Abstract
The extent and biological impact of RNA cytosine methylation are poorly understood, in part owing to limitations of current techniques for determining the targets of RNA methyltransferases. Here we describe 5-azacytidine-mediated RNA immunoprecipitation (Aza-IP), a mechanism-based technique that exploits the covalent bond formed between an RNA methyltransferase and the cytidine analog 5-azacytidine to recover RNA targets by immunoprecipitation. Targets are subsequently identified by high-throughput sequencing. When applied in a human cell line to the RNA methyltransferases DNMT2 and NSUN2, Aza-IP enabled >200-fold enrichment of tRNAs that are known targets of the enzymes. In addition, it revealed many tRNA and non-coding RNA targets not previously associated with NSUN2. Notably, we observed a high frequency of C>G transversions at the cytosine residues targeted by both enzymes, allowing identification of the specific methylated cytosine(s) in target RNAs. Given the mechanistic similarity of cytosine RNA methyltransferases, Aza-IP may be generally applicable for target identification.
Although cytosine methylation is most commonly studied in DNA, it is also found in RNA1. As with DNA, cytosine RNA methylation occurs at the C5 position (m5C). RNA methylation has been detected in both prokaryotic and eukaryotic non-coding RNAs (ncRNAs) such as tRNA and rRNA1 . Recent high-throughput RNA methylation profiling by bisulfite sequencing in HeLa cells verified and extended the repertoire of m5C modifications in RNA2, motivating a more thorough examination of the scope (cell types and developmental contexts) and functions of RNA methylation.
The m5C-RNA methyltransferases (m5C-RMTs) have been subdivided into six families based on structural and functional properties: RsmB/Nol1/NSUN1, RsmF/YebU/NSUN2, RlmI, Ynl022, NSUN6 and DNMT21. Only DNMT2-family enzymes are ‘single-cysteine type’; similar to DNA-MTases, they utilize a single cysteine in their catalytic pocket3, whereas the other m5C-RMT family enzymes utilize two cysteines4. Here we focus on DNMT2 and NSUN2, as they represent one member of each family that is either highly studied (DNMT2) or highly disease relevant (NSUN2).
DNMT2 functions primarily, if not exclusively, as a m5C-RMT, with three verified tRNA targets: tRNAAsp, tRNAGly and tRNAVal 3, 5-7. Most organisms lacking DNMT2 lack obvious phenotypes8, although DNMT2-deficient zebrafish display developmental perturbations6. Notably, DNMT2 activity attenuates tRNA cleavage during stress conditions, and promotes response to RNA viruses in Drosophila7, 9. NSUN2 also methylates cytosines in tRNAs as well as the ncRNA subunit of Rnase P and possibly mRNA substrates2, 10, 11, but the links between particular NSUN2 targets and cellular functions are unknown. NSUN2 has been associated with Myc-induced proliferation of cancer cells12, mitotic spindle stability 13, infertility in male mice, and the balance of self-renewal and differentiation in skin stem cells14. Notably, in humans NSUN2 mutations cause an autosomal recessive syndrome characterized by intellectual disability and mental retardation15-17. Furthermore, tRNA cytosine methylation by both Dnmt2 and Nsun2 in mice increases tRNA stability and steady-state protein synthesis10.
In principle, RNA targets of m5C-RMTs could be identified by deep RNA bisulfite sequencing of cell lines or tissues in which a particular m5C-RMT has been knocked down or knocked out2. However, this approach is labor intensive and requires effective enzyme knockout methods. In addition, in cases in which other enzymes are redundant with the m5C-RMT under study, targets of the m5C-RMT of interest may be missed. Although such an approach could identify candidate targets of the m5C-RMT, it could not distinguish between direct and indirect targets. Lastly, this approach would require extremely deep sequencing to reveal modifications on RNAs of low abundance or low methylation penetrance. To circumvent these and other issues, we developed Aza-IP, a technique that enriches the direct RNA targets of specific m5C-RMTs and identifies the precise cytosine(s) targeted by the enzyme.
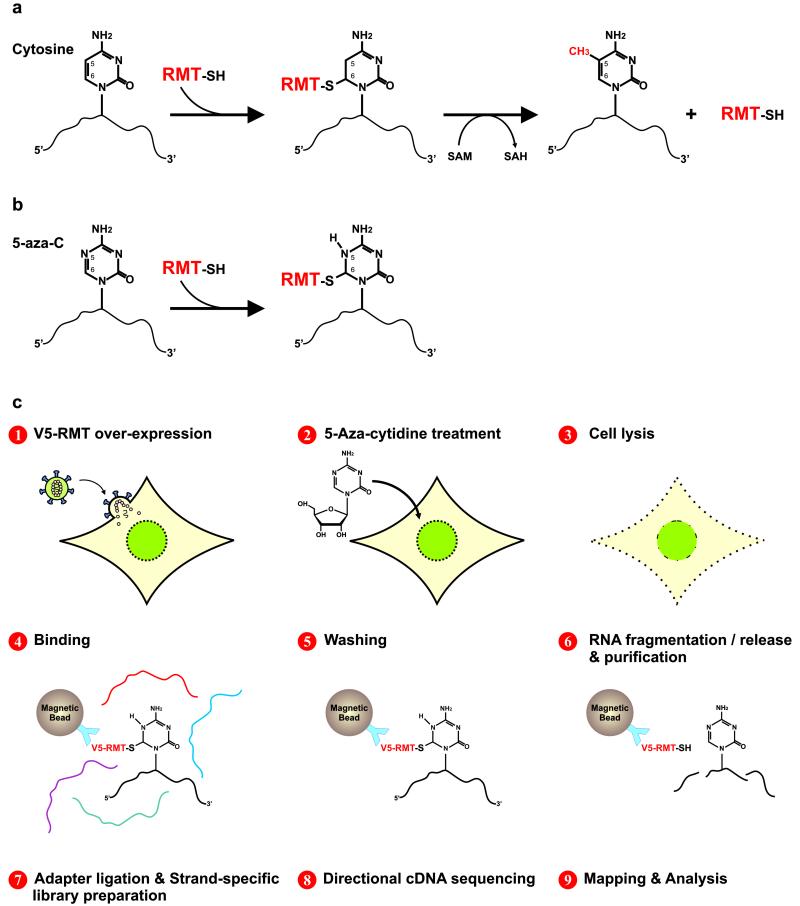
Like m5C-DNMTs, all m5C-RMTs tested to date form a covalent enzyme-substrate intermediate with their target1. Specifically, the sulfur atom of a cysteine residue in the m5C-RMT catalytic domain covalently bonds to the C6 position of the base in the target RNA. Covalent linkage precedes methylation, which occurs by enamine methylation of the C5 position of the target cytosine using the methyl donor S-Adenosyl Methionine (SAM). Free enzyme is regenerated by subsequent beta-elimination1 (Fig. 1a).
Figure 1.
RNA cytosine methylation mechanism and Aza-IP experimental design. (a) Schematic of m5C-RMTs catalyzing methylation of carbon 5 (C5) of cytosine. First, the enzyme forms a covalent thioester bond, connecting the cysteine residue of its catalytic domain to the C6 position of the target cytosine, forming an RMT-RNA adduct. Next, the RMT transfers a methyl group from cofactor S-Adenosyl Methionine (SAM) to the C5 of the target cytosine. The enzyme is then released from the adduct by β-elimination. Methylated RNA and S-Adenosyl-L-Homocysteine (SAH) are the product and byproduct of this reaction, respectively. (b) 5-azacytidine (5-aza-C) is a mechanism-based suicide inhibitor that traps the enzyme by forming a stable RMTRNA adduct. (c) Schematic representation of the Aza-IP technique.
This catalytic mechanism is disrupted by the suicide inhibitors 5-azacytidine (5-aza-C) and 5-aza-2′deoxycytidine (5-aza-dC)18. These cytidine analogs are randomly incorporated by RNA and DNA polymerases into nascent RNA or DNA molecules, respectively. Owing to nitrogen substitution at C5, RMT and DNMT enzymes remain covalently bound to the target RNA or DNA molecule (Fig. 1b), thereby depleting cells of the endogenous enzymes and resulting in hypomethylation of RNA and DNA18-21. Thus, in the presence of 5-aza-C, even over-expression of an m5C-RMT should result in only a small amount of short-lived free active enzyme, greatly reducing concern that enzyme over-expression will result in the methylation of non-physiological, irrelevant targets. Given their effectiveness in enzyme depletion, 5-aza-C and 5-aza-dC are currently used for variety of experimental and clinical applications, including linkage of a DNMT to a known target DNA in vivo22, visualization and monitoring of DNMTs in living cells23 and inducing DNA hypomethylation of tumor cells in patients24.
Aza-IP involves nine steps: 1) Expression of an epitope-tagged m5C-RMT derivative in cells (or use of an antibody capable of immunoprecipitating the endogenous RNA-bound enzyme), 2) cell growth in the presence of 5-aza-C, which is incorporated at low/moderate levels into nascent RNA, 3) cell lysis, 4) immunoprecipitation of the m5C-RMT of interest, a portion of which is covalently attached to target RNAs bearing 5-aza-C, 5) stringent washing to remove RNA contaminants, 6) RNA fragmentation, release and purification, 7) ligation of adaptor oligos to the RNA, and creation of a cDNA library in a manner that enables strand-specific assignments, 8) cDNA sequencing (50-bp single end), and 9), mapping and analysis of sequence reads to define RNA targets and site of cross-linking/catalysis (Fig. 1c, Online Methods, and Supplementary Information).
We chose HeLa cells due to their favorable growth properties and ease of lentiviral infection. We over-expressed V5-tagged (epitope derived from simian virus 5) human DNMT2 (test) or V5-tagged DsRed (control) proteins using a lentiviral expression system and the relatively strong cytomegalovirus (CMV) immediate early promoter. We grew HeLa cells in 3μM 5-aza-C for a time (12hr) empirically determined as sufficient for incorporation of 5-aza-C into nascent RNA and efficient RNA target detection (note: 3-5μM 5-aza-C facilitates 5-aza-C incorporation without detectable toxicity, and 6-24 hr time ranges should be tested). Cells were lysed in a stringent denaturing buffer supplemented with an RNase inhibitor, briefly sonicated, and cleared to yield a cell lysate. The lysate was incubated with magnetic beads coated with anti-V5, and the beads were stringently washed, taking advantage of the covalent association between DNMT2 and target RNA. We then fragmented the enriched RNA molecules while they were still bound to the beads, generating RNA fragments of appropriate size (60-200bp) for the construction of sequencing libraries. These RNA fragments were then isolated, ethanol precipitated and extracted. RNA samples were then used for directional (strand-specific) cDNA library preparation and, after application of quality control procedures, the cDNA library was subjected to high-throughput sequencing using Illumina 50-bp single-end sequencing (Online Methods and Supplementary Information).
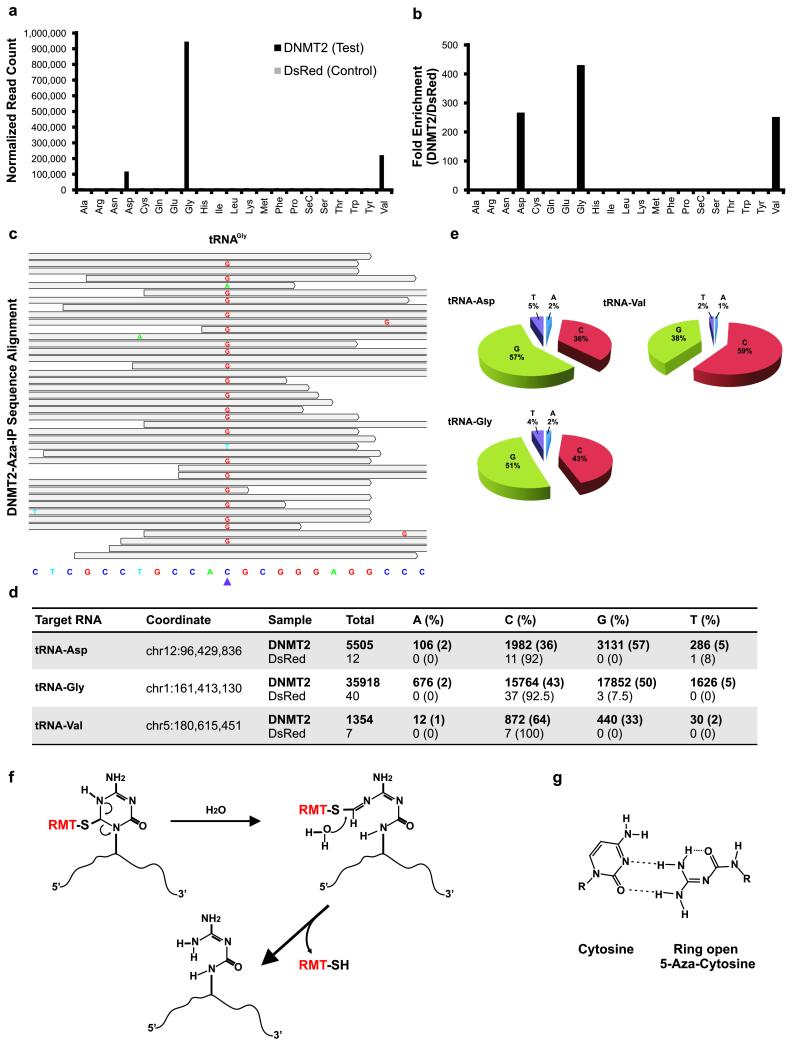
We used the USeq analysis package and Biotoolbox to identify RNAs showing statistically significant enrichment in DNMT2 immunoprecipitates (Online Methods and Supplementary Information). The known DNMT2 targets (tRNAAsp, tRNAGly and tRNAVal)7 were detected at background levels in the control V5-DsRed immunoprecipitates, but were markedly enriched 271-, 431-, and 255-fold, respectively, compared to V5-DsRed control in the V5-DNMT2 immunoprecipitates (Fig. 2a,b and Supplementary File 1). It appeared that DNMT2 did indeed release from the target 5-aza-C base during the fragmentation step, as sequence reads covering the known target cytosine (C38) were plentiful and not depleted relative to reads covering the flanking sequences. Beyond these three tRNAs, we found no comparable enrichment of other ncRNAs (rRNAs, snRNAs, snoRNAs, scRNAs, miRNAs), or mRNAs, with two preliminary exceptions; the KRT18 mRNA and the KRT18 psuedogene mRNA displayed moderate enrichment and a C>G transversion (a purine replacement of a pyrimidine, explained below). (Supplementary File 2, and Supplementary Fig. 4). Although we did not rigorously validate these mRNA enrichments in vivo, we instead used KRT18 RNA fragments and tRNAs to reveal that DNMT2 activity requires a CpG within a tRNA-type stem-loop junction (Supplemental Information and Supplementary Figures 1-7). In aggregate these findings indicate that, Aza-IP enriched three known tRNA targets of DNMT2.
Figure 2.
Aza-IP analysis of DNMT2 RNA targets. (a) Graph depicts normalized reads mapping to each tRNA in the V5-DNMT2 (test) and V5-DsRed (control) datasets (one replicate of each shown). Each tRNA is designated by a three letter amino acid abbreviation. (b) Fold enrichment was calculated from the data shown in (a) by dividing the normalized RPKM (Reads Per Kilobase per Million mapped reads) values for each tRNA type in the V5-DNMT2 dataset by the values in the V5-DsRed dataset. (c) A representative snapshot from the Integrative Genomics Viewer (IGV, BROAD Inst.) browser depicting a subset of the sequencing reads mapped to a tRNAGly locus (chr1:161,413,119-161,413,141, human genome version 19 (hg19) bottom) at base pair resolution. The grey bars span the start/stop of individual sequencing reads mapped to the locus. The mismatched nucleotides are shown with colored letters and the matched nucleotides are hidden (grey). The purple arrowhead points to the tRNAGly C38 nucleotide (chr1:161,413,130). (d) Summary of the base distribution at the known DNMT2 target sites in tRNAAsp, tRNAGly and tRNAVal. The coordinate indicates the genomic location of the target cytosine in the human genome and the raw numbers are reported for both the V5-DNMT2 and V5-DsRed Aza-IP datasets. (e) Pie graphs showing the base distributions at the target nucleotide in the mapped reads. The numbers for the tRNAs are averaged over all of the annotated tRNA loci of same type in the human genome showing coverage over the target nucleotide (C38). (f) Schematic representation of the RMT-induced ring opening and RMT-RNA dissociation model. This model was proposed by Jackson-Grusby et al.25 for mammalian DNA cytosine methyltransferases and is adapted here for RMTs. RMT covalent linkage to the C6 position of 5-aza-C induces the rearrangement and ring opening and results in dissociation of the RMT from the target RNA molecule. (g) Base-pairing behavior of ring-open 5-aza-C. The ring-open 5-aza-C prefers to pair with cytosine and is therefore read as guanosine after RT-PCR and sequencing.
A majority of the reads that mapped to the three known target tRNAs contained a single nucleotide polymorphism (SNP) present solely at the known DNMT2 target cytosine (C38) (Fig. 2c-e and Supplementary File 1). Typically, the base change involved transversion to guanosine; however adenosine and thymine were also observed at levels well above the estimated error rate of sequencing (~0.5%). Transversion was observed at the target cytosine but almost never (<1%) at other cytosines in all three DNMT2 target tRNAs. Over 200 m5C sites have been identified in different human tRNAs and their isoacceptors or isodecoders2 (tRNAs that accept the same amino acid but have different anticodons, or have identical anticodons but different body sequences, respectively). However, only C38 and not the other sites bearing m5C in these three tRNAs exhibited transversion in our datasets. Thus, Aza-IP does not simply cause mutations at cytosines, or at m5C; rather, transversion is observed specifically at DNMT2 target sites in the RNA enriched by our technique. These are true transversions, not SNPs, because standard high-throughput RNA sequencing profiles from HeLa cells revealed cytosine at C38 in >99% of the reads (data not shown). Notably, previous work with DNMTs revealed C>G transversion occurring within DNA following the growth of cells in 5-aza-C; the authors proposed that covalent attachment of the DNMT to the target 5-aza-C enables the opening of the 5-aza-C ring and its subsequent pairing with cytosine at replication25(Fig. 2f,g).
To validate the DNMT2 targets identified by Aza-IP, we performed transcriptome-wide RNA bisulfite sequencing in WT and Dnmt2-knockout MEFs; we used MEFs because knockdown of DNMT2 in HeLa could not be validated with current antibodies. We adapted existing methods and criteria (coverage>10 reads, methylation level >20%)2, defined the significantly-methylated cytosines, and compared the WT and Dnmt2-knockouts datasets (Supplementary File 3). In Dnmt2-knockout but not WT MEFs, we observed a total loss of RNA methylation at the three known DNMT2 tRNA targets identified by Aza-IP; other sites of RNA methylation were not affected. However, the site of methylation observed in human KRT18 mRNA is not conserved in the mouse KRT18 mRNA.
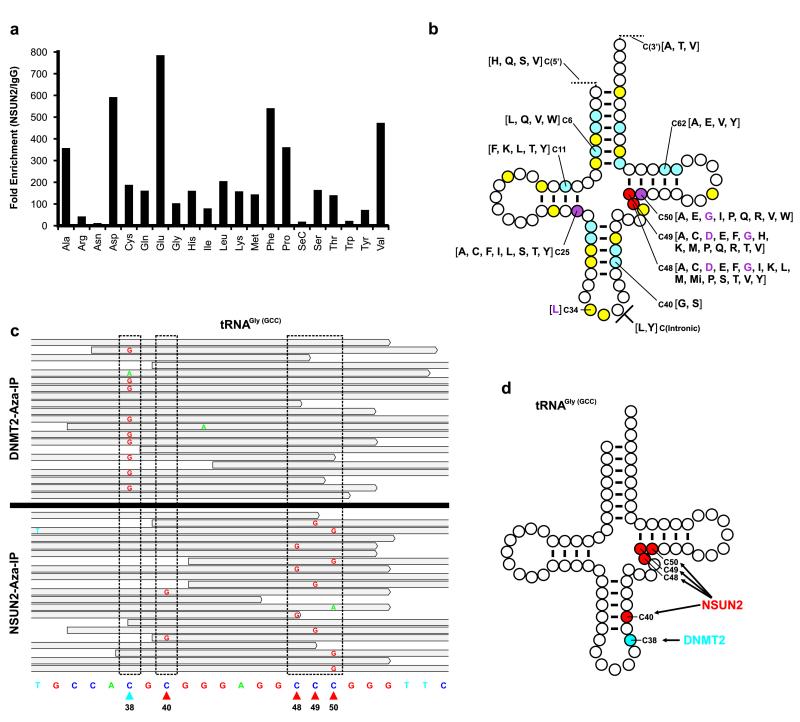
We then applied Aza-IP to identify RNA targets of the ‘two cysteine’ human enzyme NSUN2. Previous studies identified three direct tRNA targets of human NSUN2 (tRNALeu (C34), tRNAAsp (C48, 49) and tRNAGly (C48, 49, 50))2, 11, 26, four direct tRNA targets of mouse Nsun2 (tRNALeu (C34); tRNAAsp (C48, 49), tRNAGly (C40, 48, 49, 50) and tRNAVal (C48, 49))10, and seven additional target cytosines in the tRNA targets for yeast Trm4/Nsun227, 28 (Fig. 3b and Supplementary Fig. 8). We expressed V5-NSUN2 (with modifications see Online Methods) in HeLa cells, and performed two anti-V5 biological replicates (Rep1 and Rep2), and one IgG only (control) replicate, of Aza-IP. Rep1, Rep2 and IgG only experiments yielded 55,180,207, 56,051,845 and 57,133,775 mapped filtered reads, respectively.
Figure 3.
Aza-IP analysis of NSUN2 RNA targets. (a) Graph depicts fold enrichment of human tRNAs in V5-NSUN2 immunoprecipitates. Fold enrichment was calculated by dividing the normalized RPKM (Reads Per Kilobase per Million mapped reads) values for each tRNA type in the V5-NSUN2 replicate datasets (combined) by the values in the IgG control dataset. Each tRNA is designated by a three letter amino acid abbreviation. (b) A ‘standardized’ tRNA summarizing the human NSUN2 target cytosines revealed by Aza-IP in HeLa cells. Cytosines are color-coded based on the number of tRNA types that we found to be NSUN2 target sites: yellow, one tRNA type; blue, 2-5 tRNA types; purple, 6-9 tRNA types; and red >10 tRNA types. For each position, the individual tRNAs are designated by their single letter amino acid abbreviation, grouped in square brackets; purple letters refer to previously known NSUN2 target sites in the designated tRNA types (in human)2, 11, 26. For clarity of presentation, only selected positions are depicted (See Supplementary File 5 for all tRNAs, their isoacceptors/isodecoders, and target positions). (c) Integrative Genomics Viewer (IGV) browser snapshots of a random subset of the sequencing reads mapped to a tRNAGly(GCC) locus (chr17:8,029,095-8,029,117) from the separate DNMT2 (top) or NSUN2 Aza-IP (bottom) datasets. Stippled boxes show locations that meet target criteria with either enzyme. Arrowheads at bottom depict the sole DNMT2 target site (blue) or the four NSUN2 target sites (red). (d) A ‘standardized’ tRNAGly(GCC) with all five known m5C bases depicted, all five of which (and no other resident cytosine) were specifically and selectively identified by Aza-IP of DNMT2 or NSUN2.
We applied three criteria for identifying candidate NSUN2 RNA targets: RPKM (>3), enrichment (replicates/control >3-fold and FDR<0.01) and transversion frequency (>4%, and P<0.01). Our mapping and analyses enabled attribution of reads to particular isoacceptors/isodecoders, and we provide in the Supplementary Tables the precise human genome coordinate (hg19) of the methylated cytosine and p-values for transversion (Supplementary Files 4 & 5). Almost all tRNAs were enriched in both replicates of anti-V5-NSUN2 immunoprecipitates (tRNAAsn and tRNASeC excepted), with many enriched >100-fold (Fig. 3a). The diversity of candidate NSUN2 RNA targets and candidate target sites required a rigorous statistical approach. Therefore, we utilized VarScan, a package for analyzing sequence variants in parallel sequencing data, to define locations where C>G transversions were both frequent (≥4%) and highly significant (P<0.01) in both anti-V5-NSUN2 replicates, but not in the IgG only control. We also applied VarScan to RNA-seq datasets of HeLa cells, to filter out SNPs. Transversion was clear and significant (Fig. 3b and Supplementary File 5) at the known human NSUN2 tRNA targets (tRNALeu (C34), tRNAAsp (C48, 49) and tRNAGly (C48, 49, 50))2, 11, 26, as well as at C48, C49 and C50 within most tRNAs (tRNAAsn and tRNASeC excepted), greatly expanding the known repertoire. Importantly, although the target tRNAs of DNMT2 were robustly enriched within anti-V5-NSUN2 immunoprecipitates, we did not observe C>G transversion at the DNMT2 target site (C38) within the anti-V5-NSUN2 immunoprecipitates, highlighting the specificity of both the enzyme and the Aza-IP technique (Fig. 3c,d). Furthermore, we identified a large number of additional candidate NSUN2 target sites within particular tRNAs; these sites were not previously described in any organism (P<0.01) (Fig. 3b and Supplementary File 5). Moreover, we observed significant transversion sites within introns, and also upstream and downstream, of particular pre-processed tRNAs (P<0.01) (Fig. 3b and Supplementary File 5).
Notably, within a particular RNA, the extent of C>G transversion was proportionally lower when there were multiple target sites. For example, tRNAGly(GCC), bears one DNMT2 target site (C38) and four NSUN2 target sites (C40, 48, 49 & 50) (Fig. 3d); here, the transversion frequency at C38 with DNMT2 was comparable to the sum at the four NSUN2 target sites (Fig. 3c and Supplementary Fig. 9), consistent with covalent linkage of the m5CRMT to only one target site in any individual isolated tRNA.
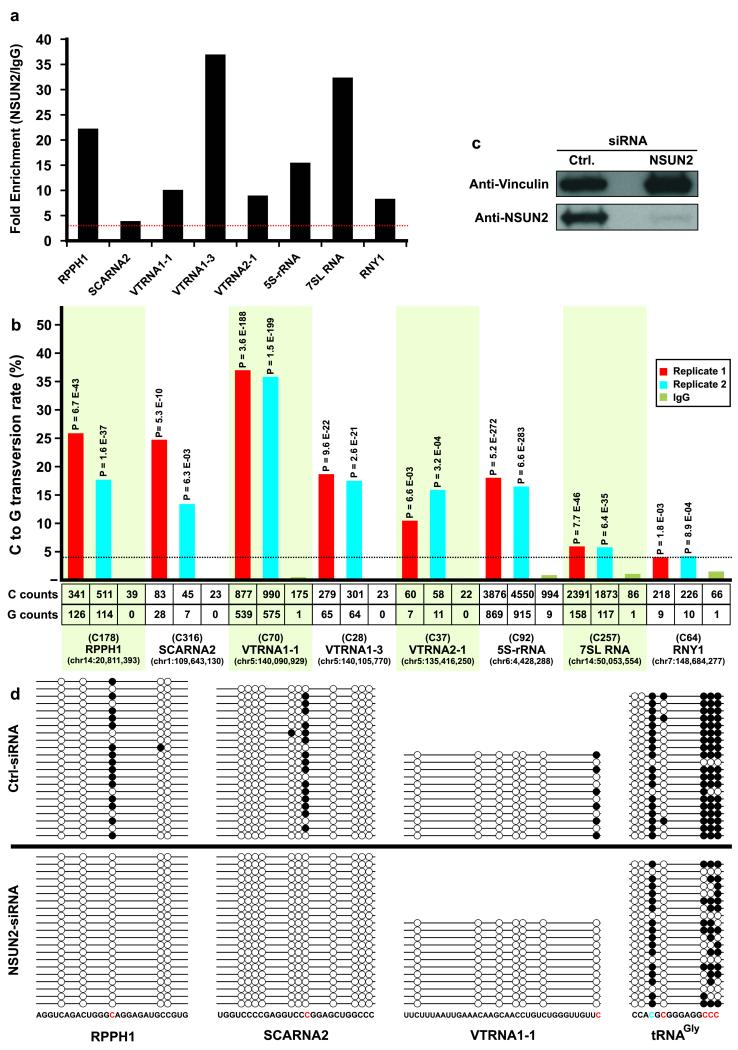
Human NSUN2 has been reported to methylate cytosines in four other RNAs: ribosomal RNA12, the RNA subunit of RNaseP (RPPH1) and two mRNAs (CINP and NAPRT1)2. Our Aza-IP analyses yielded eight candidate NSUN2 target ncRNAs: 5S rRNA, RPPH1, vault RNAs (VTRNA1-1, VTRNA1-3 and VTRNA2-1), the small cajal body-specific RNA 2 (SCARNA2, a C/D box SNORNA), a Y RNA (RNY1) and the signal recognition particle RNA (7SL RNA) (Fig. 4a). All of these RNAs contain one very clear and significant target site (Fig. 4b), and most contain one or more additional sites that also pass all three thresholds in both anti-V5-NSUN2 replicates but not the IgG only control (Supplementary File 5); these findings suggest that, as with tRNAs, most ncRNA targets have more than one NSUN2 target cytosine. Notably, Aza-IP analysis did not detect enrichment of CINP or NAPRT1 mRNAs in anti-V5 NSUN2 immunoprecipitates.
Figure 4.
New ncRNA targets and sites for human NSUN2, and validation through siRNA knockdown and bisulfite sequencing. (a) Graph depicts fold enrichment of human ncRNAs in V5-NSUN2 immunoprecipitates. Fold enrichment was calculated by dividing the normalized RPKM (Reads Per Kilobase per Million mapped reads) values for each ncRNA in the V5-NSUN2 replicate datasets (combined) by the values in the IgG control dataset. The red horizontal dotted line shows the 3-fold enrichment cut-off criteria. (b) C>G transversion at particular sites within ncRNAs enriched in anti-V5-NSUN2 immunoprecipitates. For each of the eight enriched ncRNAs, we depict the single target site with the highest C>G transversion (statistics for other candidate sites in Supplementary File 5). The bar graph shows the C>G transversion occurrence (% of total) in each V5-NSUN2 Aza-IP replicate and IgG only control; the horizontal dotted line shows the 4% transversion cut-off. P-values calculated by VarScan are indicated above columns. Provided underneath are the total number of sequenced C and G nucleotides (counts), the base position of the target cytosine within the ncRNA, and its encoded location in the genome (hg19). (c) Extracts (60μg of protein was loaded on the gel) of HeLa cells expressing NSUN2 or non-specific (control) siRNA pools were probed with anti-hNSUN2 or anti-Vinculin (control). (d) Total RNA was extracted from HeLa cells treated with NSUN2 or control siRNAs; RNA was subjected to bisulfite treatment, PCR amplification, cloning and sequencing of several clones per sample (see Online Methods). Candidate target sites of NSUN2 (in red; C178 in RPPH1, C316 in SCARNA2, C70 in VTRNA1-1, and C40, C48, C49 and C50 in tRNAGly(GCC)) and DNMT2 (in blue; C38 in tRNAGly(GCC)) were analyzed.
To validate the NSUN2 target RNAs identified by Aza-IP, we used siRNA to knock down NSUN2 expression (Fig. 4c) and then tested the location and extent of methylation on selected candidate target RNAs by conventional RNA bisulfite sequencing (Fig. 4d). Here, we tested RNAs at the top (tRNA), middle (RPPH1) and bottom (SCARNA2 and VTRNA1-1) of our enrichment results. In control siRNA-treated HeLa cells, all tested RNA candidates displayed a methylation (cytosine retention) at the precise site(s) predicted by the Aza-IP transversion, but not at other flanking cytosines. We noted a marked diminishment or elimination of methylation at these sites in NSUN2 siRNA-expressing HeLa cells, validating the involvement of NSUN2 in target methylation (Fig. 4d). For several reasons, we suggest that diminishment rather than elimination in tRNAGly methylation is predicted. Our protocol analyzes methylation at day 6, whereas NSUN2 protein knockdown via siRNA requires several days to reach >90% reduction; also tRNAs are long lived (~30-60 hours), so those tRNAs made 2 days prior will still be largely present and methylated. Also, tRNAs, given their exceptional enrichment in Aza-IP, may be preferred by NSUN2 over the ncRNAs and may compete more effectively for the remaining NSUN2 pool. Regardless, these siRNA experiments validate the conclusions of the Aza-IP analysis.
The NSUN2 candidate ncRNA targets identified here include RNAs with central functions in the processing, folding and modification of other ncRNAs (RPPH1, Y RNA, SCARNA2), RNAs important for protein synthesis and trafficking (5S rRNA and 7SL RNA) and RNAs involved in multidrug resistance and other processes (Vault RNAs)29, 30. Notably, all of the NSUN2 targets revealed by Aza-IP are either transcribed by RNA Pol III29 in the nucleolus (SCARNA2 excepted30), or function in the nucleolus (SCARNA2)30, where NSUN2 is known to reside12, 31. The biological functions of RNA methylation at these sites remain to be explored, but they could affect RNA structure, association with ribonucleoprotein complexes, or complex activity. Taken together, our work greatly increases the set of target site candidates for NSUN2 that should be considered as possible contributors to enzyme function, including in pathologies related to cancer, stem cells, and intellectual disability12, 14-17.
Comparisons of our NSUN2 datasets to the recent RNA bisulfite sequencing (RBS-seq, with ABI sequencing platform) dataset from HeLa cells2 reveals high overlap, with the minor deviations likely resulting from the high filtering thresholds utilized in the RBS-seq method. Going forward, we envision Aza-IP and RBS-seq as complementary approaches. However, as Aza-IP enriches target RNAs (revealing low-copy RNAs), identifies only direct targets, and reveals the precise methylation sites in these targets (even in situations of m5C-RMT redundancy and/or low methylation penetrance), we intend to apply Aza-IP to discover new m5C-RMT targets and to validate these using focused RBS-seq or other approaches. Finally, additional mechanism-based ‘adduct-IP’ trapping techniques (using other nucleotide analogues32) may help identify the targets of other RNA-modifying enzymes.
ONLINE METHODS
Note: here we provide the details for conducting Aza-IP with NSUN2, which is a more advanced procedure than the initial procedure used for DNMT2. For Aza-IP with DNMT2, please see the Supplementary Information.
Vector construction
Total RNA was extracted from HeLa cells using Trizol (Invitrogen) and first-strand cDNA synthesis was performed with SuperScript®III First-Strand Synthesis System (Invitrogen), using Oligo(dT) (Invitrogen), according to the manufacturer’s protocol. An NSUN2 clone bearing a V5 tag was obtained by PCR using specific primer sets and HeLa cDNA as template in a 2-step PCR format. The primer sets (Supplementary Table 2) replaced the first (ATG) codon with an AgeI restriction site, and inserted the Kozak consensus sequence containing the start codon (CACCATGG), and the sequence corresponding to the V5 tag (GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG) at the 5′ end of the amplicon. The primers also placed a NheI restriction site at the 3′ end of the amplicon, right after the stop codon. Validated PCR products were double-digested with AgeI and NheI, and cloned into an AgeI/NheI-modified pPR-lentiviral plasmid (a gift from Dr. Vicente Planelles, Utah). Virus production used standard packaging and VSVG envelope plasmids, HEK-293-FT cells (Invitrogen) transfection with polyethylenimine (Polysciences), harvesting, and titration on HeLa cells (using the EGFP marker) by flow-cytometry. Expression of V5-tagged NSUN2 was confirmed by western blot and the concentrated viral particles were stored at −80°C.
Lentiviral infection
Forty 100mm plates (total, for replicates and one control) were each seeded with two million HeLa cells, followed by infection 24hr later with concentrated virus (calculated MOI=1) mixed with DMEM (3ml, each plate, Invitrogen) supplemented with 10% FBS and 4μg/ml (final concentration) Polybrene (Millipore). 18 hours later, cells were washed with 1X PBS, trypsinized using TrypLE™ Express (Invitrogen), pooled and dispensed into sixty 150mm plates.
5-Azacytidine treatment
After 14 hours growth, media was replaced with DMEM media containing freshly-prepared 5-Azacytidine (Sigma, 5μM final), incubated for 12hr, followed by a second media exchange again with freshly-prepared 5-Azacytidine (5μM final), and incubation for another 12 hours, followed by harvesting (see below).
Preparing the pre-clearing beads
For each replicate (1 & 2) and IgG control experiments, 750μl of Dynabeads® Pan Mouse IgG (Invitrogen) were washed in 1ml of diluted modified RIPA buffer [50mM Tris PH 7.5, 1% Nonidet P-40 (NP-40), 0.1% sodium deoxycholate, 0.025% SDS, 1mM EDTA, 150 mM NaCl + Protease Inhibitor (PI) cocktail] supplemented with 5mg/ml protease free bovine serum albumin (BSA) (Sigma) three times (for 2 minutes each), and re-suspended in 1.5ml of diluted modified RIPA buffer + BSA + 20μl of RNasIN (Promega).
Preparing the antibody-coated beads
For each experiment, 1.5ml of the Dynabeads® Pan Mouse IgG were split into two 1.5ml tubes (750μl each) and washed three times (for 2 minutes each) with 1ml of diluted modified RIPA buffer + BSA. Next the beads of each tube were re-suspended in 1.5ml of diluted modified RIPA buffer + BSA + 45μg of the Invitrogen’s mouse anti-V5 antibody (for replicate 1 & 2) or IgG (for control) and incubated at room temperature, rotating for 2 hours. The beads of each tube were then washed three times (2 minutes each) with 1ml of diluted modified RIPA buffer + BSA, and then re-suspended in 1.2 ml of diluted modified RIPA buffer + BSA + 15μl of RNasIN.
Cell lysis and solubilization
Following 24 hours of 5-Azacytidine treatment, cells from each plate were washed with 1X PBS and trypsinized with 5ml of TrypLE™ Express, and quenched with 5ml of complete DMEM media. The contents of all sixty 150mm plates were pooled, spun at 2000 rpm at 4°C for 10min, washed with 15 ml of 1X PBS, spun again at 2000 rpm at 4°C for 5min, and pellets were flash frozen in liquid nitrogen and stored at −80°C. Cells were thawed in 6ml of modified RIPA buffer [50mM Tris PH 7.5, 1% Nonidet P-40 (NP-40), 0.2% sodium deoxycholate, 0.05% SDS, 1mM EDTA, 300 mM NaCl + PI cocktail] supplemented with 30μl of RNaseIN, and pipetted to homogeneity. The cell lysates (6ml) were sonicated using a Misonix Sonicator XL2020, 6 pulses of 30 seconds (setting #4 (0.9 ON time/0.1 OFF time)) interposed with 30 seconds on ice. After sonication the lysates were pooled, mixed with 18ml of dilution buffer [50mM Tris buffer (pH 7.5), 1% Nonidet P-40 (NP-40)]. The 2-fold diluted lysate was dispensed into thirty-six 1.5ml eppendorf tubes (1ml each), spun at 14,000 rpm at 4°C for 10 min, transferred to clean 1.5ml tubes and kept on ice.
Immunoprecipitation
The 36 tubes were split into 3 groups (12 tubes each), the first and second groups were considered replicates (1 & 2), and separately mixed with anti-V5 coated beads. The third group was considered ‘control’, and mixed with IgG-coated beads. For each 1ml of the lysate, 125μl of the pre-clearing beads were added, incubated at RT with rotation for 2hr. Using a magnetic stand, the supernatant was transferred to a new 1.5ml tube, and 200μl of V5-coated beads (or IgG) were added and rotated for 4hr at RT. Beads were collected by the magnet, washed with 850μl of modified RIPA + PI (3 times, 5 minutes each at RT), re-suspended in 500μl of modified RIPA+ PI and transferred to a clean 0.5ml eppendorf tube. The beads (in 12 separate tubes) were collected and the supernatants discarded.
RNA fragmentation
For each sample (0.5ml), 7.5μl of the RNA fragmentation reagent (Ambion) was mixed with 67.5μl of RNase free ddH2O (Ambion) in one tube (total of 75μl per tube), and in another tube 7.5μl of the fragmentation stop solution (Ambion) was mixed with 7.5μl of RNase free ddH2O (total of 15ul per tube). For the fragmentation, 75μl of the diluted RNA fragmentation reagent was added to the sample, incubated at 94°C for exactly 5min in a thermocycler, chilled on ice for 1min, and terminated by adding 15μl of the diluted stop solution.
Ethanol precipitation and RNA extraction
After fragmentation, the supernatants derived from each replicate were collected and pooled in a clean 1.5ml tube (total of 1080μl). Next 40μl of the 15mg/ml GlycoBlue (Ambion) and 104μl of 3M Sodium Acetate (Ambion) were mixed with the fragmented RNA and the mixture was split into three 1.5ml tubes (346μl each). Then, 865μl absolute ethanol was added to each tube and the tubes were incubated at −80°C overnight, and spun at maximum speed at 4°C for 20 min. The pellets were washed once with 1ml of 70% ice cold ethanol, air dried and dissolved in total of 40μl RNase free ddH2O (for all three of the tubes per experiment) and pooled in a single tube. Next 1ml of Trizol reagent was added and the RNA was extracted according to the manufacturer’s protocol.
Library preparation and high-throughput sequencing
Libraries involved Illumina’s directional mRNA-Seq protocol involving the ligation of adapters to fragmented RNAs followed by reverse transcription, PCR and sample clean-up using AMPure beads (Beckman Coulter Genomics). Libraries were subjected to 50-cycle single-end high-throughput sequencing using Illumina’s HiSeq 2000 system.
Computational analytical methods
Sequenced reads were aligned to the H. sapiens Feb 2009 genome build plus all known and theoretical splice junctions derived from Ensemble transcripts (see “MakeTranscriptome” application from open source USeq package33). Sequence alignments was performed using the commercial Novoalign package (http://www.novocraft.com) with options to allow gaps and mismatches, reporting 18bp or larger inserts and reporting all of the reads mapped to the repeats and generating SAM-formatted alignment files. The “RNASeq” application within USeq was used to define enriched regions and obtain the RPKM (Reads Per Kilobase per Million mapped reads) values, and to make BAM/BAI files for visualization. The Integrative Genomics Viewer (IGV)34 was used to visualize the alignment files and inspect the mapped reads at base pair resolution. Next, a combination of SAMtools (mpileup function)35 and VarScan (mpileup2cns function)36 were used to identify the cytosines showing the significant C>G transversion signatures (FDR<0.01 & transversion frequency > 4%) within the RNAs identified as significantly enriched by RNASeq application (FDR<0.01, Fold enrichment >3 & RPKM>3). For NSUN2 tRNA target sites, to convert the genomic coordinates of the candidate sites to the tRNA nucleotide numbers according to the standard tRNA numbering system37, the candidate sites within the sequenced reads (visualized in IGV) were compared to the tRNA alignments from Genomic tRNA Database38 for Homo sapiens genome (hg19 - NCBI Build 37.1 Feb 2009) and the corresponding residue numbers were deduced and reported according to the canonical tRNA cloverleaf secondary structure and numbering system37. To filter for possible C>G SNPs in the HeLa transcriptome, available RNA Seq datasets were used for comparison. Datasets are publicly available through GEO with the following accession numbers: GSE38957 and GSE44359.
RNAi-mediated hNSUN2 knockdown
For each sample, 3×105 HeLa cells were seeded in a single well of a 6-well plate. The next day, cells were transfected with 60pmol of Dharmacon’s siGENOME Human NSUN2 siRNA – SMARTpool (M-018217-01-0005) or siGENOME Non-Targeting siRNA Pool #1 (D-001206-13-05) using Lipofectamine RNAiMAX transfection reagent (Invitrogen). After 72 hours, cells of each group were passaged and transfected with 120pmol of the same siRNA pools. The cells were harvested 72 hours after the second transfection and subjected to RNA and protein extraction. Protein extracts were evaluated by western blotting for knockdown efficiency by immuno-blotting with hNSUN2 polyclonal (Prteintech-20854-1-AP) or hVinculin monoclonal (Sigma-V9131) antibodies.
Conventional RNA bisulfite sequencing
Purified RNA from hNSUN2 or control siRNA knockout HeLa cells were subjected to DNase treatment and fragmentation. For bisulfite treatment 5μg of the fragmented RNA was dissolved in 45μl of RNase free ddH2O and added to 240μl of de-ionized formamide, mixed well and incubated at 95°C for 5min then placed on ice for 2 min. Next 3μl of the 100mM hydroquinone and 312μl of 5M sodium bisulfite (pH 5) were added and the mixture was incubated at 50°C, rotating. After 16 hours, the mixture was cleaned up using Illustra NAP-10 Columns (GE Healthcare Life Sciences) and de-sulfonation was performed for 2hrs at 37°C in 1M Tris buffer pH 9.0, followed by ethanol precipitation. The first-strand DNA synthesis and PCR amplification was performed using specific primer sets (see Supplementary Table 6) designed for the bisulfite-converted tRNAs. The PCR product was cloned using TOPO TA Cloning Kit (Invitrogen) and the resulting plasmids purified from several clones were sequenced. The sequences were aligned with the corresponding tRNA sequences and the maintained cytosines (representing methylated cytosines) are depicted in the figures as closed circles.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cedric Clapier (help on hDNMT2 protein expression and purification), Kunal Rai (help on the MTase assay set-up), Vicente Planelles (gift of the lentiviral expression construct), Carlos Maximiliano Rêgo Monteiro Filho and Somaye Dehghanizadeh (help on lentiviral protein expression, assays, and IP experiments) and Jiafeng Xu and Archana Yerra (help on data not shown). We thank Brian Dalley & Nicole Moss (library preparation and sequencing), Timothy Parnell, David Nix, Brett Milash, Ying Sun & Kenneth Boucher (help and advice on analysis) and the Center for High Performance Computing (CHPC), especially Wim R. Cardoen. We thank Cynthia J. Burrows for advice on reaction mechanisms, and David A. Jones, Cynthia J. Burrows and Darrell R. Davis for many helpful comments. This work was supported by the Howard Hughes Medical Institute, the Samuel Waxman Foundation, and NCI CA24014 (for core facilities).
Footnotes
AUTHOR CONTRIBUTIONS V.K., contributed to experimental design and approaches, performed all experiments and analyses, and helped write the paper; B.C, contributed to experimental design and approaches, data interpretation, and wrote (with V.K.) the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jurkowski TP, et al. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14:1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- 5.Goll MG, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 6.Rai K, et al. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2009;119:35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 9.Durdevic Z, et al. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013 doi: 10.1038/embor.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuorto F, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 11.Brzezicha B, et al. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA) Nucleic Acids Res. 2006;34:6034–6043. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Hussain S, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco S, et al. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7:e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasi-Moheb L, et al. Mutations in NSUN2 Cause Autosomal-Recessive Intellectual Disability. Am J Hum Genet. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan MA, et al. Mutation in NSUN2, which Encodes an RNA Methyltransferase, Causes Autosomal-Recessive Intellectual Disability. Am J Hum Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez FJ, et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012 doi: 10.1136/jmedgenet-2011-100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 19.Lu LW, Chiang GH, Medina D, Randerath K. Drug effects on nucleic acid modification. I. A specific effect of 5-azacytidine on mammalian transfer RNA methylation in vivo. Biochem Biophys Res Commun. 1976;68:1094–1101. doi: 10.1016/0006-291x(76)90308-9. [DOI] [PubMed] [Google Scholar]
- 20.Lu LJ, Randerath K. Effects of 5-azacytidine on transfer RNA methyltransferases. Cancer Res. 1979;39:940–949. [PubMed] [Google Scholar]
- 21.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 22.Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schermelleh L, et al. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat Methods. 2005;2:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536–546. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci U S A. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auxilien S, Guerineau V, Szweykowska-Kulinska Z, Golinelli-Pimpaneau B. The human tRNA m (5) C methyltransferase Misu is multisite-specific. RNA Biol. 2012;9:1331–1338. doi: 10.4161/rna.22180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker M, et al. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S, Wu J, Chen L, Shan G. Signals from noncoding RNAs: Unconventional roles for conventional pol III transcripts. Int J Biochem Cell Biol. 2012;44:1847–1851. doi: 10.1016/j.biocel.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Gerard MA, et al. The scaRNA2 is produced by an independent transcription unit and its processing is directed by the encoding region. Nucleic Acids Res. 2010;38:370–381. doi: 10.1093/nar/gkp988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakita-Suto S, et al. Aurora-B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18:1107–1117. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guelorget A, Golinelli-Pimpaneau B. Mechanism-based strategies for trapping and crystallizing complexes of RNA-modifying enzymes. Structure. 2011;19:282–291. doi: 10.1016/j.str.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.