Abstract
The Notch pathway has been implicated in a number of malignancies with different roles that are cell and tissue-type dependent. Notch1 is a putative oncogene in NSCLC and activation of the pathway represents a negative prognostic factor. To establish the role of Notch1 in lung adenocarcinoma we directly assessed its requirement in K-ras-induced tumorigenesis in vivo, employing an autochthonous model of lung adenocarcinoma with concomitant expression of oncogenic K-ras and deletion of Notch1. We find that Notch1 function is required for tumor initiation via suppression of p53-mediated apoptosis, through the regulation of p53 stability. These findings implicate Notch1 as a critical effector in K-ras-driven lung adenocarcinoma and as a regulator of p53 at a post-translational level. Moreover, our study provides new insights to explain, at a molecular level, the correlation between Notch1 activity and poor prognosis in NSCLC patients carrying wild type p53. This information is critical for design and implementation of new therapeutic strategies in this cohort of patients representing 50% of NSCLC cases.
Keywords: Notch1, NSCLC, p53, apoptosis, MDM2, K-ras
Introduction
Lung adenocarcinoma belongs to the histopathological class of Non-Small Cell Lung Cancer (NSCLC) and is the leading cause of cancer-related cell deaths worldwide. The Notch pathway has been implicated in a number of malignancies, with different roles that are cell and tissue-type dependent (1-6). In the context of lung cancer, recent studies show the different Notch receptors have non-overlapping functions. Previous studies have focused on Notch3, which was found overexpressed as a consequence of a chromosome 19 translocation and shown to control tumor cell apoptosis and maintenance of aldehyde dehydrogenase-expressing lung adenocarcinoma stem cells (7-9). Several studies suggest that genetic or pharmacological targeting of the Notch3 can inhibit tumor cell proliferation (8, 10, 11). Increasing evidence also implicates Notch1 as a putative oncogene in lung adenocarcinoma. Gain-of-function mutations of the NOTCH1 gene and lack of pathway attenuation due to loss of NUMB, a negative regulator of Notch, have been described in human NSCLC (12). Expression of activated intracellular Notch1 domain (N1IC) in the pulmonary epithelium of mice induced lung adenomas, which progressed to full-blown adenocarcinoma when combined with overexpression of Myc (13). Notch pathway activation has also been correlated to poor prognosis and response to therapy in NSCLC patients (14).
Activating mutations in the K-ras gene are a common event in lung adenocarcinoma, occurring in approximately a third of patients (15). Mouse models of lung adenocarcinomas driven by oncogenic K-ras expression have allowed the study of tumor initiation and progression. In particular, inducible expression of a missense mutation at codon 12 (K-rasG12D allele), results in development of lung adenocarcinomas with low invasive and metastatic potential (16). Recent evidence supports the notion of a functional interaction between Notch and Ras signaling pathways during tumorigenesis (17). Notch signaling can be activated by oncogenic Ras and is required for Ras- induced cell transformation (18). Moreover, Notch signaling is required for H-Ras and K-ras driven oncogenesis in mouse models of breast and lung cancer, respectively (19, 20). To elucidate the role of the individual Notch receptors we assessed the requirement for Notch receptors in a cell model of lung adenocarcinoma and observed a dramatic decrease in cell numbers only after Notch1 knockdown. Thus, we examined the requirement for Notch1 in lung tumorigenesis in vivo by employing a mouse model of inducible KrasG12D expression. We find that genetic inhibition of Notch1 signaling results in reduced KrasG12D-driven tumor initiation and burden. Moreover, we show that Notch1 ablation induces p53-dependent apoptosis as a consequence of increased p53 stability. These findings suggest the status of p53 has a primary impact on the effects of Notch1 signaling in lung tumorigenesis and that the outcome of targeting Notch1 in NCSLC patients would be dependent on p53 status.
Materials and methods
Mouse strains
The conditional LSL-KrasG12D (16), Notch1lox/lox (21) mice have been previously described. To initiate lung tumorigenesis, mice were infected with Adenovirus carrying a Cre-recombinase allele, as previously described (16). All studies were conducted in compliance with Wistar Institute IACUC (Institutional Animal Care and Use Committee) guidelines.
Histology and immunohistochemistry
Formalin-fixed paraffin embedded murine lung tissue was processed by standard methods and stained with hematoxylin and eosin (H&E), as previously described (22).
Cell culture conditions
A549, H460, H522, H441, H727 cell lines were acquired from the American Type Culture Collection in 2011 and authenticated periodically by DNA typing (Last test by DDC medical services 6/2013). Cells were grown in supplemented RPMI1640 medium. siRNAs for Notch1, Notch2, Notch3 and p53 were purchased from Dharmacon (Lafayette, CO) and transfected using Lipofectamine RNAiMax (Invitrogen). pcDNA3-Myr-Akt plasmid was obtained from Addgene (23) and transfected using Lipofectamine 2000 (Invitrogen).
Western blot analysis
Lung tissue samples and cell pellets were prepared as described (24). Primary antibodies employed: Notch1 and PARP-p85 (Epitomics), cleaved Caspase-3, cleaved Caspase-7, Akt, phospho-Akt S473, phospho-MDM2 S166 (Cell Signaling Technology), p53 DO-1, MDM2 Ab2 (Millipore), p21 (BD Biosciences), Tubulin and Vinculin (Sigma).
Statistical Analysis
The differences T/L ratios were assessed for statistical significance by χ2 test and for differences in tumor numbers by Wilcoxon signed rank test. All data were represented by the mean and standard deviation (SD) indicates data variability. All other analysis utilized two-tailed unpaired Student’s t test.
Cell Cycle Analysis
72h after siRNA transfection cells were harvested, washed once with PBS and fixed in cold 70% ethanol. Fixed cells were resuspended in propidium iodide buffer (50ug/ml PI, 250mg/ml RNAse A in PBS) and incubated over night at 4C. Cell cycle distribution was evaluated using Coulter Epics XL flow cytometer (Beckman). Data were analyzed using WinMDI software.
Annexin-V staining
Apoptotic cells were measured using Annexin V-FITC (BD Pharmingen) and Propidium Iodide. Briefly, cells were harvested 72h after siRNA transfection and incubated with annexin V-FITC and PI for 15′. Cells were analyzed using Coulter Epics XL flow cytometer (Beckman) data were analyzed using FlowJo software (Tree Star, Inc.).
Results
Reduced tumor initiation and burden in LSL-K-rasG12D;Notch1flox/flox mice
To clarify the role of the individual Notch receptors in the context of lung adenocarcinoma, we analyzed the effect of Notch1-3 knockdown in vitro by siRNA in A549 lung adenocarcinoma cells, which carry an oncogenic mutation of Kras. As shown in Figure 1A, all three receptors are expressed in A549 cells, albeit at different levels. We achieved complete knockdown of Notch1 and Notch2 and > 75% knockdown of Notch3. Notably, several independent Non-Targeting siRNA controls affected Notch3 expression and we therefore included cells treated with the transfection reagent only as a further control. Cell counts for four days after siRNA treatment showed only Notch1 knockdown had a significant effect on overall cell numbers (Figure 1B).
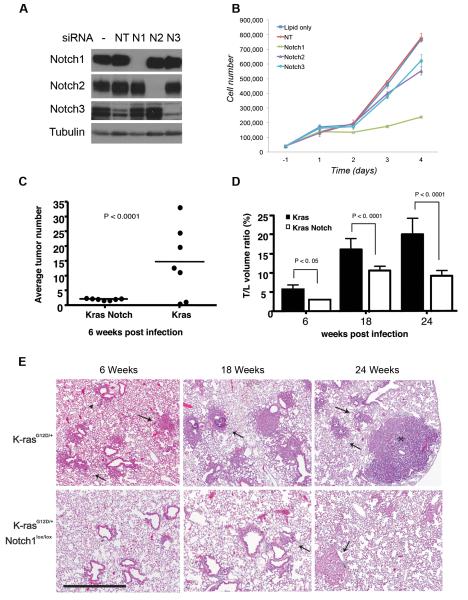
Figure 1. Role of Notch receptors in lung tumorigenesis in vitro and in vivo.
(A-B) Knockdown of Notch1-3 by siRNA in A549 cells. (A) Western blot of Notch1-3 receptors 72h after transfection. Tubulin= loading control. (B) Growth curve of A549 cells. Cells were counted in triplicate starting 24 hours after transfection. Error bars=mean ± SD. One representative experiment of 3 is shown. (C-E) Histological nalysis of lung tumorigenesis in LSL-K-rasG12D and LSL-K-rasG12D;Notch1flox/flox mice. Mice were treated with Ad-Cre (5×106 PFU) and sacrificed at the indicated time points. LSL-K-rasG12D;Notch1flox/flox,-6 weeks, n= 12. All other cohorts, n= 8. (C) Numbers of hyperplastic lesions and tumors at the 6-week time point (Wilcoxon test, Error bars=SD). (D) Average T/L ratio at the different time points. The difference in the T/L is statistically significant at all time points (χ2 test, Error bars= SD). (E) Time course progression of tumors in the LSL-K-rasG12D and LSL-K-rasG12D;Notch1flox/flox mice. Mice were sacrificed at 6, 18, and 24 weeks post infection. Representative histological findings from both groups are shown. Scale bar: 400μm. Arrowheads= hyperplasia, arrows=adenomas.
To assess directly whether Notch1 is required for the initiation and/or progression of K-ras-induced lung tumors, we employed the LSL-K-rasG12D mouse model of lung adenocarcinoma in which lung tumor initiation is achieved by infection with an adenovirus expressing Cre-recombinase (Ad-Cre), leading to removal of a transcriptional stop element and activation of an oncogenic allele of K-rasG12D under physiological control (16). Mice with a conditional loss-of-function allele of Notch1 (Notch1flox/flox) (21) were crossed to the LSL-K-rasG12D mice to generate LSL-K-rasG12D;Notch1flox/flox mice. In these mice, Cre-mediated recombination should result in the activation of the K-rasG12D allele and simultaneous Notch1 inactivation. Following intranasal administration of Ad-Cre to LSL-K-rasG12D, LSL-K-rasG12D;Notch1flox/flox or Notch1flox/flox mice, animals from each group were sacrificed at 6, 18 and 24 weeks post-infection and both tumor numbers and volumes were assessed by histological examination. The LSL-K-rasG12Dmice displayed variable rates of hyperplasia and adenomas already at the 6-week time point, with an average of approximately 14 tumors/mouse (Figures 1A and 1C). This is similar to previously reported rates of tumor numbers in this model (4, 16). In comparison, the LSL-K-rasG12D;Notch1flox/flox displayed an almost uniform distribution of approximately 2 lesions/mouse. The Notch1lfox/flox mice did not show any signs of abnormalities (Figures 1A and 1C,). Thus, loss of Notch1 impairs tumor formation initiated by oncogenic K-ras.
In addition to tumor initiation, we examined the ratio of tumor to lung volume (T/L ratio) at the various time points. In the LSL-K-rasG12D mice the T/L ratio was approximately 6% at 6 weeks, 17% at 18 weeks and rose to approximately 20% at the 24-week time point. In comparison, T/L ratios in the LSL-K-rasG12D;Notch1flox/flox mice were greatly reduced. At the 6-week time point the T/Lwas close to 4%. At 18 and 24-weeks the T/L ratio was 11% and 9.5%, respectively (Fig. 1B). Taken together, our findings indicate that loss of Notch1 significantly reduced K-ras lung tumor initiation and overall tumor burden. In contrast, heterozygous mice LSL-K-rasG12D;Notch1flox/+ did not display a significant decrease in tumor number and volume, suggesting that deletion of both Notch1 alleles is required to effectively impair tumor formation (not shown).
Histological examination of the tumors and pre-cancerous lesions that arose in the different groups indicates that they were grossly similar but arose at different times. In both groups three distinct types of lesions were found. Atypical adenomatous hyperplasia (AAH) was present at 6-week post infection in both groups of mice, although at much higher levels in LSL-K-rasG12D mice (Fig. 1C). Small papillary adenomas were evident at 6-weeks post infection in the LSL-K-rasG12D mice and larger adenomas and adenocarcinomas were present at 18 and 24 weeks. In comparison, only adenomas were apparent in the LSL-K-rasG12D;Notch1flox/flox mice at 18-week time point, and adenocarcinomas were observed at the 24-week time point (Fig. 1C). Both groups of mice also presented with epithelial hyperplasia (EH) of the bronchiole and clearly evident in the LSL-K-rasG12D mice at 6 weeks post-infection. In comparison, in the LSL-K-rasG12D;Notch1flox/flox mice, EH lesions were only evident at the 18 week time points (Not shown).
The efficiency of Cre-mediated recombination of the Notch1 allele was assessed by PCR on several tumors microdissected from the LSL-K-rasG12D;Notch1flox/flox mice from the 18-week time point. A band representing the unrecombined allele of Notch1 was still evident in the tumors, reflecting incomplete inactivation of the Notch1flox/flox alleles (Figure S1). This incomplete recombination was also evident at the protein level, as residual amounts of Notch1 protein were present in tumors isolated from the lungs of LSL-K-rasG12D;Notch1flox/flox mice (Figure 2, panel E). These findings indicate that lung tumors initiated by oncogenic K-ras are dependent on Notch1 function, and only cells that escape inactivation of both alleles of Notch1 are able to progress efficiently in tumorigenesis.
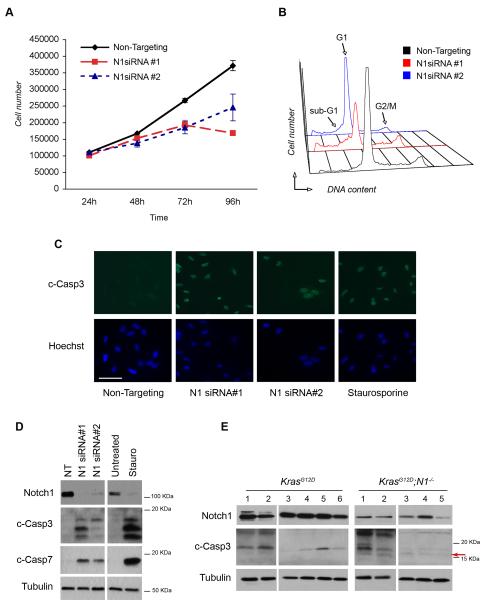
Figure 2. Notch1 ablation induces apoptosis.
(A) Growth curve of A549 cells after Notch1 knockdown with siRNA#1 and siRNA#2. Cells were counted in triplicate starting 24 hours after transfection. Error bars= mean ± SD. One representative experiment of 3 is shown. (B) Cell cycle profiles of A549 cells after Notch1 knockdown. 72h after transfection cells were fixed and stained with propidium iodide (PI) and analyzed for DNA content. The experiment was repeated 3 times. (C) Immunofluorescence staining of cleaved caspase-3 after Notch1 knockdown. Cells were fixed in paraformaldehyde and stained 72h after Notch1 siRNA#1 and siRNA#2 transfection. Treatment with staurosporine (500nM, 16h) was included as a positive control for apoptosis. Scale bar: 50μm. (D) Western blot analysis for caspase-3 and caspase-7 activation. Cells were harvested 72h after transfection. Staurosporine= positive control. (E) Western blot analysis of lung tumors from K-rasG12D and K-rasG12D;Notch1flox/flox mice at 14 weeks after Ad-Cre administration. Protein extracts were analyzed for Notch1 and cleaved caspase-3. Arrow=17KDa cleavage form that is only present in Notch1 deleted samples. Tubulin= loading control.
Notch1 controls survival of lung adenocarcinoma cells in vitro and in vivo
To investigate the mechanisms underlying reduced tumor initiation and burden observed in the LSL-K-rasG12D;Notch1lfox/flox mice we employed an siRNA approach to knockdown Notch1 expression in A549 cells. Two independent siRNA sequences, N1 siRNA#1 and N1siRNA#2, which demonstrated >90% and approximately 50% efficiency in reduction of Notch1 protein levels, respectively, were selected (Fig. 2D). A549 cells were transfected with Notch1 siRNA or Non-targeting siRNA control and counted for 4 consecutive days after transfection to assess effects on cell proliferation. Both Notch1 siRNA treatments resulted insignificantly reduced cells numbers compared Non-Targeting siRNA transfected cells (Fig. 2A). Analysis of cell cycle distribution by propidium iodide (PI) staining showed Notch1 ablation induced the appearance of a fraction of cells with a sub-G1 DNA content, suggestive of apoptotic cell death (figure 2B and Table 1). Importantly, the extent of cell cycle perturbation correlated with the efficiency of Notch1 knock-down by the two different siRNAs, with a stronger impact elicited by siRNA#1 compared to a milder phenotype induced by siRNA#2. To directly assess whether knockdown of Notch1 induced apoptosis, caspase-3 cleavage was examined by immunofluorescence and western blotting using an antibody specific to the cleaved forms of caspase-3. Intense staining for cleaved caspase-3 was observed in the nuclei of siRNA-transfected cells, as well as in cells treated with staurosporine to induce apoptosis (positive control) (Figure 2C). Western blot analysis also confirmed caspase-3 activation and cleavage of caspase-7 (Fig. 2D). To further confirm the observations suggesting induction of apoptosis in A549 cells after Notch1 knockdown reflect the events occurring in vivo, we assessed caspase-3 activation in protein extracts from lung tumors dissected from LSL-K-rasG12D and LSL-K-rasG12D;Notch1flox/flox mice. Caspase-3 is synthesized as an inactive pro-enzyme of approximately 32 kDa and is processed by proteolytic cleavage in cells undergoing apoptosis generating two active subunits, p17 KDa and p12 KDa (25, 26). Partially processed but still active caspase-3 forms are also present, which retain part (p19) or all (p20) of the pro-domain (27-29). Western blot with a cleaved caspase-3 antibody specific for the cleaved forms showed that tumors from K-rasG12D animals display a baseline level of caspase-3 activation, as indicated by the presence of a 19 KDa fragment. However, the K-rasG12D;Notch1flox/flox tumors additionally displayed the 17 KDa cleavage form, which corresponds to fully cleaved caspase-3. (Fig. 2E, indicated by the red arrow). Note that the antibody used does not recognize pro-caspase-3 or p12. These results suggest that diminished levels of Notch1 expression lead to increased cell death in the A549 lung adenocarcinoma cells and in vivo, providing a mechanism to explain the impaired tumor initiation and progression observed in the LSL-K-rasG12D;Notch1flox/flox mice.
Table 1. Notch1 ablation induces apoptosis (related to Figure 3B).
Percent of A549 cells in phases of the cell cycle after Notch1 knockdown. 72h after transfection cells were fixed and stained with propidium iodide (PI) and analyzed for DNA content. The experiment was repeated 3 times.
| G1 | S | G2/M | Sub-G1/Apop | |
|---|---|---|---|---|
| Non-targeting | 63.2 | 14 | 15.1 | 7.9 |
| N1siRNA#1 | 39.4 | 15 | 16.9 | 29.1 |
| N1siRNA#2 | 63.7 | 9.9 | 10.3 | 16.4 |
Onset of apoptosis after Notch1 ablation is mediated by stabilization of p53
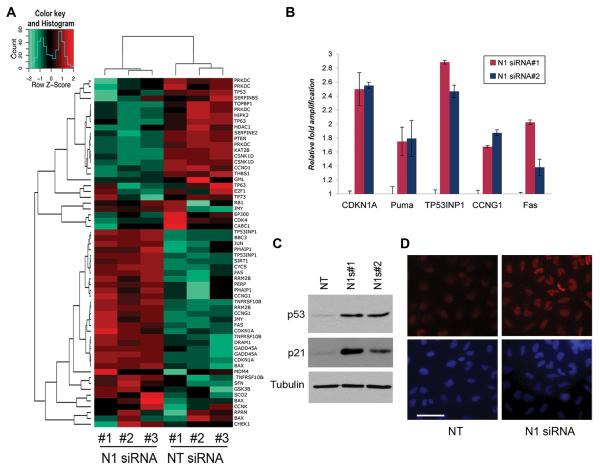
Notch1 Intracellular Domain (N1IC) is a transcriptional regulator and activation of the Notch1 receptor ultimately results in modulation of transcription of several target genes. To test the transcriptional effects of Notch1 ablation in A549 cells we performed an expression profiling microarray experiment in which we compared the global transcriptional profile of cells transfected with Notch1 siRNA#1 to that of cells transfected with non-targeting siRNA control. Functional classification of the genes regulated by Notch1 siRNA revealed a significant enrichment in cell cycle-G2/M checkpoint regulation, apoptosis and a characteristic p53 signature (Fig. 3A). Transcriptional regulation of a number of p53-target genes, including CDKN1A, Puma, TP53INP1, CCNG1 and Fas, was further validated by quantitative PCR in cells treated with both Notch1 siRNAs (Fig. 3B). Importantly, the transcription level of p53 itself was not altered after Notch1 knockdown, leading us to hypothesize that Notch1 might affect the p53 pathway by regulating p53 protein levels. To test this hypothesis, protein extracts from cells transfected with both Notch1 siRNAs were analyzed by western blotting. Indeed p53 protein levels are significantly upregulated by Notch1 ablation. Furthermore, we confirmed upregulation of p21 protein, a major p53 target, (Fig. 3C). The transcriptional profiles and analysis of p53 protein expression suggest that inhibition of Notch1 is associated with p53 protein stabilization in the A549 cells. Immunfluorescence staining of p53 in A549 cells confirmed increased expression in Notch1 siRNA#1-treated cells compared to Non-targeting siRNA and also demonstrated increased p53 nuclear localization, supporting the idea that Notch1 controls p53 stability and localization (Figure 3D).
Figure 3. Notch1 ablation stabilizes p53 protein levels.
(A) Expression profiles of A549 cells transfected with Non-Targeting siRNA or Notch1 siRNA#1. Three independent RNA were analyzed by microarray and target genes were clustered by IPA software. Panel shows regulation of p53 target genes. (B) RT-qPCR validation of select targets from the microarray data using independent cDNA samples from cells transfected with both Notch1 siRNAs. Error bars= mean ± SD of triplicates. Experiment was repeated twice with similar results. (C) Western blot for p53 and p21 expression of A549 cells 72h after transfection with both Notch1 siRNAs. (D) Immunfluorescence of A549 cells transfected with Non-Targeting siRNA or Notch1 siRNA#1. Cells were fixed with paraformaldehyde and stained with anti-p53 and Hoechst. One representative field is shown for each condition. Scale bar: 50μm.
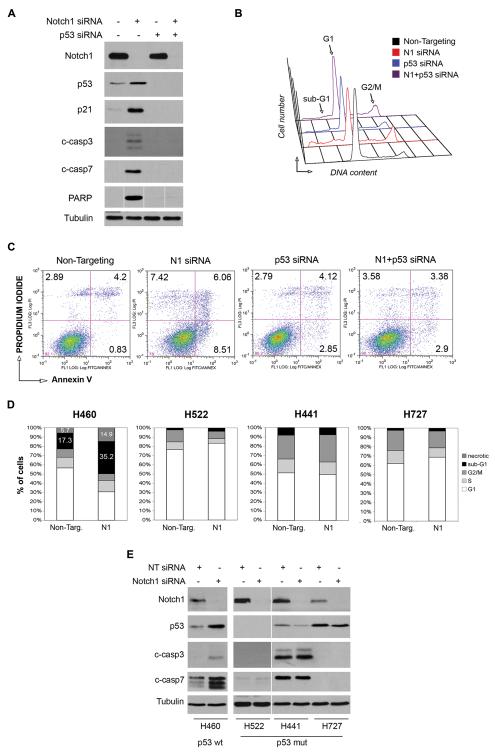
To confirm that p53 stabilization mediates the apoptosis induced by Notch1 ablation and rule out that involvement of p53 is a secondary effect, we knockdown p53 and assessed whether this rescued the apoptotic phenotype. As described above, knockdown of Notch1 induced p53 stabilization, p21 upregulation, caspase-3 and 7 and PARP cleavage (Fig. 4A, second lane) and PI staining profiles confirmed the partial G2/M block of cell cycle and the appearance of the sub-G1 peak (Fig. 4B and Table 2). We also analyzed Annexin-V expression by FACS and found the fraction of Annexin-positive cells, representing early and late apoptotic cells, increased from 5.03% in Non-Targeting cells to 14.57% after Notch1 knockdown (Fig. 4C). As expected, knockdown of p53 alone abrogated p21 expression and had little effect on activation of caspases and PARP (Fig.4A, third lane). Unexpectedly, p53 siRNA alone induced a partial accumulation of A549 cells in G2/M phase and a low level of apoptosis as indicated by the presence of a sub-G1 peak (Figure 4B and Table 2). A modest but reproducible increase in the percentage of Annexin-positive cells compared to Non-Targeting transfected cells was also observed (Figure 4C). The simultaneous ablation of p53 and Notch1 completely abrogated p21 induction and caspase-3, caspase-7 and PARP cleavage that are caused by knockdown of Notch1 alone (Figure 4A, fourth lane). Furthermore, analysis of the cell cycle distribution indicated that the fraction of A549 cells in sub-G1 phase was reduced from 16.4% to 9.2%, corresponding to the effect of p53 knockdown alone (Figure 4B, Table 2). Likewise, the fraction of Annexin V-positive cells was reduced from 14.6% in cells with Notch1 knockdown to 6.3% in cells with combined knockdown of Notch1 and p53, the latter value corresponding to the apoptotic rate of p53 siRNA-only cells (Figure 4C). These data demonstrate that p53 knockdown can rescue the apoptotic phenotype induced by Notch1 ablation.
Figure 4. p53 stabilization mediates apoptosis induced by Notch1 ablation.
(A-C) Rescue of Notch1 siRNA-induced apoptosis by p53 siRNA in A549 cells. All experiments were performed 72h after transfection. (A) Western blot of cells transfected with Notch1 siRNA#1, p53 siRNA or both, compared to Non-Targeting siRNA cells. Protein extracts were analyzed for p53 and p21 expression, caspase-3, caspase-7 and PARP cleavage. Tubulin= loading control. (B) Cell cycle analysis. Cells were fixed and stained with propidium iodide (PI) for DNA content. (C) FACS analysis of apoptosis by Annexin-V staining. Cells were fixed and stained with Annexin-V and PI. Numbers indicate the percentage of cells in the respective quadrant. Experiments were repeated three times, one representative experiment is shown. (D-E) Effect of Notch1 knockdown in NSCLC cell lines H460, H522, H441 and H727 (D) Cells were transfected with Notch1 siRNA#1 and stained with PI at 72h post transfection. Percentages of cells in the different phases of cell cycle plotted for each cell line. (E) Western blot analysis of Notch1 and p53 expression and caspase3 and caspase7 cleavage after Notch1 knockdown. Tubulin= loading control.
Table 2. p53 stabilization mediates apoptosis induced by Notchl ablation (related to Figure 4B).
Percent of A549 cells in phases of the cell cycle after Notch1 or/and p53 knockdown. 72h after transfection cells were fixed and stained with propidium iodide (PI) and analyzed for DNA content. The experiment was repeated 3 times.
| G1 | S | G2/M | Sub-G1/Apop | |
|---|---|---|---|---|
| Non-targeting | 70.9 | 13.2 | 14.4 | 1.39 |
| N1 siRNA | 43.2 | 12.9 | 21.6 | 16.4 |
| p53 siRNA | 57.7 | 13.2 | 18.3 | 9.2 |
| N1+p53 siRNA | 51.4 | 15.1 | 22.1 | 9.2 |
We next confirmed that the involvement of p53 in programmed cell death as a consequence of Notch1 knockdown is a general feature of NSCLC cell lines and not specific to A549 cells. We transduced Notch1 siRNA into other NSCLC cell lines harboring wild type or mutated p53 alleles and evaluated the cell cycle distributions. Importantly, knockdown of Notch1 induced apoptosis only in H460 cells, which carry wild type p53 (30), as indicated by doubling of the population of cells in the sub-G1 fraction (Figure 4D). Concurrent with this, activation of caspase-3 and caspase-7 was also observed in the H460 cells (Figure 5E). In contrast, no apoptosis was observed in the H522, H441 and H727 cells, all carrying mutant p53 (Figure 5D and 5E). These findings further demonstrate that apoptosis induced by Notch1 knockdown in NSCLC cells is p53-mediated and requires functional p53.
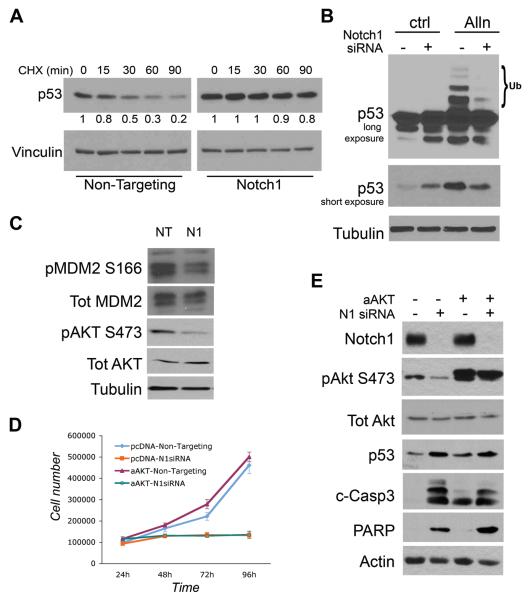
Figure 5. Notch1 controls p53 stability and MDM2 phosphorylation independently of Akt.
(A) 72h post transfection with Non-Targeting or Notch1 siRNA A549 cells were treated with CHX (10 μg/ml) for the indicated times. Whole cell protein extracts were analyzed by Western blot for p53. Vinculin= loading control. (B) Notch1 and Non-Targeting siRNA transfected cells were treated with ALLN (50 μM) for 6h. Protein extracts were analyzed by Western blot for expression of Notch1 and p53. Vinculin= loading control. (C) Non-Targeting and Notch1 siRNA-treated cells were analyzed by Western blot for expression and phosphorylation status of MDM2 and Akt. Tubulin= loading control. (D) A549 cells stably transfected with active Akt (aAkt) were transfected with Non-Targeting and Notch1-siRNA. Cells were counted in triplicate starting 24h post transfection. Error bars= mean ± SD. (E) Cells treated as in (D) were analyzed by western blot with the indicated antibodies 72h after siRNA transfection.
Loss of Notch1 results in increased p53 stability
To establish the increase in p53 levels after Notch1 knockdown is dependent on the regulation of p53 protein stability, we treated A549 cells with cyclohexamide (CHX), an inhibitor of de novo protein synthesis. Non-targeting and Notch1 siRNA transfected cells were harvested at different time points after CHX treatment and levels of p53 protein were monitored by western blotting. As expected, in the Non-Targeting treated cells a greater than 75% reduction in p53 levels was observed after 90 minutes of treatment whereas virtually no change in p53 levels was detected in Notch1 knockdown cells within the same time frame, indicating that Notch1 ablation results in an increase in p53 stability (Figure 5A). The levels of p53 are tightly regulated by rate of degradation, mediated by the ubiquitin pathway (31). To test whether Notch1 is able to affect p53 ubiquitination we treated the cells with the proteosomal inhibitor ALLN, which blocks the degradation of ubiquitinated proteins and leads to an increase in the level of ubiquitinated p53. While A549 control cells treated with ALLN showed a typical pattern of ubiquitinated p53, ubiquitinated p53 was almost completely absent in cells transfected with Notch1 siRNA before ALLN treatment (Figure 5B). Moreover, a shorter exposure (Figure 5B, third row) shows that p53 is stabilized by ALLN treatment in control cells but not in Notch1 knockdown cells (compare fourth lane to second lane), suggesting that the majority of p53 molecules accumulated after Notch1 ablation are not protected from degradation when the proteasome is inhibited. Taken together these experiments show that Notch1 favors p53 proteasomal degradation by positively regulating its ubiquitination.
The MDM2 protein is the primary regulator of p53 stability promoting its ubiquitination and degradation (32, 33). To investigate the involvement of MDM2 in the observed stabilization of p53 we assessed the expression levels and activation of MDM2 after Notch1 knockdown. As shown if Fig.5C, MDM2 phosphorylation on Ser 166 is decreased after Notch1 knockdown whereas the overall expression level is not altered (MDM4/HDMX expression is also not affected, not shown). A link between Notch signaling and regulation of MDM2 has been previously demonstrated in T-ALL through Notch1 transcriptional regulation of PTEN, a negative regulator of Akt activity (34). Akt in turn activates MDM2 by phosphorylation at Ser-166 and -186, resulting in diminished cellular levels and transcriptional activity of p53 (35-37). Indeed we observed a decrease in phosphorylation of Akt at Ser473 in Notch1 knockdown cells, suggestive of a reduced activation of PI3K/Akt signaling (Figure 5C). However, overexpression of an activated form of Akt did not rescue the decrease in cell number caused by knockdown of Notch1 (Fig. 5D), nor was it able to revert p53 stabilization, caspase-3 activation and PARP cleavage (Fig. 5E). These results indicate that while Notch1 levels might affect Akt activity, this is likely not the mechanism through which Notch1 regulates p53 stability and lung adenocarcinoma cell survival.
Discussion
The Notch receptors are emerging as important players in the pathogenesis of lung adenocarcinoma. Recent studies in which pan-Notch inhibition was achieved by gamma-secretase inhibitors (GSIs) indicate that repression of Notch signaling can impair tumorigenesis in vivo (20). However, the use of GSIs is associated with severe intestinal toxicity in patients, due to the simultaneous inhibition of multiple Notch receptors (38). Thus, there is a strong rationale for targeting the Notch receptors individually. Previous work focused on Notch3 in lung cancer. However these studies were limited, to our knowledge, to in vitro or xenograft models. More recently, hyperactivation of the Notch1 pathway has been described as a common event in human NSCLC, due to activating mutations in the Notch1 gene or loss of the negative regulator Numb (12). The mechanisms through which Notch1 exerts its function in lung tumorigenesis are unclear.
To examine which Notch receptor/s has a relevant role in lung tumorigenesis we first evaluated the requirement for Notch1-3 in vitro and found Notch1 was the most suitable candidate for further in vivo studies. Through Notch1 ablation in vivo, we observed a dramatic decrease in tumor initiation and burden in an autochthonous mouse model of lung adenocarcinoma, demonstrating Notch1 is required for K-ras-induced lung adenocarcinoma. In the cases where tumors did arise in the LSL-K-rasG12D;Notch1flox/flox mice, further analysis indicated these tumors likely “escaped”, through incomplete inactivation of the Notch1flox/flox allele. Interestingly, a recent study employing the LSL-K-rasG12D model combined with conditional activation of a dominant-negative inhibitor of Notch activity (DN-MAML) found no evidence of inhibition of K-ras-induced lung adenocarcinoma in vivo (39). Given our findings showing that low levels of Notch1 expression are sufficient to allow “escape” of tumors, it is likely that the limited inhibition of Notch activity offered by of DN-MAML was unable to attenuate K-ras driven lung tumorigenesis.
To identify mechanisms underlying the requirement for Notch1 in lung adenocarcinoma we used A549 in which siRNA-mediated ablation of Notch1 resulted in cell death. Importantly, we were able to detect increased rates of apoptosis in vivo by analyzing tumor samples from KrasG12D and KrasG12D;Notch1flox/flox lungs for caspase-3 activation and identifying a band corresponding to fully cleaved caspase-3 only in tumors from these mice. Notch1 intracellular domain is a transcriptional regulator that functions in a tissue and cell type-specific manner. Gene expression analysis was used to gain insights into the molecular alterations occurring after Notch1 ablation in A549 cells. We did not find significant changes in expression levels of the Notch target genes traditionally used as readouts of Notch activation (e.g. Hes and Hey families). This is not totally surprising; indeed growing evidence suggests that different Notch receptors regulate these genes in a context and tissue-specific manner. For example, the inhibition of Notch signaling in the pancreas does not lead to decreased Hes1 expression (4, 40) and similarly, overexpression of N1ICD (Notch1 Intra Cellular Domain) in mouse lungs does not induce Hes1 gene expression (13).
The gene expression approach allowed us to uncover a characteristic p53 signature in response to Notch1 knockdown, coherent with A549 cells expressing functional p53 (41). Importantly P53 mRNA levels were not altered, excluding the possibility that Notch1 controls p53 expression at the transcriptional level. We found indeed that p53 protein levels were increased and the vast majority of the protein was localized in the nucleus (Figure 3), indicating stabilization and activation in response to Notch1 knockdown. A causative role for p53 activation in the observed cell death phenotype was established as concomitant knockdown of p53 and Notch1 was sufficient to rescue the cells from apoptosis and revert the effect of Notch1 ablation, suggesting that Notch1 controls survival of A549 cells in a p53-dependent manner. To rule out the possibility of a cell-type specific phenomena, we confirmed p53 stabilization and induction of apoptosis in a second NSCLC cell line retaining wild type p53, whereas no cell death was observed in several cell lines with either mutant or non-functional p53 alleles.
The possibility that p53 stabilization was a consequence of DNA damage activation after Notch1 ablation was ruled out, as we did not observe accumulation of γH2AX DNA damage foci in Notch1 siRNA-transfected cells (Fig. S2). Therefore, we focused on the regulation of p53 stability and found that Notch1 knockdown results in a dramatic increase in p53 half-life and a significant impairment of p53 ubiquitination. MDM2 is the major regulator of p53 stability and can also regulate p53 function at additional levels by blocking its transcriptional activity (42, 43) and promoting export of p53 into the cytoplasm (44). Notch1 has been reported to physically interact with p53 and MDM2 (45, 46) and could potentially participate in the regulation of the MDM2/p53 interaction. Co-immunoprecipitation experiments performed after Notch1 deletion did not support this hypothesis, as we did not observe a decrease in p53 co-pulled down with MDM2 after Notch1 siRNA (Fig. S3). Tumor cells that retain wild type p53 often display deletion of the cell cycle regulator ARF, one of the products encoded in the CDKN2A locus, which binds to MDM2 and promotes its degradation thus blocking its ability to mediate ubiquitination of p53 (47). We have previously shown that Notch1 can suppress p53 during lymphomagenesis, through repression of the ARF-MDM2-p53 axis (48). This is not likely the case in NSCLC cells because A549 and H460 cell lines do not express ARF as a consequence of CDKN2A deletion (49-51) and show a high level of MDM2 activity, which allows the cells to keep p53 activity at a minimum. Therefore, the requirement of a functional ARF to control apoptosis and p53 stability in response to Notch1 ablation can be ruled out in our system.
Notch signaling has been previously shown to increase MDM2 nuclear localization and activity through AKT-mediated phosphorylation (34-36). While we observed a decrease in Akt activation after Notch1 knockdown, the forced activation of the Akt pathway in A549 cells failed to rescue the cell death induced by Notch1 knockdown, indicating the Akt pathway does not likely mediate the effects of Notch1 ablation in these cells (Fig.5D,E). It should also be noted that previous work suggests Notch1 signaling is activated and controls cell survival via Akt activation specifically under hypoxic conditions (52). However, we did not observe differences in Notch1 activation between hypoxic or normoxic conditions (Fig. S4). Based on these results, we propose that Notch1 regulates MDM2 activity through mechanisms that are ARF- and Akt-independent.
Finally, our data show that control of survival of lung adenocarcinoma cells by Notch1 requires a functional p53. Recent data have described a correlation between the levels of activated NOTCH-1 and poor prognosis in a cohort of NSCLC patients, but only in patients with wild type p53 (12). Our combined findings demonstrate that p53 status represents a critical determinant in selection of therapeutic strategies for lung adenocarcinoma patients.
Supplementary Material
Acknowledgments
We thank Drs. Maureen Murphy, Dario Altieri and Michael May for helpful discussions and reagents. This work was supported by grant CA124495 (J.L.K).
Footnotes
The authors declare no conflict of interest exists.
References
- 1.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 2.Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 92:411–55. doi: 10.1016/S0070-2153(10)92013-9. [DOI] [PubMed] [Google Scholar]
- 3.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 11:338–51. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 4.Hanlon L, Avila JL, Demarest RM, Troutman S, Allen M, Ratti F, et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer research. 70:4280–6. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 6.Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer research. 2003;63:8323–9. [PubMed] [Google Scholar]
- 7.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–7. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 8.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer research. 2007;67:8051–7. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer research. 70:9937–48. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, et al. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer research. 2005;65:3555–61. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, Mernaugh R, Yi F, Blum D, Carbone DP, Dang TP. Targeting specific regions of the Notch3 ligand-binding domain induces apoptosis and inhibits tumor growth in lung cancer. Cancer research. 70:632–8. doi: 10.1158/0008-5472.CAN-09-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–8. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer research. 71:6010–8. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 116:5676–85. doi: 10.1002/cncr.25551. [DOI] [PubMed] [Google Scholar]
- 15.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–39. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- 18.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 19.Kiaris H, Politi K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, et al. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maraver A, Fernandez-Marcos PJ, Herranz D, Canamero M, Munoz-Martin M, Gomez-Lopez G, et al. Therapeutic Effect of gamma-Secretase Inhibition in Kras(G12V)-Driven Non-Small Cell Lung Carcinoma by Derepression of DUSP1 and Inhibition of ERK. Cancer cell. 2012;22:222–34. doi: 10.1016/j.ccr.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–58. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 22.Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, et al. Requirement for Rac1 in a K-ras Induced Lung Cancer in the Mouse. Cancer Res. 2007;67:8089–94. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 23.Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2110–5. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene. 2012 doi: 10.1038/onc.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. The Journal of biological chemistry. 1994;269:30761–4. [PubMed] [Google Scholar]
- 26.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 27.Boulakia CA, Chen G, Ng FW, Teodoro JG, Branton PE, Nicholson DW, et al. Bcl-2 and adenovirus E1B 19 kDA protein prevent E1A-induced processing of CPP32 and cleavage of poly(ADP-ribose) polymerase. Oncogene. 1996;12:529–35. [PubMed] [Google Scholar]
- 28.Garcia-Calvo M, Peterson EP, Rasper DM, Vaillancourt JP, Zamboni R, Nicholson DW, et al. Purification and catalytic properties of human caspase family members. Cell death and differentiation. 1999;6:362–9. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 29.MacFarlane M, Cain K, Sun XM, Alnemri ES, Cohen GM. Processing/activation of at least four interleukin-1beta converting enzyme-like proteases occurs during the execution phase of apoptosis in human monocytic tumor cells. J Cell Biol. 1997;137:469–79. doi: 10.1083/jcb.137.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somasundaram K, El-Deiry WS. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–57. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 31.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 32.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 33.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 34.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11598–603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–82. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 37.Malmlof M, Roudier E, Hogberg J, Stenius U. MEK-ERK-mediated phosphorylation of Mdm2 at Ser-166 in hepatocytes. Mdm2 is activated in response to inhibited Akt signaling. The Journal of biological chemistry. 2007;282 doi: 10.1074/jbc.M604953200. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 39.Osanyingbemi-Obidi J, Dobromilskaya I, Illei PB, Hann CL, Rudin CM. Notch signaling contributes to lung cancer clonogenic capacity in vitro but may be circumvented in tumorigenesis in vivo. Mol Cancer Res. 2011;9:1746–54. doi: 10.1158/1541-7786.MCR-11-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazur PK, Einwachter H, Lee M, Sipos B, Nakhai H, Rad R, et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2010;107:13438–43. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 42.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 43.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–60. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 2001;12:175–86. [PubMed] [Google Scholar]
- 45.Kim SB, Chae GW, Lee J, Park J, Tak H, Chung JH, et al. Activated Notch1 interacts with p53 to inhibit its phosphorylation and transactivation. Cell Death Differ. 2007;14:982–91. doi: 10.1038/sj.cdd.4402083. [DOI] [PubMed] [Google Scholar]
- 46.Pettersson S, Sczaniecka M, McLaren L, Russell F, Gladstone K, Hupp T, et al. Non-degradative ubiquitination of the Notch1 receptor by the E3 ligase MDM2 activates the Notch signalling pathway. The Biochemical journal. 2013;450:523–36. doi: 10.1042/BJ20121249. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 48.Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005;65:7159–68. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 49.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O’Meara S, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Molecular cancer therapeutics. 2006;5:2606–12. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishizaki M, Sasaki J, Fang B, Atkinson EN, Minna JD, Roth JA, et al. Synergistic tumor suppression by coexpression of FHIT and p53 coincides with FHIT-mediated MDM2 inactivation and p53 stabilization in human non-small cell lung cancer cells. Cancer research. 2004;64:5745–52. doi: 10.1158/0008-5472.CAN-04-0195. [DOI] [PubMed] [Google Scholar]
- 51.Lu W, Lin J, Chen J. Expression of p14ARF overcomes tumor resistance to p53. Cancer research. 2002;62:1305–10. [PubMed] [Google Scholar]
- 52.Eliasz S, Liang S, Chen Y, De Marco MA, Machek O, Skucha S, et al. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene. 29:2488–98. doi: 10.1038/onc.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.