Abstract
Vertebrate 4.1 proteins have a spectrin-actin-binding (SAB) domain, which is lacking in all the invertebrate 4.1 proteins indentified so far, and it was therefore proposed that the SAB domain emerged with the advent of vertebrates during evolution. Here we demonstrated for the first time that amphioxus (an invertebrate chordate) protein 4.1, though lacking a recognizable SAB, was able to bind both spectrin and actin, with a binding capacity comparable to that of human protein 4.1. Detailed structure-activity analyses revealed that the unique domain U2/3 was a newly identified SAB-like domain capable of interacting with spectrin and actin, suggesting the presence of a “cryptic” SAB domain in amphioxus 4.1 protein. We also showed that amphioxus 4.1 protein gene was the common ancestor of vertebrate 4.1 protein genes, from which 4.1R, 4.1N, 4.1G, and 4.1B genes originated. This work will encourage further study on the structure-activity of invertebrate 4.1 protein and its interacting proteins.
Metazoan cell membranes are supported by a two-dimensional protein network which lays closely apposed to the cytoplasmic surface face of the lipid bilayer. It is known as the “membrane-associated cytoskeleton”1 and made up of three key components: spectrin, actin, and protein 4.1 (also called band 4.1 or simply 4.1). Protein 4.1 and a fourth cytoskeletal protein, called ankyrin, provide two links that function independently to tether the network to transmembrane proteins.
Vertebrates have four highly conserved genes encoding protein 4.1; their protein products are known as 4.1R (red cell type), 4.1N (neuron type), 4.1G (general type), and 4.1B (brain type). Protein 4.1R is the first member of protein 4.1 identified from red blood cells in 19792, and is thus regarded as the prototypical member of protein 4.1 family. Protein 4.1R is predominantly expressed in red blood cells, although it is expressed in many other cell types. The other three proteins are essentially absent from red cells, but present widely in other cell types3. All four proteins have a common organization of domains (Fig. 1a), such as the N-terminal domain FERM (four point one protein-ezrin-radixin-moesin), spectrin-actin-binding domain (SAB), and C-terminal domain (CTD). Adjacent to the FERM domain is a FERM-adjacent (FA) domain4. Both FERM and CTD domains can bind transmembrane proteins, and SAB domain provides the cytoskeletal linkage, except for that of protein 4.1N, which is so divergent that it does not bind spectrin and actin5. FA domain is the site of phosphorylation by a number of protein kinases, including protein kinase C. Interspersed between the common domains are nonconserved sequences that display far less identity between the individual proteins. They are called unique domains: U1 (also known as “headpiece”), U2, and U3. Sequences of U1, U2, and U3 are distinct from each other and their functions remain poorly defined6.
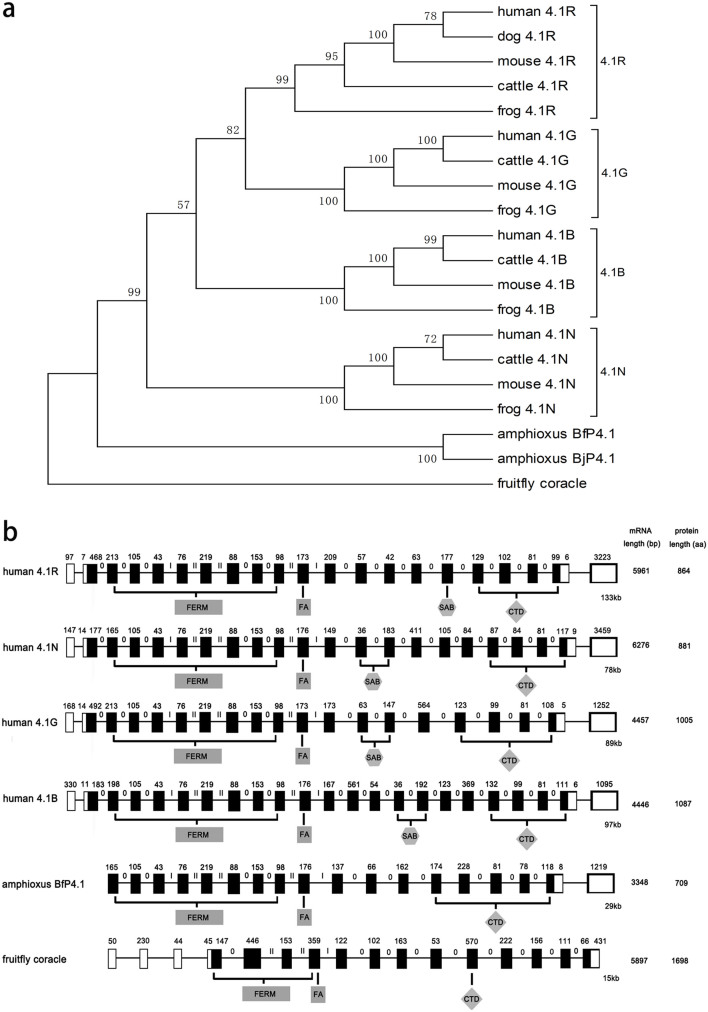
Figure 1. Domain structures of protein 4.1 family.
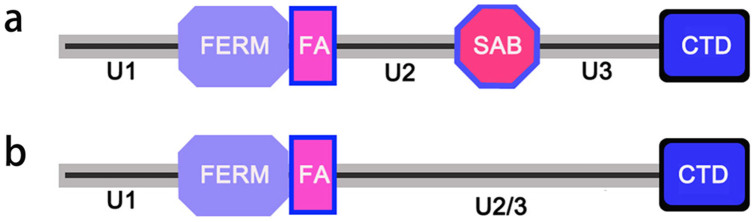
(a) Protein 4.1 in vertebrates; (b) Protein 4.1 in amphioxus.
Accumulating data show that protein 4.1 emerged early in evolution; all known invertebrate genomes have a single copy of protein 4.1 genes7. For example, a protein 4.1 gene has been found in sea anemone Nematostella vectensis, although it lacks the SAB domain4,7. Similarly, protein 4.1 homologue found in Drosophila does not have the SAB domain either8. Recently, we have isolated a cDNA in the basal chordate amphioxus (Branchiostoma japonicum) coding for protein 4.1, named BjP4.1, which possesses both the common domains FERM, CTD, and FA, and the unique domains U1, and U2/3, but not the SAB domain (Fig. 1b). These data together indicate that the SAB domain is a late evolutionary development9,10. The questions thus arise as to when the SAB domain of protein 4.1 emerged in evolution, and whether invertebrate protein 4.1 can bind spectrin and actin. To our best knowledge, no study has ever been conducted to examine the interaction of invertebrate protein 4.1 with spectrin and actin. Additionally, in contrast to the enormous progresses on vertebrate protein 4.1 structure-function study, little information as such is available for invertebrate protein 4.1. The aims of this study were therefore to explore whether amphioxus protein 4.1 can bind to spectrin and actin, and if so, which site would be essential for the binding to spectrin and actin.
Results
Sequence and phylogeny of BjP4.1
The cDNA of BjP4.1 obtained contained a longest open reading frame which codes for a protein of 529 amino acids with a predicted molecular mass of approximately 59.9 kDa. Sequence comparison revealed that the predicted protein BjP4.1 was 47.0% to 60.8% identical to the orthologs in fruit fly and vertebrates (Supplementary Figure S1 and Supplementary Table 1). BLASTp searching at NCBI showed that BjP4.1 has an intact FERM domain at residues 42 to 315, a FA domain at residues 325 to 370 and a CTD domain at residues 439 to 526, but it lacks a recognizable SAB domain (Fig. 1b). Therefore, BjP4.1 has a canonical domain combination, U1-FERM-FA-U2/3-CTD, characteristic of known invertebrate protein 4.1 members.
To investigate the relationship between BjP4.1 and other 4.1 proteins, a phylogenetic tree was constructed using the amino acid sequences of BjP4.1 and other representative 4.1 proteins (Supplementary Table 1) by neighbor joining method (Fig. 2a). It was found that BjP4.1 formed an independent cluster with Florida amphioxus protein 4.1, which is positioned at the base of vertebrate 4.1 proteins. This well reflects the established phylogeny of chosen organisms, suggesting that amphioxus protein 4.1 might be the prototype of vertebrate 4.1 proteins 4.1R, 4.1G, 4.1B and 4.1N.
Figure 2. Phylogenetic tree and genomic structures.
(a) Phylogenetic tree of 4.1 proteins. The phylogenetic tree was constructed by the neighbor-joining method within the package MEGA 5.0 software package using 1000 bootstrap replicates. Note that amphioxus protein 4.1 was positioned at the base of vertebrate protein 4.1R, 4.1G, 4.1B, and 4.1N. (b) Genomic structures of protein 4.1 genes in human, amphioxus and fruit fly. The open and black rectangles represent non-coding exons and coding exons respectively. The horizontal lines between two rectangles represent intron. The numbers above the rectangles represent the nucleotide number of each exon. The Roma numbers above the horizontal lines show the intron phase.
A search of the recently completed draft assembly and automated annotation of the Florida amphioxus B. floridae genome revealed the presence of a genomic DNA sequence (ESTEXT_FGENESH2_PG.C_ 710159) of protein 4.1 (http://genome.jgi-psf.org//Brafl1/Brafl1.home.html). The single B. floridae protein 4.1 cDNA encodes a protein with about 89.6% identity to BjP4.1 at amino acid level, suggesting that BjP4.1 is highly conserved in interspecies. Analysis of the genomic structure exhibited that B. floridae 4.1 protein gene consists of 18 exons interspaced by 17 introns, and ranges about 29 kb in length (Fig. 2b). The introns all begin with GT and end with an AG dinucleotide, sequences thought necessary for correct RNA splicing of various eukaryotic genes11. It was notable that human 4.1R, 4.1N, 4.1G, and 4.1B genes contained 21 exons, 22 exons, 20 exons and 23 exons, respectively. For human 4.1 proteins and BjP4.1, their FERM domains were both encoded by 8 exons, FA domains by 1 exon, and CTD domains by 4 exons. In addition, the exon-intron splice sites of both human 4.1 proteins and BjP4.1, including FERM, FA and CTD domains, are conserved (Fig. 2b), i.e. they all have the same phase introns12. These indicate that the primary composition of human 4.1 proteins and amphioxus protein 4.1 is highly conserved across humans and basal chordate, although amphioxus protein 4.1 lacks the SAB domain.
Tissue-specific expression
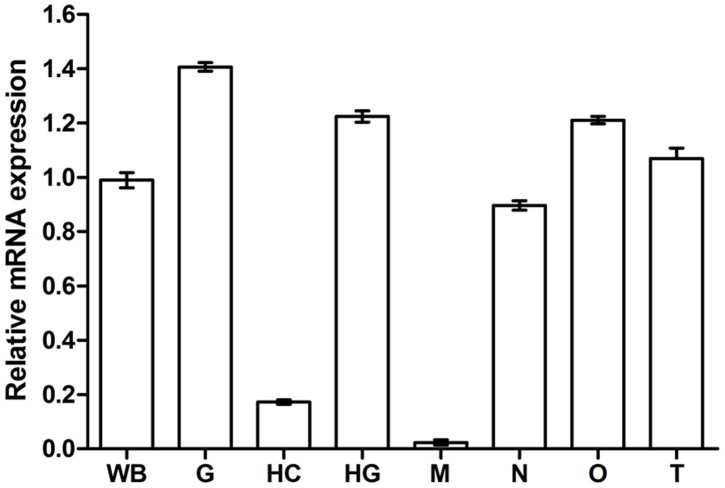
qRT-PCR assay was used to examine the expression pattern of Bjp4.1 in the different tissues of amphioxus, including the hepatic caecum, hind-gut, gill, muscle, notochord, testis and ovary. The dissociation curve of amplification product showed only a single peak, indicating that the amplification was specific (data not shown). As shown in Fig. 3, Bjp4.1 mRNA was predominantly detected in the hind-gut, gill, notochord, testis and ovary, and at a lower level it was present in the hepatic caecum. Little expression of Bjp4.1 was measured in the muscle. These show that Bjp4.1 is expressed in a tissue-specific fashion in B. japonicum.
Figure 3. Expression pattern of Bjp4.1 in B. japonicum.
WB, whole body; G, gill; HC, hepatic caecum; HG, hind-gut; M, muscle; N, notochord; O, ovary; T, testis.
Binding of rBjP4.1 to spectrin and actin
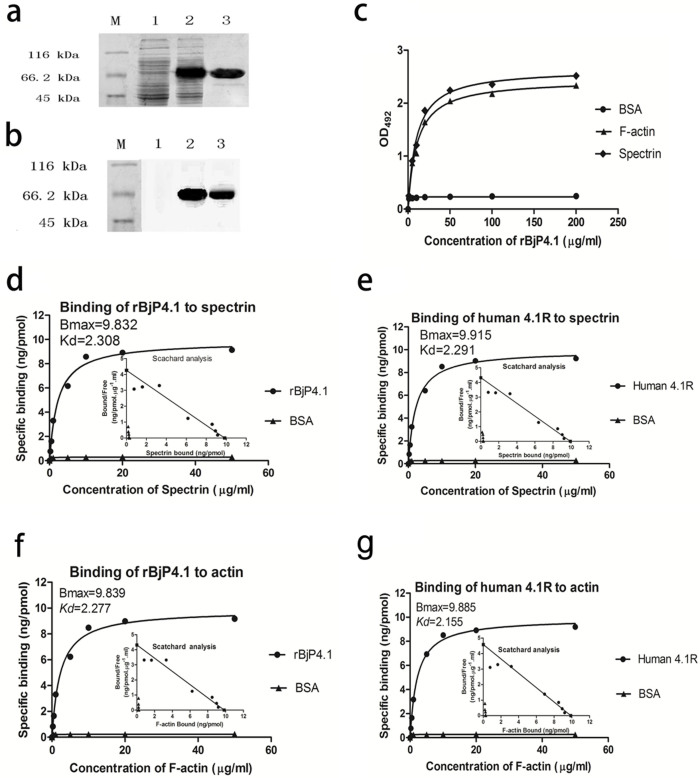
An expression vector, including the cDNA encoding BjP4.1 and a 5′ additional His6 tag, was constructed and transformed into Escherichia coli cells. The recombinant protein rBjP4.1 was induced by isopropyl β-D-thiogalactoside and was purified by affinity chromatography on a nickel-nitrilotriacetic acid resin column. SDS-PAGE followed by staining with Coomassie Brilliant Blue R-250 demonstrated the presence of a single band of approximately 66 kDa (Fig. 4a), corresponding to the expected size. Additionally, Western blotting showed that the purified protein reacted with human EPB41 protein polyclonal antibody (Inc:sc-25967; Santa Cruz Biotechnology, California, USA), indicating that Bjp4.1 was correctly expressed (Fig. 4b).
Figure 4. Interaction of rBjP4.1 with spectrin and F-actin.
The purified rBjP4.1 was analysed by SDS-PAGE (a) and Western blotting (b). M, protein markers; Lane 1, total cellular extracts from E. coli BL21 containing pET28a/Bjp4.1 before induction; Lane 2, total cellular extracts from IPTG-induced E. coli BL21 containing pET28a/Bjp4.1; Lane 3, rBjP4.1 purified on Ni-NTA resin column. c, Binding of rBjP4.1 to spectrin and actin measured by ELISA. Spectrin and actin were applied to the wells of a 96-well microplate, air-dried overnight at room temperature, and followed by ELISA. BSA instead of spectrin or actin was used as control. d–g, Saturation curves of the binding of rBjP4.1 and recombinant human 4.1R to spectrin and actin. rBjP4.1 and human 4.1R dissolved in re-distilled water was applied to the wells of a 96-well microplate, air-dried overnight at room temperature, and followed by Scatchard analysis. BSA instead of rBjP4 or recombinant human 4.1R was used as control.
To test if rBjP4.1 can bind to spectrin and actin, an ELISA was carried out first. It was found that rBjP4.1 was capable of binding to both spectrin and F-actin in a dose-dependent manner (Fig. 4c), while BSA was not, indicating that the interaction of rBjP4.1 with spectrin and actin was specific. To characterize the affinities of rBjP4.1 to spectin and actin, the Scatchard analysis was performed. It was shown that the Bmax (ng/pmol protein) and Kd values (μg/ml) of the affinities of rBjP4.1 to spectrin and F-actin were about 9.832 and 2.308, and 9.839 and 2.277 (Fig. 4d and e), respectively. It was clear that the binding of rBjP4.1 to spectrin and F-actin was specific and saturable. Notably, the Bmax and Kd values of the affinities of recombinant human 4.1R to spectrin and F-actin were about 9.915 and 2.291, and 9.885 and 2.155 (Fig. 4f and g), individually. In comparison, the Bmax and Kd values of the affinities of rBjP4.1 to spectrin and F-actin were closely comparable to those of recombinant human 4.1R. Consistent with the ELISA results, no specific binding of BSA to spectrin and F-actin was detected by Scatchard analysis. These data denote that like human 4.1R, rBjP4.1 is able to specifically bind to spectrin and actin.
Affinity of truncated BjP4.1 peptides to spectrin and actin
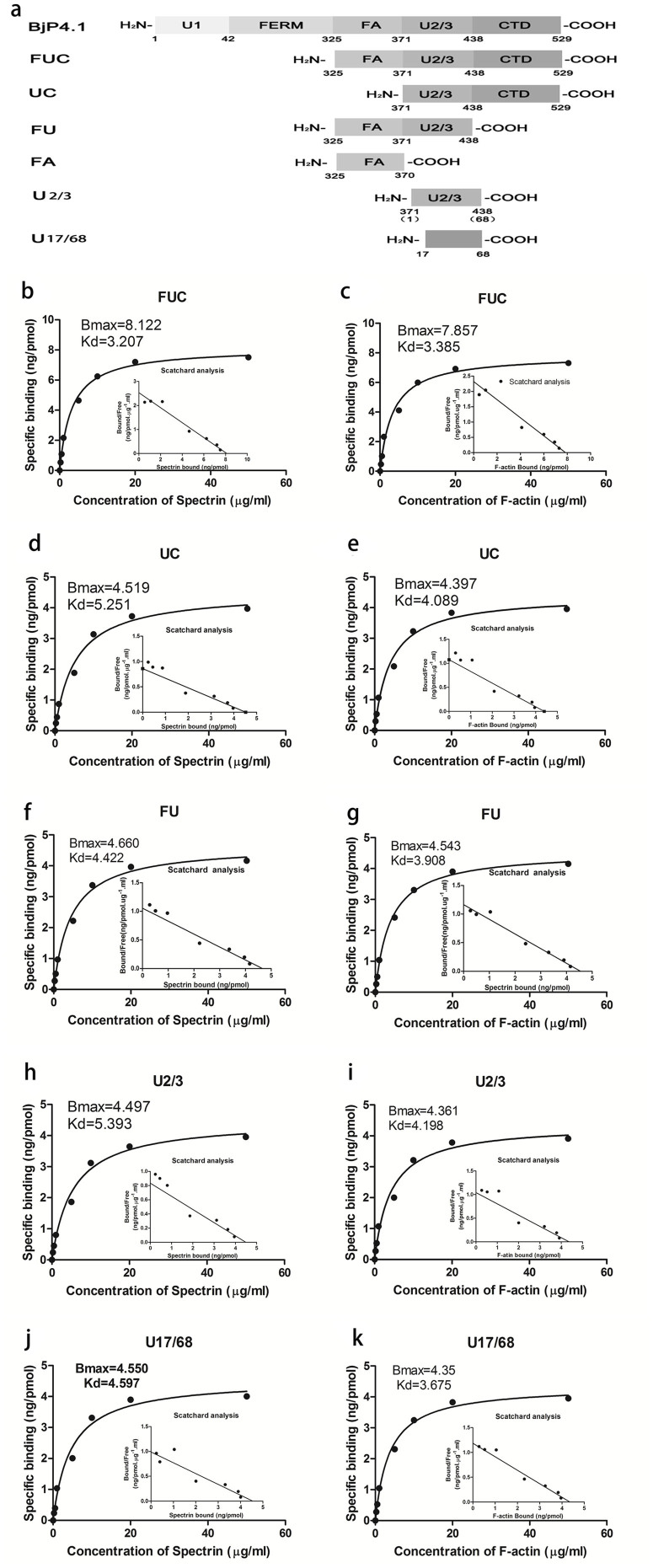
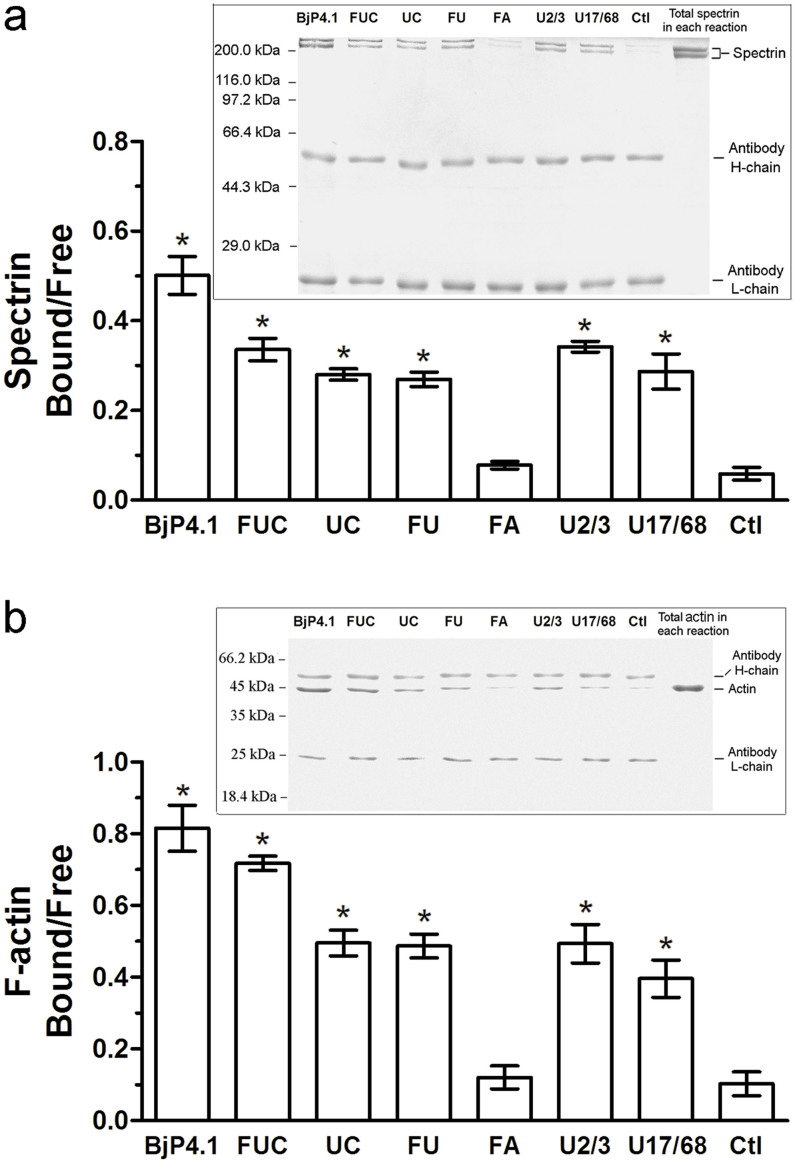
To determine the structure-activity relationship, various truncated BjP4.1 peptides (FUC, UC, FU, FA, U2/3 and U17/68), with the specific domains deleted (Fig. 5a), were expressed in E. coli, purified (Supplementary Figure S2) and subjected to binding analysis. As shown in Fig. 5, the truncated peptide FUC had an affinity to spectrin and actin, which is comparable to full rBjP4.1. Similarly, the truncated peptides UC, FU, U2/3, and U17/68 retained strong affinities to spectrin and actin, although their binding activities were conspicuously reduced by the deletion of U1-FERM-FA, U1-FERM plus CTD, U1-FERM-FA plus CTD, and the N-terminal 16 residues of U2/3 (Fig. 5). By contrast, the truncated peptide FA had no affinity to spectrin and actin (data not shown). Specifically, the Bmax and Kd values of the affinities of FUC, UC, FU, U2/3 and U17/68 to spectrin and actin were ~8.122 and ~3.207, and ~7.857 and ~3.385 (Fig. 5b and c), ~4.519 and ~5.251, and ~4.397 and ~4.089 (Fig. 5d and e), ~4.66 and ~4.422, and ~4.543 and ~3.908 (Fig. 5f and g), and ~4.497 and ~5.393, and ~4.361 and ~4.198 (Fig. 5h and i) as well as ~4.550 and ~4.597, and ~4.35 and ~3.675 (Fig. 5j and k), respectively.
Figure 5. Interaction of truncated BjP4.1 peptides with spectrin and actin.
(a) A diagram showing BjP4.1 truncation. (b–k) Saturation curves of the binding of truncated BjP4.1 peptides FUC, UC, FU, U2/3, and U17/68 to spectrin and actin. The truncated peptides dissolved in re-distilled water were applied to the wells of a 96-well microplate, air-dried overnight at room temperature, and followed by Scatchard analysis.
Coimmunoprecipitation assay was also used to examine the interactions of rBjP4.1 and its truncated BjP4.1 peptides with spectrin and actin. An anti-His-tag antibody was utilized to immunoprecipitate rBjP4.1 and its recombinant truncated peptides, and the amount of cosedimenting spectrin or actin was quantified to estimate the binding capacity. Consistent with the Scatchard analysis results above, rBjP4.1 displayed a strong affinity to spectrin and actin, and the truncated peptides FUC, UC, FU, U2/3 and U17/68 also retained considerable affinities to spectrin and actin (Fig. 6a and b). In contrast, the FA peptide did not bind spectrin and actin (Fig. 6a and b).
Figure 6. Coimmunoprecipitation of spectrin and actin with rBjP4.1 and its truncated peptides.
Incubation of 1 μM spectrin (a) or actin (b) with various recombinant peptides (0.8 μM) was conducted as described under “Methods”. Incubation of the anti-His-tag monoclonal antibody with spectrin or actin alone was the negative control. The symbol * shows a significant difference from control (p<0.05). The corresponding Western blots were shown as inserts, respectively.
These data together indicated that (1) the affinities of BjP4.1 to spectrin and actin were only slightly reduced by the deletion of U1-FERM domains; (2) the affinities of FUC to spectrin and actin were further reduced by the deletion of FA although it itself did not bind to spectrin and actin; (3) the affinities of U2/3 to spectrin and actin were very similar to those of FU and UC, i.e. the deletion of FA and CTD had little influence on the affinities of U2/3 to spectrin and actin; and (4) the affinities of U2/3 to spectrin and actin was not affected by the deletion of its N-terminal 16 residues. It is clear that although the presence of U1-FERM-FA contributes to the affinities of BjP4.1 to spectrin and actin, the U2/3 domain of BjP4.1 is the primary structure capable of interacting with spectrin and actin, and the deletion of its N-terminal 16 residues has little effect on the interaction of U2/3 with spectrin and actin.
U2/3 is a competitive inhibitor of spectrin- and actin-4.1R interaction
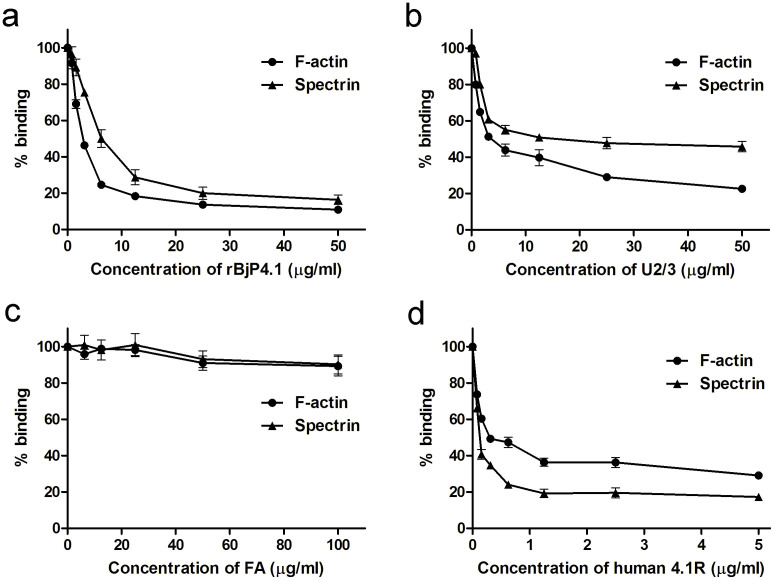
To further confirm the specific binding of U2/3 to spectrin and actin, we performed an ELISA to check if U2/3 can competitively inhibit the binding of spectrin and actin to human 4.1R. Spectrin and actin were both pre-incubated with increasing concentrations of U2/3 and then probed over immobilized human 4.1R. It was found that ~50% inhibition of the binding of spectrin and actin to immobilized human 4.1R was achieved at 12.5 and 3.13 μg/ml of U2/3, respectively (Fig. 7). Similarly, the full length rBjP4.1 caused ~50% inhibition of the binding of spectrin and actin to immobilized human 4.1R at 6.25 and 3.13 μg/ml, and almost complete inhibition of the binding of spectrin and actin to human 4.1R at 50 μg/ml. By contrast, FA did not inhibit the binding at all even at a concentration of 100 μg/ml. Furthermore, human 4.1R could also significantly inhibit the binding of spectrin and actin to immobilized U2/3 (Fig. 7d), with ~50% inhibition of the binding of spectrin and actin to U2/3 achieved at 0.1 and 0.3 μg/ml of human 4.1R, respectively. Taken together, these results suggest that U2/3 is able to compete with human 4.1R for binding to spectrin and actin and vice versa, and the U2/3 domain of BjP4.1 is indeed the primary structure capable of interacting with spectrin and actin.
Figure 7. Competition ELISA.
Spectrin and F-actin were pre-incubated with increasing doses of rBjP4.1 (a), U2/3 (b) or FA (c), and allowed to react with recombinant human 4.1R immobilized on a 96-well ELISA plate. In addition, spectrin and F-actin were also pre-incubated with increasing doses of human 4.1R (d), and allowed to react with U2/3 immobilized on a 96-well ELISA plate. Bound spectrin or actin was detected as described under “Methods”, and the levels were expressed as a percentage of binding in the absence of any compound.
Affinity of U2/3-derived peptides to spectrin and F-actin
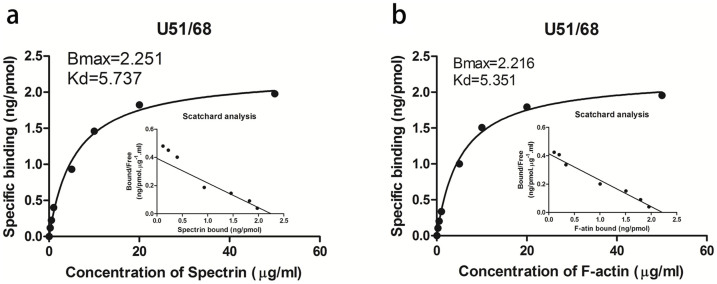
To pinpoint the key region of U2/3 (which consists of 68 residues) involved in binding to spectrin and actin, the different peptides, including U1/16, U17/33, U34/50 and U51/68, were synthesized and subjected to the ELISA. It was found that the peptides U1/16, U17/33 and U34/50 had little affinities to spectrin and actin (Supplementary Figure S3). By contrast, U51/68 retained some affinities to spectrin and actin. As shown in Fig. 8, the Bmax and Kd values of the affinities of U51/68 to spectrin and actin were ~2.251 and ~5.737, as well as ~2.216 and ~5.351, that were about half of those of U2/3. These show that the C-terminal region of U2/3 consisting of 17 residues of U2/3 is the minimum structure which is still capable of binding to spectrin and actin.
Figure 8. Saturation curve of the binding of the synthesized peptide U51/68 to spectrin and actin.
The peptide dissolved in re-distilled water was applied to the wells of a 96-well microplate, air-dried overnight at room temperature, and followed by Scatchard analysis.
Discussion
Protein 4.1 evolved to protect cell membranes against mechanical stresses and to organize membrane microstructure. In vertebrates, protein 4.1 functions to link transmembrane proteins with the underlying spectrin/actin cytoskeleton via the SAB domain. In this study we have cloned and characterized the first cDNA of a protein 4.1 in the cephalochordate amphioxus. The deduced 529-amino-acid long protein, BjP4.1, has a N-terminal FERM domain and a CTD domain, but not the recognizable SAB domain, which are typical of known invertebrate protein 4.1 members. The phylogenetic analysis showed that BjP4.1 is likely to be the common ancestor of vertebrate 4.1 proteins, from which the vertebrate four genes encoding 4.1R, 4.1N, 4.1G, and 4.1B originated, possibly via gene duplication. This appears further strengthened by the presence of a single protein 4.1 gene (ESTEXT_FGENESH2_PG.C_ 710159) in the B. floridae genome.
As protein 4.1 genes from B. japonicum and B. floridae are highly conserved, sharing more than 89% identity at amino acid levels, the genomic structure of B. floridae protein 4.1 gene thus allows us to compare the amphioxus gene structure with that of vertebrate (human) protein 4.1 genes. As in human 4.1R, 4.1N, 4.1G and 4.1B genes, both FERM and FA as well as CTD domains in amphioxus 4.1 are encoded by 8 exons, 1 exon and 4 exons, respectively. Additionally, FERM-, FA- and CTD-encoding domains in both human and amphioxus are identical. Furthermore, human 4.1R, 4.1N, 4.1G and 4.1B as well as amphioxus 4.1 genes all possess the same phase introns. These show that the primary composition of human 4.1 proteins and amphioxus 4.1 protein is highly conserved throughout chordate evolution in terms of both exon-intron structure and sequence homology, hinting at the clue that 4.1 gene transcription is regulated similarly in cephalochordate and vertebrates.
SAB domain functions to link transmembrane proteins with the underlying spectrin/actin cytoskeleton. It has been proposed that protein 4.1-spectrin-actin interactions arose with the vertebrates9. This seems to be supported by the reports that all the protein 4.1 members identified in invertebrates including sea anemone, fruit fly and ascidian lack a recognizable SAB domain4,7. Surprisingly, we demonstrate here that amphioxus protein 4.1, BjP4.1, though lacking a recognizable SAB domain, is capable of binding to both spectrin and actin, with a binding capacity comparable to that of human protein 4.1. This reveals the presence of a “cryptic” spectrin-actin-binding activity in amphioxus protein 4.1, i.e. the capacity to bind spectrin and actin acquired with the advent of basal chordate. This suggests that the vertebrate-like membrane-associated cytoskeleton already evolved in the invertebrate species such as amphioxus. Whether the similar structure with spectrin-actin-binding activity is present in other invertebrate protein 4.1 remains to be determined.
We then underwent to map the structure for the spectrin-actin-binding activity in BjP4.1. It is found that the truncated peptide FUC with U1-FERM depleted retains the spectrin-actin-binding capacity comparable to that of full BjP4.1, suggesting the existence of the spectrin-actin-binding activity in FA-U2/3-CTD domains. It is further shown that the peptide FA has no affinity to spectrin and actin, while U2/3 (U1-FERM-FA plus CTD deleted) retains a considerable spectrin-actin-binding activity, which is similar to that of the peptide UC (U1-FERM-FA deleted). These suggest that the U2/3 domain is the primary structure for interaction of BjP4.1 with spectrin and actin, i.e. the U2/3 is a newly identified domain analogous to the SAB in vertebrate 4.1 proteins. The biochemical characterization of a SAB-like domain in amphioxus protein 4.1 will make it possible to identify additional interacting proteins.
To pinpoint the key region of U2/3 (which consists of 68 residues) involved in binding to spectrin and actin, the different peptides, including U1/16, U17/33, U34/50 and U51/68, are synthesized and subjected to the binding assay. It is demonstrated that neither U1/16 nor U17/33 nor U34/50 have the ability to bind spectrin and actin, but U51/68 still retains some affinity to spectrin and actin. It is clear that the C-terminal 17 amino acids (PPTVEVKTETVKYVASDE) in U2/3 are the minimum sequence which retains the capacity to interact with spectrin and actin.
In summary, this study highlights the presence of an novel SAB-like domain in amphioxus 4.1 protein, via showing that amphioxus 4.1 protein binds both spectrin and actin, and that the U2/3 is the primary region capable of interacting with spectrin and actin. It also suggests that amphioxus 4.1 protein gene represents the common ancestor of vertebrate 4.1 protein genes, from which vertebrate 4.1 protein genes originated. Taken together, these imply that the SAB domain probably evolved with the advent of chordates.
Methods
Cloning and sequencing of cDNA
Total RNAs were extracted with Trizol (TAKARA) from the amphioxus B. japonicum and polyA+ RNA was purified using polyA tract mRNA isolation system II (Promega) according to the manufacturer's instructions. The first-strand cDNA was synthesized with reverse transcription system (Promega) using oligo d(T) primer. The specific primers (upstream primer 5′-ATGCCGTCGGAGGAGGGTGTTA-3′, and downstream primer 5′-GCTGTCTTCATGCTGACTCTCTAC-3′) were designed based on the putative protein 4.1 found in the Florida amphioxus B. floridae genome database (http://genome.jgi-psf.org//Brafl1/Brafl1.home.html)13. To obtain the full sequence of B. japonicum protein 4.1 gene (Bjp4.1), PCR was performed using the first-strand cDNA as template.
The cDNA sequence obtained was analyzed for coding probability with the DNATools program. Comparison against the GenBank protein database was performed using the BLAST network server at NCBI14. Molecular mass (MW) and isoelectric point (pI) were determined using ProtParam (http://www.expasy.ch/tools/protparam.html). Nuclear localization signal was predicted by the PSORT II (http://psort.hgc.jp/form2.html). Protein domains were analyzed using the SMART program (http://smart.embl-heidelberg.de/). Multiple protein sequences were aligned using the MegAlign program by the CLUSTAL method in DNASTAR software package15. Phylogenetic tree was constructed by the neighbor-joining method16 within the package MEGA 5.0 software package using 1000 bootstrap replicates17. The exon-intron organization of amphioxus protein 4.1 gene was derived from NCBI database (http://www.ncbi.nlm.nih.gov/) and Ensembl database (http://www.ensembl.org).
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was used to examine the expression profile of Bjp4.1 in the different tissues of B. japonicum. Total RNAs were prepared with Trizol (Invitrogen) from the whole animals and the different tissues including the hepatic caecum, hind-gut, gill, muscle, notochord, testis, and ovary. After digestion with RQ1 RNase-free DNase (Progema) to eliminate the genomic contamination, the RNAs were used to synthesized cDNAs with reverse transcription system using oligo d(T) primer, and the cDNAs were used as template. The PCR primer sets specific for Bjp4.1 (upstream primer 5′-GGTCAAATATGTGGCCTCCGATG-3′, and downstream primer 5′-TGTGTGGTGGTTGTCGTGGAGATAG-3′) and β-actin (upstream primer 5′-TTCCAGCCTTCATTCCTCG-3′, and downstream primer 5′-CGGTGTTGGCGTACAGGTC-3′) were designed using primer 5.0 program. After qualification of the cDNA template, qRT-PCR was performed on ABI 7500 real-time PCR system (Applied Biosystems, USA) as described by Wang et al18,19. The β-actin gene was chosen as the reference for internal standardization.
Expression and purification of recombinant BjP4.1
The complete coding region of Bjp4.1 was amplified by PCR with the upstream primer 5′-GCGAATTCATGCCGTCGGAGGAGGGTG-3′ (EcoRI site is underlined), and the downstream primer 5′-GGAAGCTTGCTGTTGGGTGTCGTCAGTCGTC-3′ (HindIII site is underlined). The PCR product was digested with EcoRI and HindIII, respectively, and sub-cloned into the plasmid expression vector pET28a (Novagen) previously cut with the same restriction enzymes. The identity of the insert was verified by sequencing and the plasmid was designated pET28a/Bjp4.1.
The cells of Escherichia coli BL21 were transformed with the plasmid pET28a/Bjp4.1. A single positive colony screened was cultured overnight in LB (Luria–Bertani) broth containing kanamycin (50 μg/ml). The culture was diluted 1:100 in LB broth and subjected to further incubation at 37°C for 5 h. Isopropyl-β-D-thiogalactoside (IPTG)-induced expression and purification of the recombinant protein were performed as described by Fan et al20. The eluted samples were analyzed by SDS/PAGE (12% gel), followed by staining with Coomassie Brilliant Blue R-250. Protein concentrations were determined by the method of Bradford using bovine serum albumin as a standard.
Western blotting
The lysates of IPTG-induced E. coli BL21 cells with pET28a/Bjp4.1 and the recombinant BjP4.1 (rBjP4.1) purified were resolved by SDS/PAGE (12% gel with a 5% spacer gel). The lysates of non-induced E. coli BL21 cells with pET28a/Bjp4.1 were also analysed as a negative control. The blots were performed and analyzed as described by Liu and Zhang21. The reaction was quenched with distilled water after the bands became visible.
Assay for binding of rBjP4.1 to spectrin and actin
Spectrin (Sigma-aldrich, St. Louis, USA) was dissolved in PBS, giving a concentration of 40 μg/ml. Similarly, 40 μg/ml of actin (Cytoskeleton Inc., Denver, USA) was prepared according to the kit instruction. An aliquot of 50 μl (2 μg) of spectrin or F-actin was applied to each well of a 96-well microplate, air-dried overnight at room temperature, and incubated at 60°C for 30 min to fix the ligand. After blocking with 200 μl of 20 mg/ml BSA in 10 mM PBS (pH7.4) at 37°C for 2 h and washing with 200 μl of 10 mM PBS containing 1% Tween 20, each well was added 50 μl of different concentrations (0, 1, 5, 10, 20, 50, 100, and 200 μg/ml) of rBjP4.1, incubated at room temperature for 3 h, and added 100 μl of anti-His-tag mouse monoclonal antibody (Cwbio, Beijing, China) diluted to 1:5000 with 10 mM PBS (pH 7.4) containing 1% milk powder. After incubation at 37°C for 2 h, the wells were each added 100 μl of anti-mouse IgG/HRP antibody (Cwbio, Beijing, China) diluted to 1:2000 with 10 mM PBS containing 1% milk powder, incubated at 37°C for 2 h, and then added 75 μl of 0.4 mg/ml O-phenylenediamine (Amresco, Solon, USA) in 51.4 mM Na2HPO4, 24.3 mM citric acid, and 0.045% H2O2 (pH 5.0). After reaction at 37°C for 20 min, 25 μl of 2 mM H2SO4 was added to each well to terminate the reaction, and absorbance at 492 nm was monitored by a microplate reader (Genios Plus, Tecan). For control, BSA was used instead of spectrin or F-actin.
For Scatchard analysis, rBjP4.1 was dissolved in re-distilled water, giving a concentration of 40 μg/ml. An aliquot of 50 μl (2 μg) rBjP4.1 solution was applied to each well of a 96-well microplate, air-dired overnight at room temperature, and incubated at 60°C for 30 min to fix rBjP4.1. After blocking with 200 μl of 20 mg/ml BSA in 10 mM PBS (PH 7.4) at 37°C for 2 h, each well was washed with 200 μl of 10 mM PBS containing 1% Tween 20, added 50 μl of spectrin solution or F-actin solution with different concentrations (0, 0.25, 0.5, 1, 5, 10, 20, 50 μg/ml) of spectrin or actin, and incubated at room temperature for 3 h. After washing four times with 200 μl of 10 mM PBS containing 1% Tween 20, the wells were each added 100 μl of anti-spectrin antibody (Cwbio, Beijing, China) or anti-actin antibody (Bios, Beijing, China), that were both diluted to 1:1000 with 10 mM PBS (pH 7.4) containing 1% milk powder, incubated at 37°C for 2 h, washed, and then processed as above. For controls, BSA (negative control) and recombinant human 4.1R (positive control) were used instead of rBjP4.1 and processed similarly. The equilibrium association constant (Kd) and apparent maximum number of bindings (Bmax) were determined according to the Scatchard plot using the software GraphPad Prism 5.01.
Expression of truncated rBjP4.1 and binding assay
To search for the core region for interaction of BjP4.1 with spectrin and actin, the structure-activity relationship was explored. The N-terminal U1-FERM domains, N-terminal U1-FERM-FA domains, N-terminal U1-FERM domains plus C-terminal U2/3-CTD domains, N-terminal U1-FERM domains plus C-terminal CTD domains as well as N-terminal U1-FERM-FA domains plus C-terminal CTD domain were deleted from BjP4.1, respectively. The upstream primers used were: 5′-GGAATTCAGCAAAATCTTGCGCCT-3′, 5′-GGAATTCATGGACGCAGCTCCT-3′, 5′-GGAATTCAGCAAAATCTTGCGCCT-3′, 5′-GGAATTCAGCAAAATCTTGCGCCT-3′ and 5′-GGAATTCATGGACGCAGCTCC-3′ (EcoRI site is underlined); and the downstream primers used were: 5′-CCTCGAGTCAGTCGTCAGTCGTCT-3′, 5′-CCTCGAGTCAGTCGTCAGTCGTCT-3′, 5′-CCTCGAGTCAGCTCTTGCTGGAGAA-3′, 5′-CCTCGAGTCACTCATCGGAGGC-3′ and 5′-CCTCGAGTCACTCATCGGAGGC-3′ (XhoI site is underlined). Similarly, the N-terminal 16 residues of U2/3, which are more divergent as revealed by phylogenetical analysis, were deleted. The upstream and downstream primers used were: 5′-GGAATTCGACTTGTCAAAGCTCCC-3′ (EcoRI site is underlined) and 5′-CCTCGAGTCACTCATCGGAGGC-3′ (XhoI site is underlined). Construction of the expression vector plasmid, transforming into E. coli BL21 as well as expression and purification of recombinant truncated proteins were all performed as described above. The plasmids constructed (See Fig. 5a) were verified by sequencing and designated FUC (U1-FERM domains deleted), UC (U1-FERM-FA domains deleted), FA (U1-FERM domains plus U2/3-CTD domains deleted), FU (N-terminal U1-FERM domains plus C-terminal CTD domains deleted), U2/3 (U1-FERM-FA domains plus CTD domain deleted), and U17/68 (N-terminal 16 residues of U2/3 domain deleted), individually. Assay for binding of the truncated proteins to spectrin and actin was performed as above.
Coimmunoprecipitation assay
An aliquot of 1 μM of spectrin or F-actin was mixed in 20 μl reaction volume with BjP4.1 or its various recombinant truncated peptides (0.8 μM) in PBST (pH 7.4, containing 1% Tween 20 and 0.3 M NaCl) at 25°C for 60 min with gentle shaking, respectively. The anti-His-tag mouse monoclonal antibody (Cwbio, Beijing, China) was subsequently added (final concentration: 1 μM) and the incubation continued at 25°C for 60 min. A total of 20 μl of protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology), blocked by BSA previously, was added to the reaction mixture and incubated at 25°C for 30 min with gentle shaking. After centrifugation at 1000 g at room temperature for 1 min, the agaroses were washed five times with PBST and two times with PBS. The precipitated proteins were eluted from the agaroses with 30 μl of SDS/PAGE loading buffer and analysed by SDS/PAGE (8% gel for spectrin-group and 12% gel for actin-group). The immunoblotting analysis was carried out as above. The bound fractions were counted using SensiAnsys software. Incubation of the monoclonal antibody with spectrin or actin alone followed by interaction with protein A/G PLUS-Agarose, was the negative control.
Competition ELISA
To investigate if the U2/3 domain of BjP4.1 could compete with human 4.1R for binding to spectrin and F-actin and vice versa, a competition ELISA was preformed. Briefly, each well of plates was coated with 50 μl of 10 μg/ml recombinant human 4.1R, and incubated with 50 μl of spectrin (5 μg/ml) or F-actin (5 μg/ml) supplemented with different concentrations (0–50 μg/ml) of full-length rBjP4.1, U2/3 or FA at 25°C for 3 h. Bound spectrin or actin was detected as above. Reversely, to examine human 4.1R mediated the inhibition of spectrin and actin binding to immobilized U2/3, ELISA plate was coated with 50 μl of 40 μg/ml U2/3, and incubated with 50 μl of spectrin (10 μg/ml) or F-actin (10 μg/ml) supplemented with different concentrations of recombinant human 4.1R (0–5 μg/ml) at 25°C for 3 h. Bound spectrin or actin was detected as above.
Synthesis of U2/3-derived peptides and binding assay
As U17/68 retained considerable capacity to bind spectrin and actin (see below), the different peptides derived from U2/3, i.e. U1/16, U17/33, U34/50, and U51/68, were synthesized by Sangon Biotech Co. Ltd (Shanghai, China), and subjected to the ELISA as above.
Statistical analysis
All the experiments were conducted three times. Statistical analyses were performed using the computer program SPSS 13.0. The statistical significance of difference between mean values was calculated by analysis of variance, and the difference at p < 0.05 was considered significant.
Author Contributions
S.Z. conceived the experiments. L.W., Y.W. and Z.G. took the experimental data. L.W., Z.L. and Z.G. analyzed the data. S.Z., L.W., Z.L. and Z.G. wrote the paper. All authors joined the discussion and provided comments.
Supplementary Material
Functional characterization of protein 4.1 homolog in amphioxus: Defining a cryptic spectrin-actin-binding site
Acknowledgments
This work was in part supported by a grant of Natural Science Foundation of China (31172071) to SCZ.
References
- Bennett V. & Baines A. J. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev 81, 1353–1392 (2001). [DOI] [PubMed] [Google Scholar]
- Leto T. L. & Marchesi V. T. A structural model of human erythrocyte protein 4.1. J Biol Chem 259, 4603–4608 (1984). [PubMed] [Google Scholar]
- Taylor-Harris P. M. et al. Cardiac muscle cell cytoskeletal protein 4.1: analysis of transcripts and subcellular location–relevance to membrane integrity, microstructure, and possible role in heart failure. Mamm Genome 16, 137–151 (2005). [DOI] [PubMed] [Google Scholar]
- Baines A. J. A FERM-adjacent (FA) region defines a subset of the 4.1 superfamily and is a potential regulator of FERM domain function. BMC Genomics 7, 85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimm J. A., An X., Nunomura W. & Mohandas N. Functional characterization of spectrin-actin-binding domains in 4.1 family of proteins. Biochemistry 41, 7275–7282 (2002). [DOI] [PubMed] [Google Scholar]
- Baines A. J. The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life. Protoplasma 244, 99–131 (2010). [DOI] [PubMed] [Google Scholar]
- Hoover K. B. & Bryant P. J. The genetics of the protein 4.1 family: organizers of the membrane and cytoskeleton. Curr Opin Cell Biol 12, 229–234 (2000). [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Dawson I. A. & Artavanis-Tsakonas S. A Drosophila homologue of membrane-skeleton protein 4.1 is associated with septate junctions and is encoded by the coracle gene. Development 120, 545–557 (1994). [DOI] [PubMed] [Google Scholar]
- Baines A. J. Evolution of the spectrin-based membrane skeleton. Transfus Clin Biol 17, 95–103 (2010). [DOI] [PubMed] [Google Scholar]
- Bateman A. et al. The Pfam protein families database. Nucleic Acids Res 30, 276–280 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F. & Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A 75, 4853–4857 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. & Deutsch M. Association of intron phases with conservation at splice site sequences and evolution of spliceosomal introns. Mol Biol Evol 16, 1528–1534 (1999). [DOI] [PubMed] [Google Scholar]
- Putnam N. H. et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Chenna R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31, 3497–3500 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K. & Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5, 150–163 (2004). [DOI] [PubMed] [Google Scholar]
- Sanderson M. J. & Wojciechowski M. F. Improved bootstrap confidence limits in large-scale phylogenies, with an example from Neo-Astragalus (Leguminosae). Syst Biol 49, 671–685 (2000). [DOI] [PubMed] [Google Scholar]
- Wang S., Zhang S., Zhao B. & Lun L. Up-regulation of C/EBP by thyroid hormones: a case demonstrating the vertebrate-like thyroid hormone signaling pathway in amphioxus. Mol Cell Endocrinol 313, 57–63 (2009). [DOI] [PubMed] [Google Scholar]
- Wang G., Zhang S. & Wang Z. Responses of alternative complement expression to challenge with different combinations of Vibrio anguillarum, Escherichia coli and Staphylococcus aureus: evidence for specific immune priming in amphioxus Branchiostoma belcheri. Fish Shellfish Immunol 26, 33–39 (2009). [DOI] [PubMed] [Google Scholar]
- Fan C. et al. Identification and expression of a novel class of glutathione-S-transferase from amphioxus Branchiostoma belcheri with implications to the origin of vertebrate liver. Int J Biochem Cell Biol 39, 450–461 (2007). [DOI] [PubMed] [Google Scholar]
- Liu M. & Zhang S. A kringle-containing protease with plasminogen-like activity in the basal chordate Branchiostoma belcheri. Biosci Rep 29, 385–395 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional characterization of protein 4.1 homolog in amphioxus: Defining a cryptic spectrin-actin-binding site