 1.
Cosegregation of PTPN11 mutations and Noonan phenotype. A, Restriction analysis of PTPN11 PCR product containing exon 8, showing segregation of the 922A→G mutation in the original large family with NS that shows linkage to 12q24 (Jamieson et al. 1994). The family tree is shown above. The mutation introduces an EcoRV restriction site, resulting in 246- and 366-bp products, whereas the wild-type allele remains undigested (612 bp). B, DHPLC elution profiles of PTPN11 exon 8 PCR products, showing segregation of the 923A→G change in a family inheriting Noonan-like/multiple giant-cell lesion syndrome (Bertola et al. 2001). A single peak characterizes the wild-type profile, and a variant profile, characterized by two peaks, is observed in all affected family members. The family tree is shown on the left. The gray-shaded symbol indicates an indeterminate phenotype.
Eight additional mutations were found recurrently. Among them, the 182A→G (Asp61Gly), 188A→G (Tyr63Cys), and 236A→G (Gln79Arg) substitutions in exon 3, as well as the 1510A→G (Met504Val) change in exon 13, have previously been reported as disease-causing mutations (Tartaglia et al. 2001). Interestingly, 90% of mutational events involved amino acid residues located in the N-SH2 and PTP functional domains (fig. 2). The exceptions were the 317A→C transversion, observed in two individuals with sporadic NS and in one family, predicting an Asp106Ala substitution in the peptide linking the N-SH2 and C-SH2 domains, and the 417G→C and 417G→T changes, both predicting a Glu139Asp substitution within the C-SH2 domain.
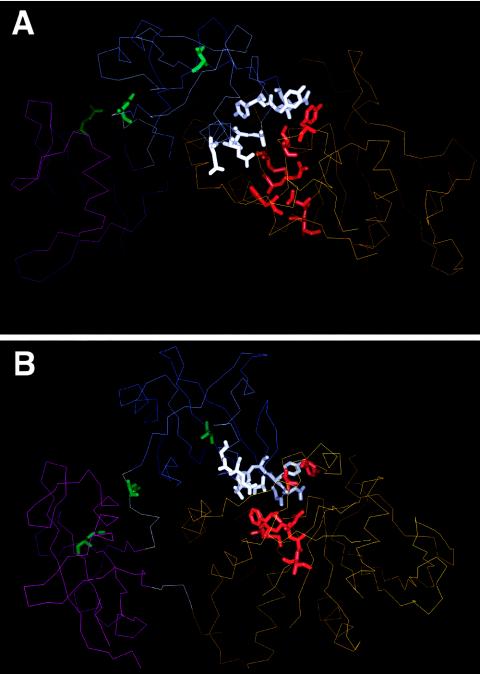
Figure 2.
Location of mutated residues in SHP-2 in the inactive conformation: Cα trace of the N-SH2 (thin blue line), C-SH2 (thin magenta line), and PTP (thin orange line) domains; and N-SH2/C-SH2 and C-SH2/PTP linkers (thin gray lines), according to Hof et al. (1998). The views represented in panels A and B are orthogonal to each other. Mutated residues are indicated with their side chains as thick lines (N-SH2 residues located in or close to the N-SH2/PTP interaction surface are shown in white; linker, C-SH2, and N-SH2-phosphopeptide binding residues are shown in green; and PTP residues located in or close to the N-SH2/PTP interaction surface are shown in red).
On the basis of the secondary structure of SHP-2 in the inactive conformation determined by Hof et al. (1998), the N-SH2 and PTP mutations were noted to cluster dramatically in specific regions of those domains. Of the 10 N-SH2 substitutions, 5 were positioned in the D′E loop and flanking βD′ and βE strands (residues 57–65), and 4 were in the βF strand and αB helix (residues 69–84). The nine PTP mutations were restricted to the DB loop (residues 277–288; n=4), βC strand (residues 303–311; n=3), and HI loop (residues 499–507, n=2). Most strikingly, all N-SH2 and PTP mutations except the Thr42Ala substitution involved residues located in or close to the N-SH2/PTP interacting surface (fig. 2).
Fifteen sequence variants that are likely to represent neutral polymorphisms were observed in subjects with NS, unaffected family members, and control individuals (table 4). These changes included 13 intronic SNPs, a single-base deletion within the 5′ UTR, and a synonymous change in exon 3.
Table 4.
Polymorphisms in the PTPN11 Gene
| Location | Nucleotide Change |
| Exon 1 (5′ UTR) | −140delGa |
| Intron 1 | +25G→C |
| Intron 1 | +54C→A |
| Exon 3 | 255C→T (His85) |
| Intron 4 | +12G→C |
| Intron 4 | +78A→G |
| Intron 7 | −21C→T |
| Intron 7 | −30T→C |
| Intron 7 | −32A→C |
| Intron 7 | −35A→C |
| Intron 7 | −132T→C |
| Intron 9 | −9C→A |
| Intron 10 | −63G→A |
| Intron 13 | +26G→A |
| Intron 15 | +40T→C |
Position refers to the A of the ATG initiator codon.
Genetic Heterogeneity in NS
PTPN11 mutations were detected in the 11 families for which the disorder appeared to be linked to the NS1 locus, including the original large family described by Jamieson et al. (1994). As anticipated, no PTPN11 sequence change was observed in the four families for which linkage to NS1 had been excluded. For the entire study group of 119, PTPN11 mutations were observed in 45%, a slightly lower incidence than had been seen in a small cohort (Tartaglia et al. 2001). PTPN11 mutations were detected in 59% of individuals with familial NS, whereas such defects were observed in only 37% of individuals with sporadic NS. This statistically significant (P<.02) difference in PTPN11 mutation prevalence suggests that the additional gene or genes responsible for NS engender incomplete penetrance or have greater adverse affects on fertility than does PTPN11.
Genotype-Phenotype Correlation
Because of the clinical heterogeneity observed in NS, we investigated possible associations between genotype and phenotype. The distribution of several major clinical features of NS, in subjects with and without mutations in PTPN11, is shown in table 5. A statistically significant association with pulmonic stenosis was found in the group with PTPN11 mutations (70.6% vs. 46.2%; P=.008). In contrast, a statistically significantly lower incidence of HCM was observed in this group (5.9% vs. 26.2%; P=.004). There was no significant difference in the prevalence of atrial and/or ventricular septal defects or other congenital heart malformations between the groups with and without PTPN11 mutations. Similarly, there was no difference in the rates of short stature, pectus deformities, cryptorchidism, or enrollment in special education (as a marker of developmental delay).
Table 5.
Clinical Features in Subjects with NS with and without PTPN11 Mutations
|
No./Total (%) of Subjects |
|||
| Clinical Feature | With PTPN11 Mutation | Without PTPN11 Mutation | Pa |
| Cardiac defects: | |||
| HCM | 3/51 (5.9) | 17/65 (26.1) | .004 |
| Pulmonic stenosis | 36/51 (70.6) | 30/65 (46.2) | .008 |
| Septal defects | 6/50 (12.0) | 11/63 (17.5) | NS |
| Short stature | 39/51 (76.5) | 45/64 (70.3) | NS |
| Special education | 11/46 (23.9) | 21/59 (35.6) | NS |
| Pectus deformities | 39/50 (78.0) | 46/61 (75.4) | NS |
| Cryptorchidism | 26/31 (83.9) | 25/35 (71.4) | NS |
“NS” indicates a difference that is not statistically significant.
The clinical manifestations of NS were compared between the cohort with N-SH2 mutations and the cohort with PTP mutations. Although this analysis had less statistical power, owing to sample size, no significant differences were identified. The phenotype observed in subjects with the common Asn308Asp substitution (n=17) was not qualitatively different from the phenotype in subjects with other mutations, except for the fact that no subject carrying the Asn308Asp change was enrolled in special education.
We identified an A→G transition at position 923 (Asn308Ser) in a family with typical features of NS associated with multiple giant-cell lesions in bone (Bertola et al. 2001). This mutation cosegregated perfectly with this phenotype (fig. 1B). One family member (III-3) had a typical NS phenotype (i.e., pulmonic stenosis, cryptorchidism, short stature, and distinctive face and chest deformity) and exhibited asymptomatic lesions in the right ramus of the mandible and in the maxilla, as well as osteolytic lesions in both humeri and in the left femur. Similarly, his affected sister (III-2) had multiple lesions in the mandible. Serum calcium, phosphate, and PTH levels were normal in both subjects. Their mother (II-2) had typical NS features but did not have any bone lesions. The same Asn308Ser mutation was observed in another family with NS that had no known bony involvement.
Discussion
Recently, we established that missense mutations in PTPN11 cause NS and implicated a gain-of-function mechanism. In the present study, we extend those initial observations in several important respects. These include a more complete assessment of the range of PTPN11 lesions causing NS, evaluation of disease penetrance, establishment of genotype-phenotype correlation, and broadening of the phenotype associated with PTPN11 mutations to include NS with multiple giant-cell lesions.
In our present analysis of a large cohort with sporadic and familial cases of NS, we estimated the PTPN11 mutation prevalence to be 45%. Although it is quite similar to the rate of 50% observed in a small cohort with NS, there are some limitations to the accuracy of this estimate. In the present study, DHPLC was used to screen the PTPN11 coding exons. Since this method is purported to have a sensitivity of 96%–100% under ideal conditions (Xiao and Oefner 2001), the true prevalence of point mutations in this cohort could be slightly higher. Second, no attempt was made to look for other types of molecular lesions that might also cause NS, such as large intragenic deletions, changes in the 3′ UTR, or promoter defects. Lastly, our current results revealed that the prevalence detected in any NS cohort would be sensitive to the composition of sporadic and familial cases. Since the subjects in our cohort were not ascertained in a cross-sectional manner (i.e., there was probably a systematic bias toward familial cases), our prevalence figure could be an overestimate. With those issues stipulated, the contribution of PTPN11 mutations to the etiology of NS appears to be ∼50%.
When these results are combined with our previous work, we have now identified 78 unrelated individuals with NS who have mutations in PTPN11. All mutations are missense changes that affect amino acid residues that are conserved among the vertebrate SHP-2 orthologs. Most altered residues are also conserved among the vertebrate SHP-1 proteins. Previously, we had argued that the NS-causing PTPN11 defects result in gain-of-function effects on SHP-2. This was supported by energetics-based structural analysis of two mutants (Tartaglia et al. 2001) and by the observation that two NS mutant alleles closely resembled two engineered SHP-2 mutants with increased phosphatase activity in vitro and inductive effects in Xenopus animal caps (O’Reilly et al. 2000). Since we have failed to identify any nonsense, frameshift, or splicing defect among 78 PTPN11 mutations, it seems highly unlikely that SHP-2 haploinsufficiency results in the NS phenotype.
The distribution of the altered amino acid residues in SHP-2 has a nonrandom pattern (table 3 and fig. 2). The vast majority of the NS mutations clustered in the N-SH2 and PTP functional domains but were not restricted to those domains, as was seen previously. The N-SH2 domain interacts with the PTP domain and binds to phosphotyrosyl-containing targets on activated receptors or docking proteins, using two separate sites. These sites show negative cooperativity, so that N-SH2 can work as intramolecular switch to control SHP-2 catalytic activity. In the inactive state, the N-SH2 and PTP domains share a broad interaction surface. More precisely, the N-SH2 D′E loop and flanking βD′ and βE strands closely interact with the catalytic cleft, blocking the PTP active site. Crystallographic data on SHP-2 in the inactive conformation revealed a complex interdomain hydrogen-bonding network—involving Asn58, Gly60, Asp61, Cys459, and Gln506—that stabilizes the protein (Hof et al. 1998). Numerous polar interactions between N-SH2 residues located in strands βF and βA, helix αB, and residues of the PTP domain further stabilize the inactive conformation. Significantly, most of the residues mutated in NS are either directly involved in these interdomain interactions (i.e., Gly60, Asp61, Ala72, Glu76, and Gln79) or in close spatial proximity to them (i.e., Tyr62, Tyr63, Thr73, Tyr279, Ile282, Phe285, Asn308, Ile309, Arg501, and Met504). This distribution of molecular lesions suggests that the pathogenetic mechanism in NS involves altered N-SH2/PTP interactions that destabilize the inactive conformation without altering SHP-2’s catalytic capability. Consistent with this view, no mutation altered Cys459 (the residue essential for nucleophilic attack), the PTP signature motif (positions 457–467), or the TrpProAsp loop (positions 423–425), which are all essential for phosphatase activity.
Three of the NS mutations affected residues outside of the interacting regions of the N-SH2 and PTP domains. One recurrent mutation affected Asp106, which is located in the linker stretch connecting the N-SH2 and C-SH2 domains. Although functional studies are required to understand the functional significance of the Asp→Ala substitution, we hypothesize that this mutation might alter the flexibility of the N-SH2 domain, thus inhibiting the N-SH2/PTP interaction. Two mutated residues, Thr42 (N-SH2 domain) and Glu139 (C-SH2 domain), are spatially far from the N-SH2/PTP interaction surfaces. In contrast to the other mutated residues, Thr42 and Glu139 are implicated in the intermolecular interactions of the SH2 domains with phosphotyrosyl-containing peptides (Lee et al. 1994; Huyer and Ramachandran 1998). Specifically, Thr42 directly interacts with the tyrosine phosphate, and Glu139 is adjacent to Arg138 and Ser140, which form hydrogen bonds to that phosphate. Since the phenotype of the subjects bearing these mutations was typical for NS, molecular characterization is needed in order to understand how defects in phosphotyrosine binding result in developmental perturbations similar to those affecting SHP-2 inactivation.
Previously, the penetrance of NS had not been addressed in a systematic fashion. Two lines of evidence now suggest that NS caused by PTPN11 mutations is almost completely penetrant. First, we analyzed 11 families, including the large kindred first described by van der Burgt et al. (1994), for which significant or suggestive linkage to the NS1 locus had been established (Jamieson et al. 1994). In each instance, a PTPN11 mutation was identified that was inherited by all of the affected individuals in the family but by none of the unaffected ones. Analysis of small kindreds harboring PTPN11 mutations revealed the same consistent pattern. Second, we genotyped a high percentage (83%) of the unaffected parents for defects discovered in their offspring with apparently sporadic NS. In only one instance was a mutation identified. Although it is possible that inclusion of “milder” cases of NS might have uncovered some instances of incomplete penetrance, the strict criteria for NS employed in the present study identified a cohort with almost 100% penetrance.
We observed a statistically significantly higher incidence of pulmonic stenosis among subjects with NS inheriting PTPN11 mutations. Experimental evidence with mice indicates that epidermal growth factor (EGF) signaling is important for semilunar valve development and that SHP-2 is a component of the EGF-mediated signal transduction pathway (Qu et al. 1999; Chen et al. 2000). A proportion of mice that are homozygous for a hypomorphic Egfr allele exhibit thickened aortic and pulmonary valve leaflets because of an increased number of mesenchymal cells. Coinheritance of a Ptpn11 knockout allele in heterozygosity results in a higher prevalence and increased severity of those valve abnormalities. These findings implicate SHP-2 in aspects of EGF-mediated semilunar valvulogenesis, such as mesenchymal transformation and proliferation, as well as leaflet remodeling. The specific manner in which the gain-of-function PTPN11 changes observed in NS result in pulmonic stenosis (but not aortic stenosis) remains to be determined. An understanding of the relatively reduced prevalence of pulmonic stenosis in other genetic forms of NS must await an elucidation of those disease genes. Similarly, the reduced prevalence of HCM in the cohort with PTPN11 mutations suggests that SHP-2 plays less of a role in cardiomyocyte proliferation than do other NS disease genes.
Finally, we demonstrated the cosegregation of a 923A→G PTPN11 mutation in a family inheriting the Noonan-like/multiple giant-cell lesion syndrome. Since a 923A→G mutation was also identified in an unrelated kindred with classic NS, additional genetic factors or events may be necessary to result in the proliferation of these giant cells. Only a single family with this rare phenotype was available for genotyping, so the percentage of cases attributable to PTPN11 defects remains to be determined. Although the Noonan-like/multiple giant-cell lesion syndrome was introduced as a distinct entity characterized by the association of some cardinal features of NS with giant-cell lesions of bone and soft tissues (Cohen et al. 1974; Cohen and Gorlin 1991), it can now be viewed as part of the NS spectrum. This result implies that mutation screening of PTPN11 for other NS-like conditions, such as the cardio-facio-cutaneous, Leopard, and Noonan/NF1 syndromes, is necessary to elucidate whether those entities are distinct nosologic conditions, allelic disorders, or extreme phenotypes of a single disorder with markedly variable expressivity.
Acknowledgments
The authors thank the subjects and families who participated in this study and the physicians who referred the subjects. Thanks are also owed to Fabien Lecaille for his helpful assistance in preparing graphical representations of SHP-2 mutations. Mariella Sorcini is thanked for her support and mentorship of M.T. This work was supported, in part, by Mount Sinai Children's Health Research Center grant 5 P30 HD 28822, by National Institutes of Health grant 5 K24 HD001294 (to B.D.G.), and by a grant from the Birth Defects Foundation (United Kingdom) and British Heart Foundation grant PG/98101 (both to S.J.).
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NS [MIM 163950] and Noonan-like/multiple giant-cell lesion syndrome [MIM 163955])
References
- Allanson JE (1987) Noonan syndrome. J Med Genet 24:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrandale JM, Gore-Willse A, Rock S, Ren J-M, Zhu J, Davis A, Livingston JN, Rabin DU (1996) Insulin signaling in mice expressing reduced levels of Syp. J Biol Chem 271:21353–21358 [DOI] [PubMed] [Google Scholar]
- Bertola DR, Kim CA, Pereira AC, Mota GF, Krieger JE, Vieira IC, Valente M, Loreto MR, Magalhaes RP, Gonzalez CH (2001) Are Noonan syndrome and Noonan-like/multiple giant cell lesion syndrome distinct entities? Am J Med Genet 98:230–234 [DOI] [PubMed] [Google Scholar]
- Bertola DR, Kim CA, Sugayama SMM, Albano LMJ, Wagenfur J, Moyses L, Gonzalez CH (2000) Cardiac findings in 31 patients with Noonan’s syndrome. Arq Bras Cardiol 75:409–412 [DOI] [PubMed] [Google Scholar]
- Brady AF, Jamieson CR, van der Burgt I, Crosby A, van Reen M, Kremer H, Mariman E, Patton MA, Jeffery S (1997) Further delineation of the critical region for Noonan syndrome on the long arm of chromosome 12. Eur J Hum Genet 5:336–337 [PubMed] [Google Scholar]
- Burch M, Sharland M, Shinebourne E, Smith G, Patton M, McKenna W (1993) Cardiologic abnormalities in Noonan syndrome: phenotypic diagnosis and echocardiographic assessment of 118 patients. J Am Coll Cardiol 22:1189–1192 [DOI] [PubMed] [Google Scholar]
- Chen B, Bronson RT, Klaman LD, Hampton TG, Wang J-F, Green PJ, Magnuson T, Douglas PS, Morgan JP, Neel BG (2000) Mice mutants for Egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nat Genet 24:296–299 [DOI] [PubMed] [Google Scholar]
- Cohen MM Jr, Gorlin RJ (1991) Noonan-like/multiple giant cell lesion syndrome. Am J Med Genet 40:159–166 [DOI] [PubMed] [Google Scholar]
- Cohen MM Jr, Ruvalcaba RHA, Graham CB, Harrison MT, Morgan AF (1974) A new syndrome simulating the Noonan syndrome, the Leopard syndrome, and hyperparathyroidism. Syndrome Ident 2:14–17 [Google Scholar]
- Cunnick JM, Meng S, Ren Y, Despont C, Wang HG, Djeu JY, Wu J (2002) Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J Biol Chem 277:9498–9504 [DOI] [PubMed] [Google Scholar]
- Feng G-S (1999) Shp-2 tyrosine phosphatase: signaling one cell or many. Exp Cell Res 253:47–54 [DOI] [PubMed] [Google Scholar]
- Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE (1998) Crystal structure of the tyrosine phosphatase SHP-2. Cell 92:441–450 [DOI] [PubMed] [Google Scholar]
- Huyer G, Ramachandran C (1998) The specificity of the N-terminal SH2 domain of SHP-2 is modified by a single point mutation. Biochemistry 37:2741–2747 [DOI] [PubMed] [Google Scholar]
- Jamieson CR, van der Burgt I, Brady AF, van Reen M, Elsawi MM, Hol F, Jeffery S, Patton MA, Mariman E (1994) Mapping a gene for Noonan syndrome to the long arm of chromosome 12. Nat Genet 8:357–360 [DOI] [PubMed] [Google Scholar]
- Kozlowski M, Mlinaric-Rascan I, Feng GS, Shen R, Pawson T, Siminovitch KA (1993) Expression and catalytic activity of the tyrosine phosphatase PTP1C is severely impaired in motheaten and viable motheaten mice. J Exp Med 178:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-H, Kominos D, Jacques S, Margolis B, Schlessinger J, Shoelson SE, Kuriyan J (1994) Crystal structure of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure 2:423–438 [DOI] [PubMed] [Google Scholar]
- Legius E, Schollen E, Matthijs G, Fryns J-P (1998) Fine mapping of Noonan/cardio-facio-cutaneous syndrome in a large family. Eur J Hum Genet 6:32–37 [DOI] [PubMed] [Google Scholar]
- Maile LA, Clemmons DR (2002) Regulation of insulin-like growth factor-1 receptor dephosphorylation by SHPS-1 and the tyrosine phosphatase SHP-2. J Biol Chem 277:8955–8960 [DOI] [PubMed] [Google Scholar]
- Marino B, Digilio MC, Toscano A, Giannotti A, Dallapiccola B (1999) Congenital heart diseases in children with Noonan syndrome: an expanded cardiac spectrum with high prevalence of atrioventricular canal. J Pediatr 135:703–706 [DOI] [PubMed] [Google Scholar]
- Maroun CR, Naujokas MA, Holgado-Madruga M, Wong A, Park M (2000) The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the Met receptor tyrosine kinase. Mol Cell Biol 20:8513–8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milarski KL, Saltiel AR (1994) Expression of catalytically inactive Syp phosphatase in 3T3 cells blocks stimulation of mitogen-activated protein kinase by insulin. J Biol Chem 269:21239–21243 [PubMed] [Google Scholar]
- Noonan JA (1968) Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am J Dis Child 116:373–380 [DOI] [PubMed] [Google Scholar]
- Nora JJ, Nora AH, Sinha AK, Spangler RD, Lubs HA (1974) The Ullrich-Noonan syndrome (Turner phenotype). Am J Dis Child 127:48–55 [DOI] [PubMed] [Google Scholar]
- O’Reilly AM, Pluskey S, Shoelson SE, Neel BG (2000) Activated mutants of SHP-2 preferentially induce elongation of Xenopus animal caps. Mol Cell Biol 20:299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Johnson MR, Melnick MB, Perrimon N (1996) The nonreceptor protein tyrosine phosphatase corkscrew functions in multiple receptor tyrosine kinase pathways in Drosophila. Dev Biol 180:63–81 [DOI] [PubMed] [Google Scholar]
- Pluskey S, Wandless TJ, Walsh CT, Shoelson SE (1995) Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J Biol Chem 270:2897–2900 [DOI] [PubMed] [Google Scholar]
- Qu C-K, Feng G-S (1998) Shp-2 has a positive regulatory role in ES cell differentiation and proliferation. Oncogene 17:433–439 [DOI] [PubMed] [Google Scholar]
- Qu C-K, Yu W-M, Azzarelli B, Cooper S, Broxmeyer HE, Feng G-S (1998) Biased suppression of hematopoiesis and multiple developmental defects in chimeric mice containing Shp-2 mutant cells. Mol Cell Biol 18:6075–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C-K, Yu W-M, Azzarelli B, Feng G-S (1999) Genetic evidence that Shp-2 tyrosine phosphatase is a signal enhancer of the epidermal growth factor receptor in mammals. Proc Natl Acad Sci USA 96:8528–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton TM, Ciruna BG, Holmyard D, Kulkarni S, Harpal K, Rossant J, Pawson T (2000) The SH2 tyrosine phosphatase Shp2 is required for mammalian limb development. Nat Genet 24:420–423 [DOI] [PubMed] [Google Scholar]
- Saxton TM, Henkemeyer M, Gasca S, Shen R, Rossi DJ, Shalaby F, Feng G-S, Pawson T (1997) Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J 16:2352–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro LM, McKenna WJ (1983) Distribution of left ventricular hypertrophy in hypertrophic cardiomyopathy: a two-dimensional echocardiographic study. J Am Coll Cardiol 2:437–444 [DOI] [PubMed] [Google Scholar]
- Shi Z-Q, Yu D-H, Park M, Marshall M, Feng G-S (2000) Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol Cell Biol 20:1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-Gerlach M, Wallasch C, Ullrich A (1998) SHP-2, SH2-containing protein tyrosine phosphatase-2. Int J Biochem Cell Biol 30:559–566 [DOI] [PubMed] [Google Scholar]
- Stofega MR, Herrington J, Billestrup N, Carter-Su C (2000) Mutation of the SHP-2 binding site in growth hormone (GH) receptor prolongs GH-promoted tyrosyl phosphorylation of GH receptor, JAK2, and STAT5B. Mol Endocrinol 14:1338–1350 [DOI] [PubMed] [Google Scholar]
- Tang TL, Freeman RM Jr, O’Reilly AM, Neel BG, Sokol SY (1995) The SH2-containing protein-tyrosine phosphatase SH-PTP2 is required upstream of MAP kinase for early Xenopus development. Cell 80:473–483 [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD (2001) Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 29:465–468 [DOI] [PubMed] [Google Scholar]
- Tsui HW, Siminovitch KA, de Souza L, Tsui FW (1993) Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat Genet 4:124–129 [DOI] [PubMed] [Google Scholar]
- van der Burgt I, Berends E, Lommen E, van Beersum S, Hamel B, Mariman E (1994) Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet 53:187–191 [DOI] [PubMed] [Google Scholar]
- van der Burgt I, Brunner H (2000) Genetic heterogeneity in Noonan syndrome: evidence for an autosomal recessive form. Am J Med Genet 94:46–51 [DOI] [PubMed] [Google Scholar]
- Xiao W, Oefner PJ (2001) Denaturing high-performance liquid chromatography: a review. Hum Mut 17:439–474 [DOI] [PubMed] [Google Scholar]