Abstract
To facilitate high-throughput biochemical analyses of membrane proteins, we have developed a novel display technology in a microarray format. Both a single-pass (CD4) and a multiple-pass (GPR77) human transmembrane proteins were engineered to be displayed in the membrane envelop of herpes simplex virions. These viruses produce large spherical virions displaying multiple copies of envelop proteins. Our aim was to engineer this virus to express these human proteins during the virus productive cycle and incorporate the human proteins into the virion during the assembly process. Another strategy presented includes engineering a fusion of glycoprotein C (gC), a major constituent of herpes simplex virus type 1 (HSV-1) virions, by hijacking the cis-acting signals to direct incorporation of the chimeric protein into the virion. The expression of the human proteins in infected cells, at the cell surface and in purified virions, is in the correct transmembrane orientation and the proteins are biochemically functional. Purified virions printed on glass slides form a high-density Virion Display (VirD) Array and the displayed proteins were demonstrated to retain their native conformations and interactions on the VirD Array judging by similar assays, such as antibody staining, as well as lectin and ligand binding. This method can be readily scaled or tailored for different modalities including to a high-content, high-throughput platform for screening ligands and drugs of human membrane proteins.
Introduction
Approximately one-third of the human proteome is comprised of membrane proteins that belong to protein families with a variety of biochemical activities, such as transporters, channels, receptors, recognition molecules, and adhesion molecules. Membrane proteins are critically important molecules for cell survival, maintenance of cell homeostasis, cell signaling, immune surveillance, molecular transport, and cell-cell communication. This class of proteins represents up to 70% of therapeutic targets for all prescribed drugs. Therefore, development of a high- throughput platform that enables profiling membrane proteins in an active conformation for their biochemical activities will have an important impact on drug discovery by streamlining small molecule screening methods. However, membrane proteins, especially those containing multi- pass transmembrane (TM) domains, are notoriously difficult to study because they have to be embedded in a membrane to maintain a native conformation and many require proper posttranslational modifications (PTMs), such as glycosylation, which occurs during transport in the cellular secretory pathway. Although membrane protein microarrays have been produced, biochemical purification using detergents limits the throughput and subsequent manipulation of such arrays1, 2.
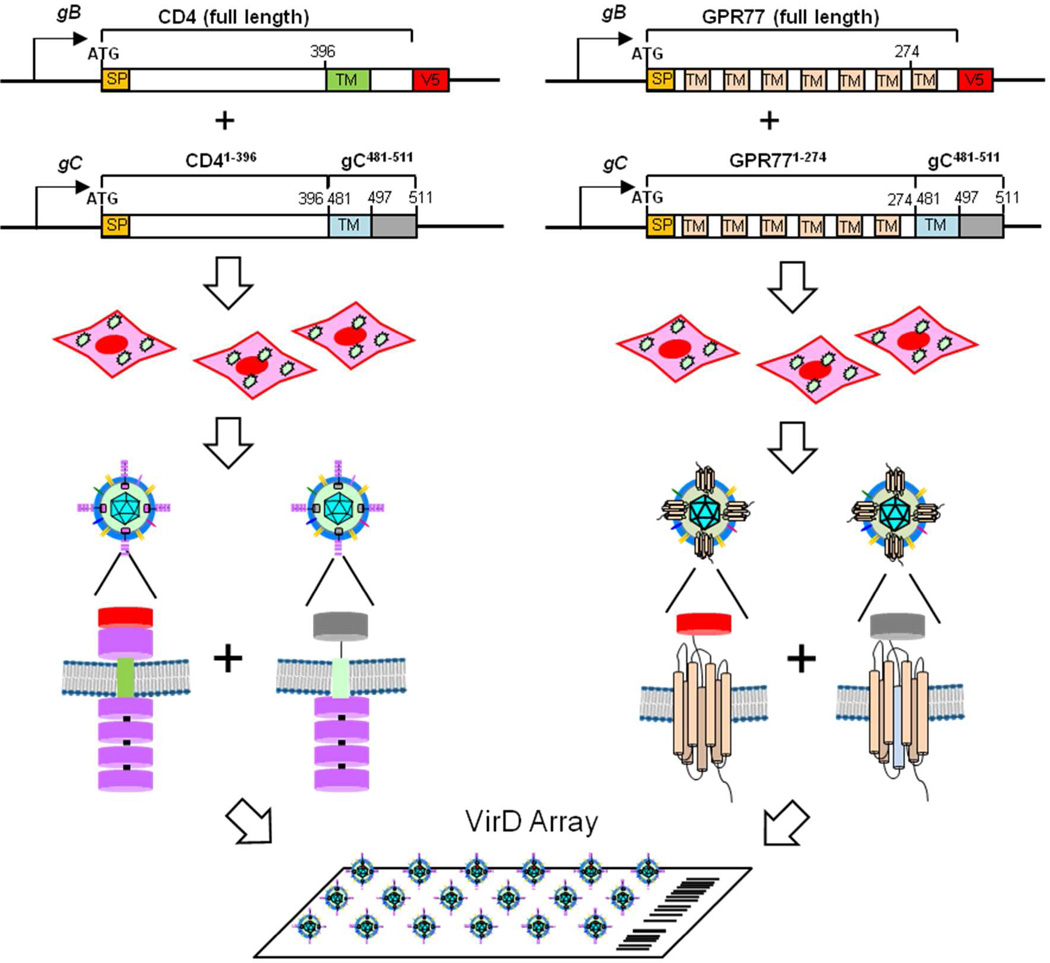
To develop a new, high-throughput platform that displays human membrane proteins in their native conformation, we made use of herpesvirus virons as a vehicle to fabricate a Virion Display microarray, dubbed VirD Array. Herpesviruses, such as herpes simplex virus type 1 (HSV-1), produce large membrane-enveloped virons that contain high copies of the viral glycoproteins, such as the three major glycoproteins gB, gC, and gD, that are distributed regularly in the circular virion structure. The DNA genome of this virus can be genetically manipulated to express foreign proteins in their native configuration or as fusions with the viral proteins. Our singular goal was to develop a method to efficiently incorporate human membrane proteins in HSV-1 virions and then to discover whether these proteins were functional in the viral membrane. We explored two strategies for the VirD array: 1) Clone a full-length human ORF at the gB locus and express the gene under control of the strong gB promoter; 2) Fuse a human ORF to the TM and cytoplasmic domains of gC and express this chimeric gene at the gC locus under the control of the gC promoter (Fig. 1). Previous studies have also utilized a gC chimera approach (hepatitis C virus glycoprotein E2) as well as expression from the gC locus (CD4) to incorporate foreign proteins in HSV-1 virions3, 4. Important for these strategies to work is the observation that the absence of either gB or gC does not affect the ability of the virus to assemble mature enveloped virions in infected cells5–7.
Figure 1.
Development of the VirD Array. Schematic of the two strategies used for the virion display system are shown. The first utilizes expression of the CD4/GPR77 molecules tagged with the V5 epitope from the gB promoter and the second uses a chimeric expression method by fusing the CD4 ectodomain/GPR77 to the TM and C-terminus of glycoprotein C. The signal peptide (SP) is shown. Recombinant HSV-1 viruses expressing these engineered human genes were used to infect mammalian cells and the viruses released from these cells which would incorporate the human membrane protein in the virion envelop were purified and printed on FAST slides.
To test the feasibility of the two approaches, we chose CD4 as a classical type I membrane protein with a single TM domain and GPR77 (a.k.a. C5L2) as a representative of the multi- spanning (seven TM regions), G-protein coupled receptor (GPCR) membrane protein. CD4 is a well-characterized membrane glycoprotein of T lymphocytes that interacts with major histocompatibility complex class II antigens and is also a receptor for the human immunodeficiency virus8. GPR77 is involved in the complement system of the innate immune response with a canonical ligand identified (i.e., complement component C5a)9. Our goal was to examine the expression and incorporation of these human membrane proteins into HSV-1 virions using the two strategies outlined above and to determine whether these human membrane proteins are maintained in their native form in purified virions immobilized on a glass surface at high density.
Experimental Section
Cells and Viruses
Vero cells, transformed Vero cell lines and human foreskin fibroblasts (HFT) were grown in minimum essential medium - alpha medium supplemented with 10% fetal calf serum (Gibco-Invitrogen) and passaged as described in Desai et al. (1998)10. HFT is an immortalized cell line that is transduced with a retrovirus expressing human telomerase11. D6 (UL27 transformed) was used as the host cell for growth of the recombinant viruses that expressed genes from the gB locus5. The A1.1 (UL27 and UL28 transformed) cell line12 was used for the marker-rescue/marker-transfer method to introduce the human genes cloned into the gB loci13 and was a kind gift from Fred Homa of the University of Pittsburgh. Stocks of the parental wild-type virus strain KOS (HSV-1) and the mutant and recombinant viruses were prepared as previously described10. All tissue culture and virus manipulation was performed in a Biosafety Level 2 facility. Infected cells and media containing infectious material was treated with 10% sodium hypochlorite. Other material utilized during the handling of virus infected cells was subjected to steam sterilization in an autoclave.
Antibodies
Antibodies reactive to human CD4 and GPR77 were purchased from Santa Cruz Biotechnology and Sigma-Aldrich, respectively. PE-conjugated anti-CD4 antibody used for flow cytometry analysis was obtained from BD Biosciences. The V5 monoclonal antibody was purchased from Invitrogen Life Technologies. Anti-HSV-1 gB antibody clone B6 and anti-gC antibodies were a kind gift from Joseph Glorioso (University of Pittsburgh). Anti-HSV-1 gD antibody DL6 was a generous gift from David Johnson (OHSC) and Gary Cohen and Roz Eisenberg (Penn University). The VP5 antibody LP12 was kindly provided by Tony Minson (University of Cambridge, UK).
Plasmids
Plasmid pKΔ4B was derived by Cai et al.14 following engineering of linker-insertion mutants in the glycoprotein B gene. DNA sequences encoding amino acids 43 through 711 of gB were deleted and a BglII restriction site added to maintain the protein reading frame14. pKΔ4B was digested with Xho1 and BglII, treated with antartic phosphatase (NEB) and ligated with an Xho1-BglII PCR fragment amplified from pKΔ4B which deletes all gB amino acids from 1–43 (gBΔSS) but retains the gB promoter sequences (Table 1). This plasmid was designated pKgBΔSS. The sequence of gB amino acids spanning 711 to 796 were deleted from pKgBΔSS by cassette PCR mutagenesis. The PCR fragment was cloned as a BglII-BamH1 into pKgBΔSS and the resulting plasmid was labeled pKgBPR. The human CD4 and GPR77 sequences were amplified from the plasmids from the Ultimate ORF collection (Life Technologies). The sequence encoding the V5 epitope was included in the reverse primer (Table 1). The final gB promoter driven gene plasmids were labeled pKgB:CD4 and pKgB:GPCR77. Sequence analysis of the different plasmids was done prior to introduction into the virus genome. Plasmids were linearized with BamH1 for homologous recombination.
Table 1.
Primer Sequences
| Primer | Sequence (5’-3’) |
|---|---|
| UL28-Xho1-F | CTTTGCCTCGGTCTACCGGTGCGGGG |
| gBΔSS-BglII-R | GGGAGATCTGAGGCGGGACTACGGGGGCCCGTCG |
| gB-797-BglII-F | GGGAGATCTGGGTGGAGGTGGAGGTTACGTCATGCG GCTGCAGAGCAAC |
| gB-nc-BamH1-R | GGGATCCCAACCGGAGGCATCCAAC |
| gB-CD4-BglII-F | GGCAGATCTACCATGAACCGGGGAGTCCCTTTTAGG |
| gB-CD4V5-BglII-R | CCCAGATCTCTACGTAGAATCTAGACCGAGGAGAGGGTTAGGGATAGGCT- TACCAATGGGGCTACATGTCTTCTGAAA |
| gB-GPR77-BglII-F | GGCAGATCTACCATGGGGAACGATTCTGTCAGCTAC |
| gB-GPR77V5-BglII-R | CCCAGATCTCTACGTAGAATCTAGACCGAGGAGAGGGTTAGGGATAGGCT- TACCAATGGGGCTACATGTCTTCTGAAA |
| gC-KAN-F | CGCTTTGCCGGGAACGCTAGCCGATCCCTCGCGAGGGGGAGGCGTCGGGC- GGCCTGGTGATGATGGCGGGATCG |
| gC-KAN-R | GGGGGGACCAAACTATATAGATATTAAAAAGGTAACGGGGGGGTCTTGCG- TCAGAAGAACTCGTCAAGAAGGCG |
| CD4-F | GGAATTCAACATGAACCGGGGAGTCCCTTTTAGG |
| CD4-R-Overlap | CCCGATTCCAATTGGCTGCACCGGGGTGGACCATGT |
| GPR77-F | GGAATTCACCATGGGGAACGATTCTGTCAGCTAC |
| GPR77-R-Overlap | CCCGATTCCAATGGGTTCAGCCCGCAGGGCCCTGGC |
| gC-F-Overlap-CD4 | CCGGTGCAGCCAATTGGAATCGGGGTTCTCGCGGCG |
| gC-F-Overlap-GPR77 | CGGGCTGAACCCATTGGAATCGGGGTTCTCGCGGCG |
| gC-R | GGGGATCCTTACCGCCGATGACGCTGC |
| gC-RedET(CD4)-F | CGCTTTGCCGGGAACGCTAGCCGATCCCTCGCGAGGGGGAGGCGTCGGGC- ACCATGAACCGGGGAGTCCCTTTTAGG |
| gC-RedET(GPR77)-F | CGCTTTGCCGGGAACGCTAGCCGATCCCTCGCGAGGGGGAGGCGTCGGGC- ACCATGGGGAACGATTCTGTCAGCTAC |
| gC-RedET-R | GGGGGGACCAAACTATATAGATATTAAAAAGGTAACGGGGGGGTCTTGCG- TTACCGCCGATGACGCTGCCGCGA |
Marker-rescue/marker-transfer assays
The marker rescue of UL28 and marker transfer of the gB:human ORF gene was accomplished using the method described in Desai et al.13. A1.1 cell monolayers (1×106) in 60 mm petri dishes were co-transfected with 25 µl of infected cell DNA (KΔ4BX) and 0.1–0.05 µg linearized plasmid DNA using the calcium phosphate precipitation method. When plaques began to appear (72 h after transfection) the cell monolayers were harvested, freeze/thawed once, sonicated and total virus progeny titered. The recombinant virus was isolated by single plaque purification on D6 cells. Additional plaque purification was carried out by limiting dilution on the D6 cell line.
Red-ET recombination
The KOS BAC37 genome15 was transferred into TOP10 cells (Stratagene) for this method. KOS BAC37 was kindly provided by David Leib, Dartmouth University, NH. The procedure to engineer gC chimera fusions into the virus genome used the Gene Bridges Red-ET method and the protocols provided16. The kanamycin cassette surrounded by gC homologous sequences was amplified using gC-Kan-F and gC-Kan-R primers and pRPSL-neo as a template. This kanamycin gene was introduced into KOS BAC37 replacing the gC gene. Colonies that grew on kanamycin plates were screened for streptomycin sensitivity before the next step. The CD4-gC and GPR77-gC fusion genes were made using Overlap PCR methods using the primers listed in Table 1. The CD4-gC and GPR77-gC chimera fusions were amplified using the RedET primers listed in Table 1 and were used to replace the kanamycin gene. Successful isolates carrying the correct chimeric genes were identified by PCR assays and the inserted gene in the BAC genome was sequenced prior to reconstitution of infectious virus.
Transfection of Bacmid DNA to re-constitute infectious virus
The KOS Bacmids carrying the glycoprotein C chimera gene fusions were prepared using the PureLink nucleic acid purification kit (Life Technologies). The Bacmid DNA was transfected into Vero cells (5 X 105) in 12 well trays using Lipofectamine 2000 reagent (Life Technologies). Plaques generally began to appear after 3 days and this infected cell lysate was used to amplify and prepare working stocks of each of the gC chimera viruses.
Western blot analysis
Infected cell extracts were resolved by SDS-PAGE in MES buffer and transferred to iBlot membranes (Life Technologies) using an iBlot apparatus (Life Technologies) according to the manufacturer's protocol. The transferred membranes were blocked with blocking buffer (TBS with 5% non-fat milk) at room temperature for an hour with gentle shaking, and then incubated with primary antibodies (1:5000 dilution in blocking buffer) at room temperature for an hour with gentle shaking. The membranes were washed for 5 min with TBS+0.1% Tween20 (TBST) buffer for 3 times with shaking. HRP-conjugated anti-mouse antibodies (GE Healthcare) were incubated on the membranes at 5,000-fold dilution in blocking buffer for an hour with gentle shaking. The membranes were washed for 5 min with TBST buffer for 3 times with shaking and incubated with ECL Plus Western Blotting detection reagents (GE Healthcare) for 5 min before signals were visualized by ImageQuant LAS 4000 imaging system (GE Healthcare).
Immunofluorescence and Confocal Analysis
HFT cells in LabTek (#1 borosilicate glass) four well chamber slides (6×105 cells) were infected at an multiplicity of infection (MOI) of 10 plaque forming units (PFU) per cell. Infected cells were washed 2X with DPBS (Dulbecco’s phosphate buffered saline), fixed with 4% paraformaldehyde in DPBS for 25 min; washed 2X with DPBS and permeabilized with 0.25% triton X-100 in DPBS for 30 min. After permeabilization, the cells were washed 2X with 3% BSA in DPBS and non-specific reactivity was blocked for 30 min in the same buffer. For cell surface labeling the detergent permeabilizaton step was omitted and the cells incubated with blocking buffer. Primary antibody was diluted in 3% BSA/DPBS and 250 µl added to each chamber well for 60 min (room temperature). Subsequently the cells were washed 3X with 3% BSA/DPBS and then incubated with Cy3-labeled secondary antibody (Jackson Laboratories) for 45 min (room temperature). The cells were then washed 3X with 3% BSA/DPBS and then incubated in Fluormount G (EMS) prior to imaging. The stained infected cells were analyzed in a Zeiss LSM 510 confocal microscope. Most images were collected with a pinhole set at 1 Airy unit.
Virion preparation
Extracellular virions were prepared from HFT cells. Generally 8.6 X 106 cells in 100 mm petri dishes were infected at an MOI of 10 PFU/cell. The culture medium was harvested at 72 h post-infection, clarified by centrifugation at 3500 rpm for 30 min at 4°C. The supernatant was layered on a 20% sucrose cushion (W/V in growth medium) and centrifuged in a Beckman SW41 (39 K for 30 min) or SW32 (24 K for 60 min). The virion pellet was resuspended in PBS at 4°C overnight and then used for subsequent analyses. For VirD Array printing the virion preparations were resuspended in PBS plus 35% glycerol. The titers of KOS (wild-type) virions, which are infectious, was monitored by plaque assays during the different purification and manipulation procedures to ensure biological integrity of the virions.
Flow-Cytometry
Extracellular virions (150 µl volumes) were incubated with PE-conjugated flow antibodies (20 µl) and incubated at room temperature (tube rocker) for one hour in the dark. The virions (volume adjusted to 500 µl with PBS) were then sedimented through 20% sucrose cushion (250 µl) in an Eppendorf tube at 16000 g for 60 min. The supernatant was discarded and the virus pellet resuspended in 200 µl PBS. The labeled virions were analyzed in a BD FACSARIA II instrument using the DIVA software (version 6.1.3).
Enzyme-linked immunosorbent assay (ELISA)
Serial dilutions of virions were incubated in Nunc MaxiSorp flat-bottom 96 well white plates. The sealed plates were incubated at 4°C for 2 days. The wells were washed with PBS + 0.02% Tween-20 (PBS+T20) 3 X for 5 min each time on a platform shaker and then blocked with 2% BSA in PBS+T20 for 60 min at room temperature. Primary antibody dilutions were made in blocking buffer generally 1:2000 to 1:250 and incubated for 60 min. Secondary HRP conjugated mouse antibody was used at a 1:1000 concentration. The plates were washed 3X with PBS+T20 for 5 min each wash after both primary and secondary antibody binding. The reaction was quantitated using SuperSignal ELISA Pico (Pierce) chemiluminescent substrate according to the manufacturer’s procedure and the plate read in a Glomax luminometer to determine relative light units (425 nm).
VirD Array fabrication
Purified virions were arrayed in a 384-well plate and spotted on FAST slides (Whatman) in a 4x4 pattern along with BSA as a negative control. The printed arrays were stored at −80°C.
Antibody assays on VirD Arrays
VirD Arrays were blocked with blocking buffer (TBS with 3%BSA) at room temperature for an hour with gentle shaking, then incubated with primary antibodies (1:1000 dilution in blocking buffer) at room temperature for an hour with gentle shaking. The arrays were washed for 5 min with TBS+0.1% Tween20 (TBST) buffer for 3 times with shaking. To visualize the presence of human or viral proteins, Cy5-labeled anti-mouse antibodies (The Jackson Laboratory) were incubated on the arrays at 1,000-fold dilution in blocking buffer. The arrays were washed for 5 min with TBST buffer for 3 times with shaking, briefly rinsed with water, and dried by spinning. The slides were finally scanned with a GenePix 4000B scanner (MDS Analytical Technologies).
Ligand binding assay on VirD Array
The VirD Array was blocked in TBST with 1% BSA for 1 h at room temperature with gentle shaking. Complement anaphylatoxins C3a (Alpha Diagnostic Intl), C4a (MyBioSource), and C5a (Abcam) were individually labeled with Cy5 NHS Easter (GE Healthcare) and incubated on the VirD Array at 1 μM in ligand binding buffer (1 mM MgCl2, 2 mM CalCl2, 0.2% BSA, and 25 mM HEPES, pH 7.4) at room temperature for 1 h with gentle shaking. The array was washed for 5 min in ice-cold washing buffer (0.5 M NaCl in 10 mM HEPES, pH 7.4) for 3 times with shaking, dried by spinning, and scanned as described above.
Lectin binding assays on VirD Arrays
VirD Arrays were blocked in PBS with 1% BSA for 1 h with gentle shaking. Lectins (EY Laboratories) were labeled with Cy5 NHS Easter (GE Healthcare) and incubated on the VirD Array at 1 μg/ml in PBS with 0.5 mM CaCl2 and 1% BSA at room temperature for 1 h with gentle shaking. The array was washed for 5 min in PBST for 3 times with shaking, dried by spinning, and scanned as described above.
Results and Discussion
Recombinant methods were used to generate four viruses (see Experimental Section for details). Viruses, gB:CD4 and gB:GPR77, express the full-length human membrane proteins at the gB locus under the control of native gB promoter. We also incorporated a V5 epitope tag at the C-termini of both proteins for biochemical detection purposes. Viruses labeled CD4-gC and GPR77-gC, express human membrane proteins fused to the gC C-terminal domain (i.e., 481 to 511 aa), which contains the TM and a short cytosolic domain. Like the human genes cloned at the gB locus, the gC chimeras were cloned at the gC locus under the control of native gC promoter (Fig.1).
The precise mechanism by which many of the HSV-1 glycoproteins are incorporated into mature virions is still not determined. The absence of either one of the major glycoproteins, gB, gC, gD or gH-gL, does not appear to affect the incorporation of the others even though these proteins function as a complex during virus entry and egress17. Glycoprotein M may play a role in the incorporation of gC in certain cell types18. The TM domain may have a role in this process as well as the cytosolic tails of these glycoproteins which could mediate virion incorporation by interaction with the underlying tegument structure19. We first attempted to make chimeras of the human proteins with gB, but none of the fusion proteins were expressed at the cell surface and as a consequence these chimera proteins were not detected in our extracellular virus particle preparations similar to the observation demonstrated for the inner nuclear membrane protein UL3420. What became apparent was the abundant stable accumulation of the human polypeptide expressed from the gB promoter in infected cells. Thus, we generated an expression module to express the native human gene as a “viral gene” with the goal that the expressed proteins would become incorporated into mature virions during virus egress and maturation. Because there was evidence in the literature that fusion of foreign genes to gC were incorporated into the virion4, 21, this type of strategy was also developed to potentially increase the virion incorporation using cis-acting signals present in those sequences.
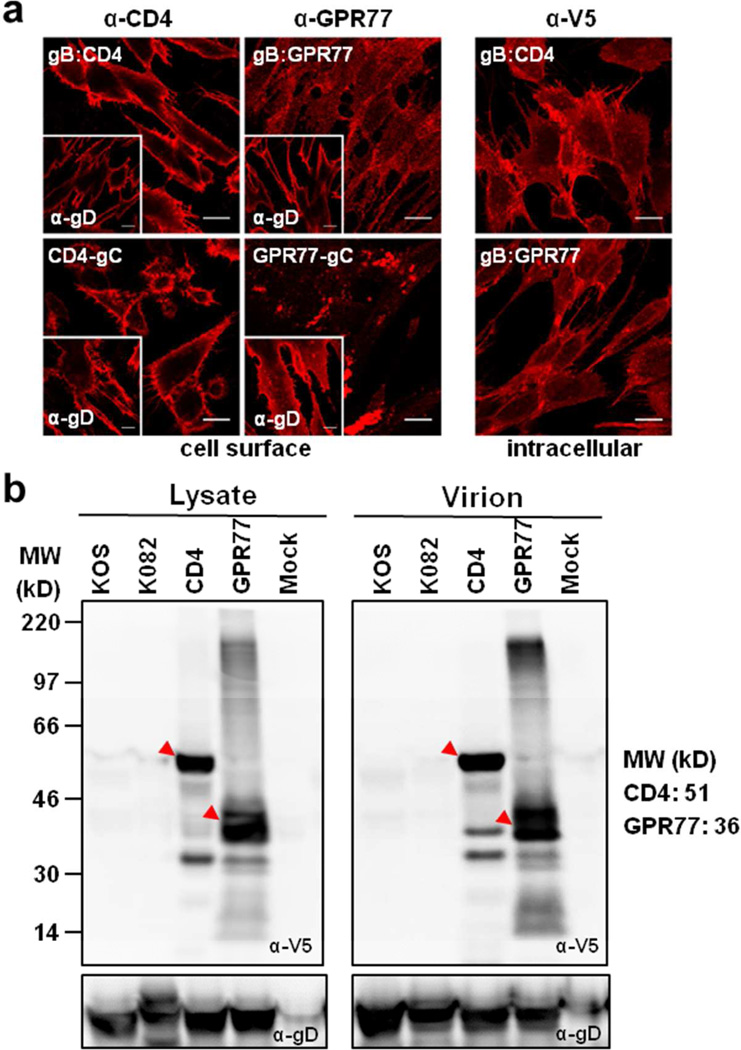
To examine whether CD4 and GPR77 were expressed and correctly processed through the secretory pathway, human fibroblasts infected with the recombinant and parental viruses were stained with antibodies against the ecto-domains of CD4, GPR77, and HSV-1 gD to visualize cell surface localization of these antigens (Fig. 2a). As expected, gD was detected on the surfaces of infected cells (Fig. 2a; insets). CD4 expressed either from the gB promoter or as a gC-chimeric protein was also detected on the cell surface as judged by the fluorescence signals. GPR77 was detected on the cell surface, but the distribution of the fluorescence was different depending on whether it was expressed from the gB promoter or as a gC-chimera. These results suggest that CD4 and GPR77, like gD, expressed off the HSV-1 genome were delivered to the surface of the plasma membrane via the canonical secretory pathway. KOS- and K082-infected cells did not react with the antibodies to CD4 and GPR77 (data not shown). The intracellular distribution of CD4 and GPR77 was examined by staining with V5 antibody following permeabilization of infected fibroblast cells (Fig. 2a; right panel). HSV-1 glycoproteins localize to nuclear, endoplasmic reticulum, Golgi and cell surface membranes during productive infection. The intracellular distribution of CD4, whether the cells were stained with anti-CD4 or anti-V5 antibodies, was similar to the intracellular distribution of gD (Fig. S1).
Figure 2.
Expression of CD4 and GPR77. a. Confocal analysis of HFT cells infected with the recombinant viruses demonstrating cell surface expression of CD4 and GPR77. The cell surface expression of gD is shown for similar infected cells in the insets of each panel. Intracellular distribution of CD4 and GPR77 expressed from the gB promoter was visualized by staining with anti-V5 antibodies following permeabilization of cells. Scale bars: 200 microns. b. Expression of CD4 and GPR77 in infected cell lysates and incorporation of these proteins in mature virions was confirmed by western blot analysis using anti-V5 antibody.
Further biochemical evidence for the expression of at least the V5-tagged human proteins was obtained using immunoblot analysis of total infected cell lysates (Fig. 2b; left panel). We also examined virion incorporation of these human membrane proteins. Wild type and gB null (K082) virions5, as well as gB:CD4, gB:GPR77 virions, were harvested from infected cell culture supernatants, clarified by low speed centrifugation and purified through a 20% sucrose cushion. These virions were analyzed using the same immunoblot methods with anti-V5 antibodies. Both gB:CD4 and gB:GPR77 virions showed strong anti-V5 reactivity at the expected molecular weights of CD4 and GRP77, while no detectable signals were observed in the other virions (Fig. 2b; right panel). Anti-gD antibodies were used as a loading control. We have also purified virus from infected cell culture supernatants using sucrose gradients and glycerol-tartrate cushions. These virions have similar virus polypeptide profiles to the particles purified using the sucrose cushion and all contained the human membrane protein, which was detected with the V5 antibody (data not shown). Together, these data confirmed that both CD4 and GPR77 were synthesized and incorporated into virions produced in infected human cells.
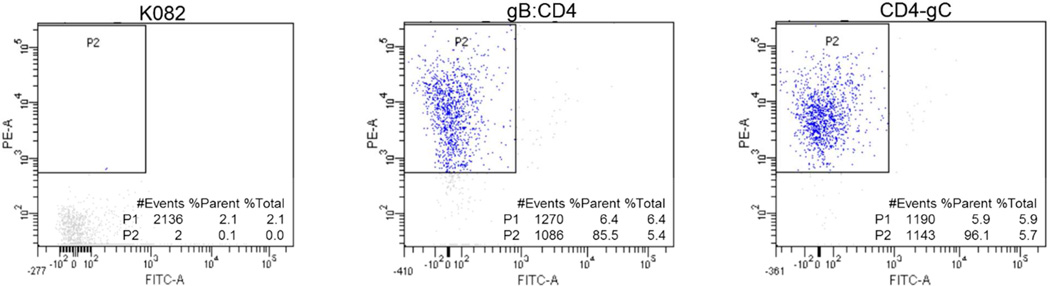
To demonstrate that human proteins could be displayed in the correct orientation after virion incorporation, we performed flow cytometry analysis of purified virions stained with PE-labeled antibodies that recognize the ecto-domain of CD4 (Fig. 3). An HSV-1 recombinant virus that incorporates the Venus fluorescent protein in the capsid was used to identify and gate purified virions (Experimental Section). K082 virions were used as a negative control for antibody binding specificity. Judging from the amounts of PE fluorescence detected within the gated virion populations, 85.5% of gB:CD4 virions were labeled with PE antibody and slightly more, 96.1% of CD4-gC virions were bound to PE antibody. These results were consistent with data from different virion preparations using similar experimental conditions. This observation was further confirmed using a standard ELISA analysis using chemilluminescent substrates for detection (Fig. S2). All virion preparations were stained with anti-gD antibodies as expected. Virions expressing CD4 or GPR77 were stained with the respective antibodies. There was little or no reactivity of the CD4 and GPR77 antibodies with the KOS or K082 virions. The signal observed with anti-gD antibodies was significantly higher because of the higher affinity of this monoclonal antibody for its antigen. Taken together, these results demonstrated that the human membrane proteins were incorporated in the correct transmembrane orientation because they reacted with antibodies that recognize the extracellular domains of these proteins. This observation suggests that the membrane proteins were embedded in the virion envelope in their native conformation.
Figure 3.
FACS analysis of the CD4 virions. The incorporation of CD4 and the conformation of the molecule in the virions was examined by labeling of purified virions with anti-CD4 (PE conjugated) antibodies followed by FACS analysis of the virions.
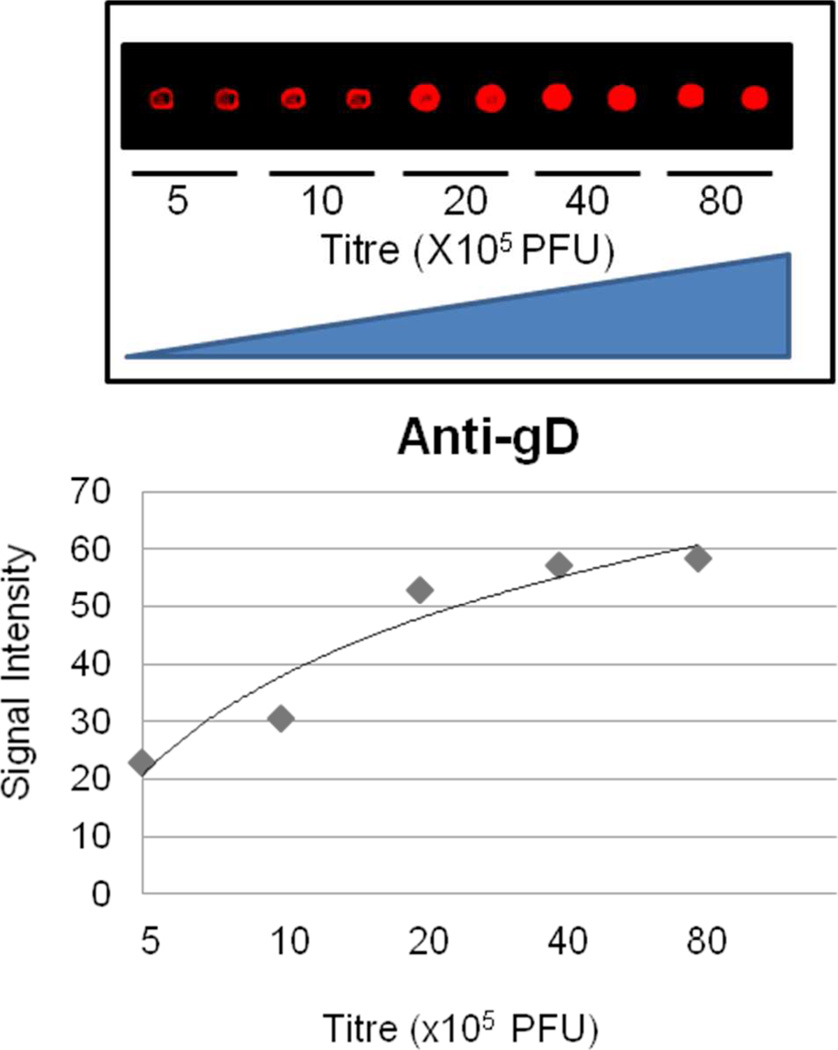
To test whether these recombinant virions could be immobilized in a microarray format at high density, while maintaining their functional integrity, we spotted them on different glass surfaces at various titers. Using wild-type KOS virions that can be titered, we typically derived virions that were at a concentration of 4×108 PFU/µl. Of this we used 0.7 nl to spot the glass surface (3 times), delivering potentially ~106 PFUs to the surface. Using anti-gD antibodies, we determined that nitrocellulose-coated slides (i.e., FAST) provided the optimal detection as low as 500,000 virions (KOS plaque forming units) per spot and the anti-gD signals started to reach saturation after the titer was increased to >4,000,000 virions (Fig. 4). Therefore, we decided to construct the VirD Array with seven different virus preparations at a titer of 8,000,000 virions (KOS plaque forming units) per spot in a 4 × 4 format. It is likely that not all of the different virion preparations will be of the same concentration because of the differences in their genetic backgrounds.
Figure 4.
Wild-type HSV-1 virions were printed on the FAST glass surface at different concentrations. Each spot represents 2.1 nl of virus suspension. The virions were detected using an anti-gD antibody against the ectodomain of this molecule. 500,000 virions (KOS plaque forming units) per spot can be easily detected by the anti-gD antibody on the VirD Arrays. The fluorescence signal begins to reach saturation after the number of virus particles printed increases to >4,000,000 virions.
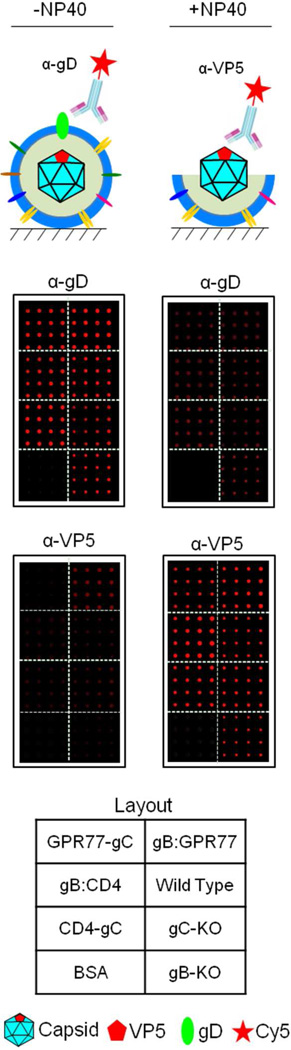
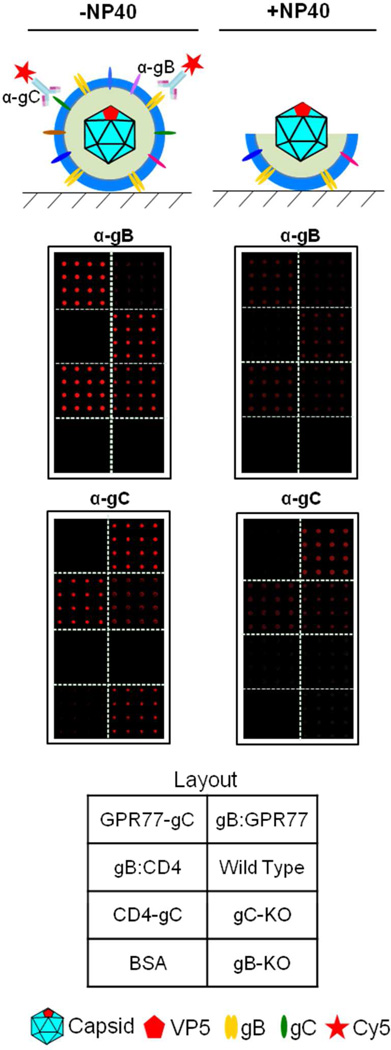
To visualize and examine the integrity of immobilized virions on glass, the arrays were stained with anti-gD ecto-domain and anti-VP5 antibodies, the latter recognizes the major capsid protein, VP5. As expected, all seven virion sectors showed strong anti-gD signals but much lower anti- VP5 signals, indicating that the vast majority of the immobilized virions were intact (Fig. 5; left panel). This conclusion was further supported by the observation that the anti-gD signals were greatly decreased on the VirD Arrays after the virion envelopes were stripped with a mild detergent treatment using NP40 (Fig. 5; right panel). In contrast, strong anti-VP5 signals were seen in all seven virion sectors following this treatment. Moreover, staining the VirD Arrays with anti-gB and -gC antibodies confirmed the expected absence of gB and gC proteins in gB:CD4/GPR77 and CD4/GRP77-gC, respectively (Fig. 6).
Figure 5.
Integrity of virions immobilized on VirD Arrays was confirmed by anti-gD and anti- VP5 antibodies. All seven virion sectors showed strong anti-gD signals but much lower anti-VP5 signals. A mild detergent treatment using 1% NP40 significantly reduced the anti-gD signals and increased the anti-VP5 signals on the VirD Arrays.
Figure 6.
The VirD arrays were incubated with anti-gB and anti-gC antibodies that bind to the extracellular domains of these proteins. Glycoprotein B was detected on wild-type (KOS), the gC null mutant and the gC chimera virion sectors as expected. Glycoprotein C was only detected on wild-type, gB null and the gB engineered virion sectors. NP40 treatment greatly reduced the anti-gB and anti-gC fluorescence signals.
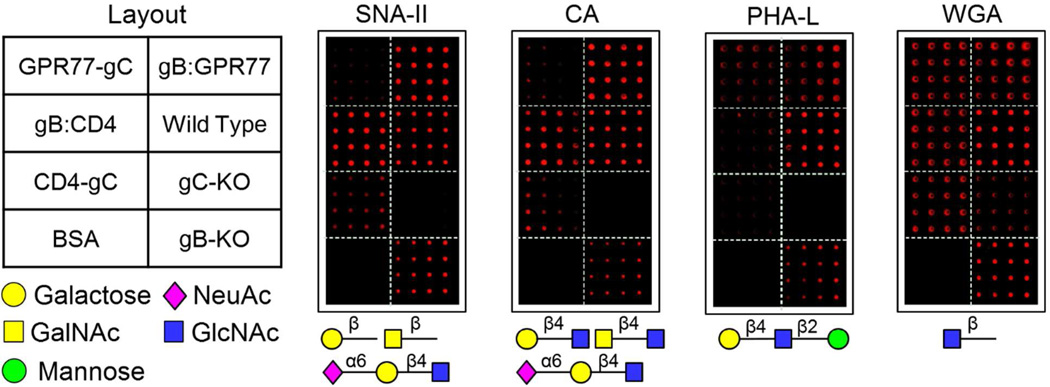
Because glycosylation is important for human membrane protein activity, we employed fluorescently labeled lectins (i.e., SNA-II, PHA-L, CA, and WGA) to profile glycan structures using the VirD Arrays22, 23. Comparison of the lectin staining patterns between wild type, gB-KO (K082) and gC-KO (gCΔ39)7 virion sectors showed that gC is more heavily glycosylated than gB, because all four lectins showed much weaker binding signals to the gC-KO virion sector (Fig. 7). This finding correlates with data showing HSV-1 gC binds to peanut lectin and Helix pomatia lectin24. A more careful analysis of lectin CA staining pattern, which recognizes Galβ(1-4)GlcNAc, GalNAcα(1-4)GlcNAc, or NeuAcα(2-6)Galβ(1-4)GlcNAc, indicated that CD4 was probably modified by these carbohydrates. This is because the CD4-gC (i.e., gC-) virion sector showed significantly higher signals than both gC-KO and GPR77-gC (i.e., gC-) virion sectors. This observation is further supported by the same SNA-II (recognizing terminal Galβ, GalNAcβ, or NeuAcα(2-6)Galβ (1-4)GlcNAc) staining pattern because it is known that SNA-II should recognize the same glycan structures or partial glycan structures as CA does based on the database of lectin specificity at Consortium for Functional Glycomics web site (http://www.functionalglycomics.org) (Fig. 7). Interestingly, a very similar glycan structure NeuAcα(2-3)Galβ(1-4)GlcNAc was previously identified on mouse CD4 expressed in CHO cells using a mass spectrometry approach, indirectly supporting our observation here8. Thus, human CD4 is very likely to be glycosylated with NeuAcα(2-6)Galβ(1-4)GlcNAc. We, however, could not definitively determine any specific glycan structures associated with GPR77, probably due to a limited number of lectins used in this study. Regardless, the above results suggest that the virion-displayed human membrane proteins were glycosylated.
Figure 7.
Lectin-glycan interactions on VirD Arrays. Fluorescently labeled lectins (i.e., SNA-II, CA, PHA-L, and WGA) were used to probe and profile glycan structures on the VirD Arrays.
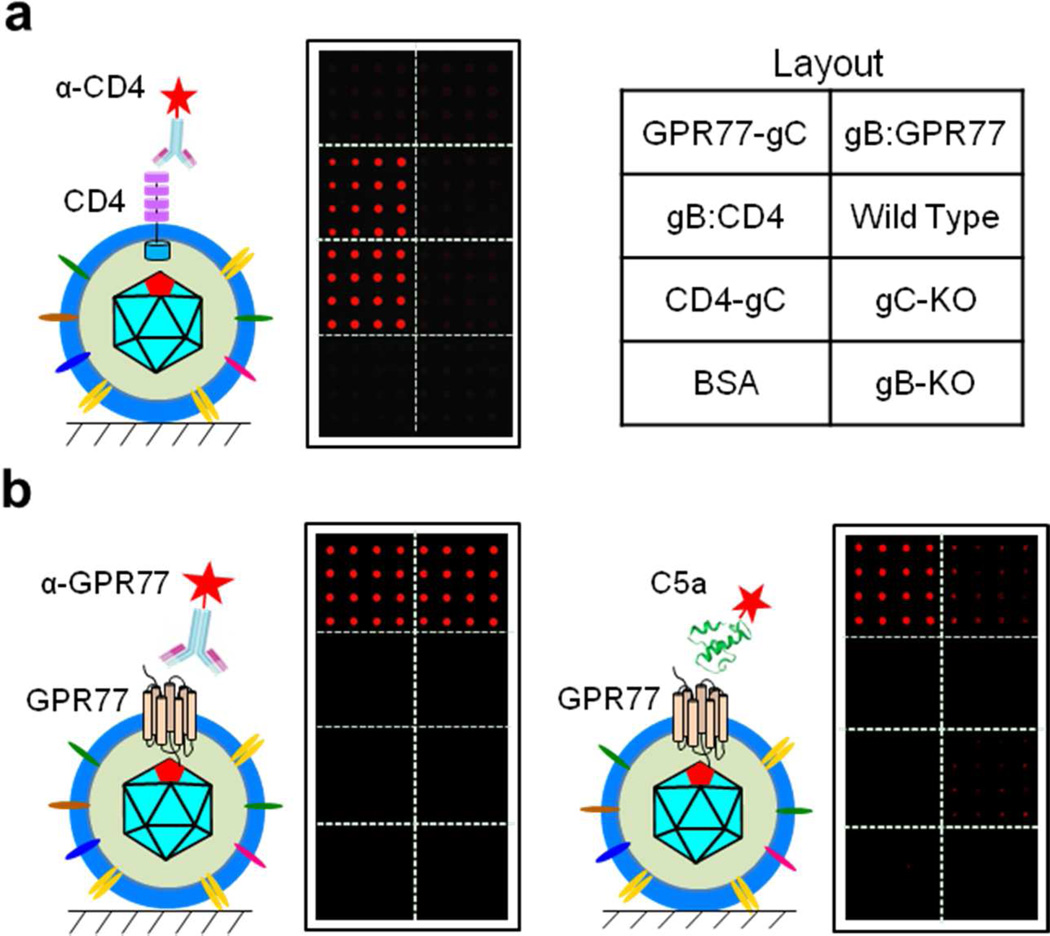
To determine whether human CD4 and GPR77 proteins displayed on the surface of virions immobilized on a glass surface were in the correct orientation, we stained the VirD Arrays with antibodies that each recognizes the ecto-domains of CD4 or GRP77 (Fig. 8). We observed strong and specific staining signals in the gB:CD4 and CD4-gC, gB:GPR77 and GRP77-gC virion sectors, respectively, indicating that these proteins are in the correct orientation and both membrane protein display strategies worked.
Figure 8.
Function and interaction assays on the VirD Arrays. a. Anti-CD4 antibody staining. Strong and specific fluorescence signals were observed only on the gB:CD4 and CD4-gC virion sectors when the arrays were incubated with antibody. b. Anti-GPR77 antibody staining and GPR77-C5a ligand interaction. The antibody binds specifically to gB:GPR77 and GPR77-gC virion sectors as judged by the fluorescence signal. The Cy5-labeled C5a ligand showed strong binding activity to the GPR77-gC virion sector and weaker binding signals to the gB:GPR77 virion sector but no detectable signals on other virion sectors.
Finally, to demonstrate that the virion-displayed human membrane proteins were functional on the glass surface, we probed the VirD Array with a fluorescently labeled canonical ligand, complement anaphylatoxin C5a, of GPR77 with a KD of 2.5 nM (see Experimental Section for more details)9. As shown in the right panel of Figure 8b, Cy5-labled C5a showed relatively strong binding activity to the GPR77-gC virion sector on the array, albeit weaker binding signals were associated with the gB:GPR77 virion sector. No detectable fluorescence signal was observed in the other virion sectors, suggesting the interactions between C5a and GPR77 on the VirD Array was specific. The other two complement anaphylatoxins, C3a and C4a, were also tested on the VirD Arrays. Neither anaphylatoxin C3a nor C4a showed any significant binding signals to the two GPR77 virion sectors on the VirD Arrays (data not shown), further confirming the specific interactions between C5a and GPR77. Taken together, these results indicate that GPR77 was displayed correctly on the virions and maintained its functional conformation on the VirD Array.
Conclusions
Display of soluble peptides or protein in various formats has been effective using different carrier systems25. In this study, we demonstrate fabrication of a VirD Array that displays human membrane proteins on the envelopes of engineered HSV-1 virions immobilized at high density on solid glass surfaces. Using antibodies and lectins we showed that two human membrane proteins, CD4 and GPR77, are in the right orientation and potentially glycosylated. We further demonstrated that virion-displayed GPR77 was in its active conformation via a binding assay with its cognate ligand C5a. The VirD Array approach has several obvious advantages: 1) Displayed human membrane proteins are embedded in human cell membranes, a more physiologically relevant environment that can help maintain their native conformation; 2) As demonstrated with GRP77, membrane proteins with multiple TM domains are likely to be folded correctly in the virion envelopes; 3) Since the virus exploits the human secretory pathways, the displayed human proteins are likely to maintain their canonical PTMs as they are transported through the secretory pathways; this was demonstrated via lectin binding assays; and 4) The VirD Array is expected to be readily transformed to a high-content platform that can display virtually all of the human membrane proteins close to their native conformation on a single glass slide.
Although only two human membrane proteins were employed to establish the VirD Array technology in this study, we envision that all of the human membrane proteins can be included to construct a comprehensive human membrane protein VirD Array. Once such a high-throughput platform is established, it will allow us to perform high-throughput screens for novel drug target identification against membrane proteins, to identify ligands of various types of receptors, and to profile PTM of membrane proteins. For example, fluorescently labeled ligands can be used to probe the VirD Arrays in order to “deorphanize” the GPCRs. Using pathogen-encoded, secreted peptides as probes, the VirD Arrays will be capable of identifying pathogenic elicitor-host receptor interactions. A more sophisticated approach is, perhaps, to engineer a reporter system expressed in the tegument of virions on the VirD Arrays so that the state (e.g., open versus closed) of a given ion channel displayed on the virions can be examined in a high-throughput fashion. When this system is coupled with drug screens, we envision the VirD Array approach will have a significant impact on identifying drug targets for GPCRs and ion channels.
Supplementary Material
Acknowledgements
We thank Poorval Joshi for helping with the molecular biology methods. We thank Wade Gibson and members of his lab for help with the RED-ET recombination and Wade for help and advice with virion purity determination. We acknowledge Yorke Zhang and Lee Blosser for help with the flow cytometry experiments. The continued support of Michael McCaffery for use of the JHU Integrated Imaging Center facilities is greatly appreciated. We also thank David Johnson, Gary Cohen, Roz Eisenberg, Joe Glorioso and Tony Minson for antibodies. Also, we thank Fred Homa, Joseph Glorioso, Myron Levine and David Leib for cell lines, viruses and BAC reagents. Research support was provided by Public Health Service grants from the National Institutes of Health, AI063182 and RC2 CA148402 (P.D.), GM076102, U54HG006434, U24CA160036, and RR020839 (H.Z.), and U54MH084691 (M.L.). Y.F. was supported in part by the China Scholarship Council.
Abbreviations
- CD4
cluster of differentiation 4
- GPR77
G protein-coupled receptor 77
- KO
knockout
Footnotes
Conflict of Interest Disclosure
The authors declare no competing financial interest.
References
- 1.Fang Y, Frutos AG, Lahiri J. J Am Chem Soc. 2002;124:2394–2395. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- 2.Tang CS, Dusseiller M, Makohliso S, Heuschkel M, Sharma S, Keller B, Voros J. Anal Chem. 2006;78:711–717. doi: 10.1021/ac051244a. [DOI] [PubMed] [Google Scholar]
- 3.Dolter KE, King SR, Holland TC. J Virol. 1993;67:189–195. doi: 10.1128/jvi.67.1.189-195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouvatsis V, Argnani R, Tsitoura E, Arsenakis M, Georgopoulou U, Mavromara P, Manservigi R. Virus Res. 2007;123:40–49. doi: 10.1016/j.virusres.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Cai WZ, Person S, Warner SC, Zhou JH, DeLuca NA. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland TC, Homa FL, Marlin SD, Levine M, Glorioso J. J Virol. 1984;52:566–574. doi: 10.1128/jvi.52.2.566-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homa FL, Purifoy DJ, Glorioso JC, Levine M. J Virol. 1986;58:281–289. doi: 10.1128/jvi.58.2.281-289.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr SA, Hemling ME, Folena-Wasserman G, Sweet RW, Anumula K, Barr JR, Huddleston MJ, Taylor P. J Biol Chem. 1989;264:21286–21295. [PubMed] [Google Scholar]
- 9.Cain SA, Monk PN. J Biol Chem. 2002;277:7165–7169. doi: 10.1074/jbc.C100714200. [DOI] [PubMed] [Google Scholar]
- 10.Desai P, DeLuca NA, Person S. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 11.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 12.Tengelsen LA, Pederson NE, Shaver PR, Wathen MW, Homa FL. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai P, Homa FL, Person S, Glorioso JC. Virology. 1994;204:312–322. doi: 10.1006/viro.1994.1536. [DOI] [PubMed] [Google Scholar]
- 14.Cai WZ, Person S, DebRoy C, Gu BH. J Mol Biol. 1988;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- 15.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. J Virol Methods. 2006;135:197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Muyrers JP, Testa G, Stewart AF. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 17.Rodger G, Boname J, Bell S, Minson T. J Virol. 2001;75:710–716. doi: 10.1128/JVI.75.2.710-716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Y, Bell S, Zenner HL, Lau SY, Crump CM. J Gen Virol. 2012;93:319–329. doi: 10.1099/vir.0.035444-0. [DOI] [PubMed] [Google Scholar]
- 19.Maringer K, Stylianou J, Elliott G. J Virol. 2012;86:12971–12982. doi: 10.1128/JVI.01913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loret S, Guay G, Lippe R. J Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laquerre S, Anderson DB, Stolz DB, Glorioso JC. J Virol. 1998;72:9683–9697. doi: 10.1128/jvi.72.12.9683-9697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao SC, Li Y, Zhou J, Qian J, Schnaar RL, Zhang Y, Goldstein IJ, Zhu H, Schneck JP. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kung LA, Tao SC, Qian J, Smith MG, Snyder M, Zhu H. Mol Syst Biol. 2009;5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundstrom M, Olofsson S, Jeansson S, Lycke E, Datema R, Mansson JE. Virology. 1987;161:385–394. doi: 10.1016/0042-6822(87)90131-0. [DOI] [PubMed] [Google Scholar]
- 25.Li M. Nat Biotechnol. 2000;18:1251–1256. doi: 10.1038/82355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.