Abstract
Usher syndrome type I is characterized by congenital hearing loss, retinitis pigmentosa (RP), and variable vestibular areflexia. Usher syndrome type ID, one of seven Usher syndrome type I genetic localizations, have been mapped to a chromosomal interval that overlaps with a nonsyndromic-deafness localization, DFNB12. Mutations in CDH23, a gene that encodes a putative cell-adhesion protein with multiple cadherin-like domains, are responsible for both Usher syndrome and DFNB12 nonsyndromic deafness. Specific CDH23 mutational defects have been identified that differentiate these two phenotypes. Only missense mutations of CDH23 have been observed in families with nonsyndromic deafness, whereas nonsense, frameshift, splice-site, and missense mutations have been identified in families with Usher syndrome. In the present study, a panel of 69 probands with Usher syndrome and 38 probands with recessive nonsyndromic deafness were screened for the presence of mutations in the entire coding region of CDH23, by heteroduplex, single-strand conformation polymorphism, and direct sequence analyses. A total of 36 different CDH23 mutations were detected in 45 families; 33 of these mutations were novel, including 18 missense, 3 nonsense, 5 splicing defects, 5 microdeletions, and 2 insertions. A total of seven mutations were common to more than one family. Numerous exonic and intronic polymorphisms also were detected. Results of ophthalmologic examinations of the patients with nonsyndromic deafness have found asymptomatic RP–like manifestations, indicating that missense mutations may have a subtle effect in the retina. Furthermore, patients with mutations in CDH23 display a wide range of hearing loss and RP phenotypes, differing in severity, age at onset, type, and the presence or absence of vestibular areflexia.
Introduction
Usher syndrome is a frequent cause of recessive syndromic deafness and is estimated to account for (a) 3%–6% of the congenitally deaf population (Vernon 1969), (b) ∼18% of those with retinitis pigmentosa (RP), and (c) >50% of the deaf-blind population (Boughman et al. 1983). The frequency of Usher syndrome has been estimated to be 3.5/100,000 in Sweden and Finland (Nuutila 1970; Grondahl 1987), 3.2/100,000 in Colombia (Tamayo et al. 1991), and 4.4/100,000 in the United States (Boughman et al. 1983).
The standard classification of Usher syndrome recognizes three distinct clinical categories (Kimberling and Moller 1995). Type I is manifested by severe to profound congenital hearing impairment, vestibular dysfunction, and retinal degeneration beginning in childhood, whereas type II is manifested by moderate to severe hearing impairment, normal vestibular function, and later onset of retinal degeneration; type III, the least common form of Usher syndrome, presents with progressive hearing loss and a variable retinal and vestibular phenotype.
At least seven different USH1 genes are responsible for the more severe, type I syndrome. These genes have been mapped to chromosomes 10q21.1, 10q22.1, 11q13.5, 11p15.1, 14q32, 17q24-25, and 21q21 (see the Hereditary Hearing Loss Homepage). There are no studies reporting clinical differences between these seven forms, but they are differentiated on the basis of either linkage analysis in informative families or mutational analyses of the genes involved. Currently, four USH1 genes have been identified. Defects in myosin VIIa, harmonin, cadherin 23, and protocadherin 15 are responsible for causing Usher syndrome type I subtypes B, C, D, and F, respectively (Weil et al. 1995; Bitner-Glindzicz et al. 2000; Verpy et al. 2000; Ahmed et al. 2001; Alagramam et al. 2001; Bolz et al. 2001; Bork et al. 2001).
In 1996, by homozygosity mapping of an inbred Pakistani family, Wayne et al. localized USH1D (MIM 601067) to the long arm of chromosome 10 (Wayne et al. 1996), a chromosomal region that also contained DFNB12 (MIM 601386), a nonsyndromic-deafness localization (Chaib et al. 1996). Mutations in CDH23 (MIM 605516) cause both Usher syndrome type ID and DFNB12. Missense mutations were found exclusively in five consanguineous families with nonsyndromic deafness (Bork et al. 2001) and in a sixth, large consanguineous family with nonsyndromic deafness (A. P. M. Brouwer, R. J. E. Pennings, M. Roeters, P. Van Hauwe, L. M. Astuto, L. H. Hoefsloot, P. L. M. Huygen, B. van den Helm, A. F. Deutman, J. M. Bork, W. J. Kimberling, F. P. M. Cremers, C. W. R. J. Cremers, and H. Kremer, unpublished data [hereafter referred to as “Brouwer et al., unpublished data”]). Splicing variants, deletions, nonsense mutations, and missense mutations of CDH23 have been identified in patients with Usher syndrome type I (Bolz et al. 2001; Bork et al. 2001; von Brederlow et al. 2002).
CDH23 has 69 exons and encodes a predicted 3,354-amino-acid protein with 27 cadherin extracellular (EC) repeats, a transmembrane domain, and a unique cytoplasmic domain. The product of CDH23 has similarity to the well-characterized protein E-cadherin; the EC repeats of E-cadherin are responsible for homophilic cell-cell adhesion, which is established by the formation of parallel stable EC-domain dimers at the same cell surface and at two opposing cell surfaces. This stability is due to interdomain rigidification of the EC domains, achieved by the binding of three calcium ions to highly conserved peptide sequences (LDRE, DXNDN, and DXD) (Nollet et al. 2000). The functions of cadherin 23 have not been defined, but it is hypothesized to be involved in the establishment of cell-cell contacts and in the organization of the EC matrix.
In the present study, we sought to further investigate the genotype/phenotype relationship, proposed by Bork et al. (2001), between CDH23 mutations and the consequent hearing, visual, and vestibular phenotype. Using a comprehensive mutation-detection strategy via heteroduplex, SSCP, and direct sequence analyses of all 69 exons from 107 probands, we were able to gain insight into the distribution and nature of diverse CDH23 mutations and their resulting phenotypes.
Subjects, Material, and Methods
Subjects and Clinical Studies
A total of 107 families with Usher syndrome and recessive nonsyndromic deafness (RNSD), comprising 281 unaffected and 173 affected members, were sampled and studied; 12 families were consanguineous. A total of 68 families (48 with Usher syndrome type ID and 20 with DFNB12) were multiplex and compatible with linkage to the USH1D region; and the remaining 39 families were characterized by sporadic cases (21 with Usher syndrome type I and 18 with RNSD). The 38 families with RNSD were drawn from an original population of 157 families referred with nonsyndromic hearing loss to Boys Town National Research Hospital. In all families, mutations in MYO7A and GJB2 were excluded by linkage and/or mutational analysis. Families were collected in the United States (57 families), Sweden (17 families), The Netherlands (10 families), Germany (10 families), Spain (3 families), Pakistan (3 families), South Africa (2 families), France (2 families), Italy (2 families), and Ireland (1 family). Several of these families were part of three previous studies (Kelley et al. 1998; Astuto et al. 2000; Bork et al. 2001). Not all individuals in the present study were sequenced for all 69 CDH23 exons.
Cases were included on the basis of the following criteria: (1) both parents had normal hearing, and (2) the proband’s hearing loss was moderate to profound. Patients with Usher syndrome meet the further criteria of presenting with evidence of RP, including extinguished or subnormal electroretinography (ERG) recordings. We refer to typical families with Usher syndrome type I as those presenting with profound deafness, RP, and absent vestibular function; atypical families are categorized as having a milder affliction in one or more of the three sensory systems (hearing, balance, and vision) involved. Two families with Usher syndrome type II were also included. Computed-tomography scans of the inner ear were also obtained for several families, and the results were normal in all cases. For each family, a pedigree was drawn. Clinical data were collected from medical histories and/or examinations, either at Boys Town National Research Hospital in Omaha or by various collaborators at their local clinical facilities. The local human-subjects committees approved the present study, and informed consent was obtained from all participants.
Blood Collection and DNA Extraction
Blood samples were collected, and genomic DNA was extracted by a Puregene kit (Gentra Systems).
PCR Conditions
Oligonucleotide primers designed by Bolz et al. (2001) and Bork et al. (2001) were used for PCR and for cycle-sequencing reactions. PCR elements were as follows: 1 × reaction buffer, 200 μM each dNTP, 2.5 mM MgCl2, 5 pmol of each forward and reverse primer, and 0.5 U of Taq DNA polymerase. PCR conditions were as follows: 95°C for 10 min, followed by 35–40 cycles of 95°C for 30 s, 55°–65°C for 30 s, and 72°C for 60 s, followed by a final extension at 72°C for 10 min.
Heteroduplex Analysis (HA)
PCR products amplified from syndromic and nonsyndromic probands and cloned control DNA were mixed, heated at 95°C for 3 min, and cooled to 25°C during a 45-min period. The reannealed reaction products were then electrophoresed through a 35-cm × 43-cm × 1-mm volume of MDE (FMC) gel, at an average of 900 V for 18–24 h, were stained with 1 μg of ethidium bromide/ml, and were visualized under UV light. RP11-472K8 BAC DNA (AC016823) was amplified for each CDH23 exon and was used as a control. Mixing with BAC control DNA prior to heteroduplex formation allows the detection of both homozygous and heterozygous mutations. A set of 96 genetically independent control samples of mixed European ancestry also was studied, to aid in differentiation between probable pathologic mutations and polymorphisms. The controls were obtained through a reading-disability research project in which no history of hearing or visual loss had been identified. If the mutation was found in one or more of the 96 control samples, it was considered to be nonpathologic.
SSCP Analysis
SSCP analysis was performed on several CDH23 exons. The amplified DNA was run on 5% polyacrylamide gels, with 10% glycerol, in 1 × Tris-borate EDTA. Samples were electrophoresed either at 45 W, in a cold room, or at 40 W, with cooling electrophoresis equipment (Gibco-BRL). A set of 96 normal independent genomic control samples was also studied, to aid in the identification of probable pathologic mutations and in the differentiation between them and polymorphisms.
DNA Sequencing
PCR products showing band shifts in SSCP and/or HA were purified and concentrated by Microcon 100 filters (Millipore) and Millipore 96-well filter plates, prior to sequencing. ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kits (Perkin Elmer) were used to generate sequence from both purified PCR products and BAC DNA templates. Five microliters of PCR products were sequenced with 4 pmol of CDH23 primers, 3 μl of ABI Ready Reaction mix, and 5 × buffer, in a total volume of 10 μl, for 35 cycles of 95°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Products of the sequencing reactions were precipitated and resuspended in a 5:1 ratio of formamide/dye loading buffer. Two microliters were loaded onto 4.8% PAGE-PLUS (Amresco) sequencing gels and were run on the ABI Prism model 377 DNA sequencer, according to ABI protocols. Sample files were generated by ABI Sequencing Analysis 3.0. Fractura 2.0 (ABI) was used to remove ambiguous sequences. Contig assembly was performed by the DNASTAR SeqMan 5.03 program. In some cases, direct sequencing could not unequivocally identify the DNA sequence change; for those cases, PCR products were cloned into a plasmid vector (TOPO TA Cloning Kit; Invitrogen), to separate individual alleles prior to sequencing.
Splicing Program
All IVS changes were analyzed by the BDGP splice-site–prediction program (available at the Berkeley Drosophila Genome Project Splice Site Prediction by Neural Network Web site). IVS sequence changes that predict a loss of splicing-acceptor or -donor activity and that are not present in control samples are considered pathologic.
Results
Mutational Analyses
Two hundred fourteen chromosomes were screened for mutations in CDH23, by a combination of heteroduplex, SSCP, and/or sequencing analyses. A total of 36 different CDH23 mutations were identified in 45 families. Tables 1 and 2 list the mutations observed in, respectively, the families with DFNB12 and the families with Usher syndrome type ID. Table 3 lists the presumed nonpathogenic variants in CDH23. Codon numbering starts with the first in-frame methionine of the human CDH23 (GenBank accession number AF312024; Bolz et al. 2001).
Table 1.
Twelve Novel Mutations in the CDH23 Gene Observed in 9 of 38 Probands with RNSD
| Nucleotide (Codon) Change | Exon | Domain | Proportion ofChromosomes | Family |
| 371A→G (D124G) | 5 | EC1 | 1/214 | 1846 |
| 1355A→G (N452S) | 13 | EC4 | 1/214 | 3055 |
| 1439T→A (L480Q) | 13 | EC5 | 1/214 | 2640 |
| 1745G→A (R582Q) | 15 | EC6 | 1/214 | 1194 |
| 3178C→T (R1060W) | 26 | EC10 | 1/214 | 3063 |
| 3557G→A (G1186D) | 29 | EC11 | 1/214 | 2640 |
| 4756G→C (A1586P) | 37 | EC15 | 2/214 | PKDF030 |
| 4783G→A (E1595K) | 37 | EC15 | 1/214 | 1566 |
| 5536G→A (D1846N) | 42 | EC17 | 1/214 | 3055 |
| 6442G→A (D2148N) | 47 | EC20 | 3/214 | 3409, 1668 |
| 7393C→T (R2465W) | 52 | EC23 | 1/214 | 1846 |
| 7823G→A (R2608H) | 54 | EC24 | 1/214 | 1668 |
Table 2.
Mutations in the CDH23 Gene Observed in 35 of 69 Probands with Usher Syndrome
| Nucleotide (Codon) Change | Exon | Domain | Proportion ofChromosomes | Family |
| Missense mutations: | ||||
| 3617C→G (P1206R) | 30 | EC11 | 1/214 | 1078 |
| 3625A→G (T1209A) | 30 | EC11/EC12a | 2/214 | 220 |
| 5237G→A (R1746Q)b | 40 | EC16/EC17a | 6/214 | 792, 809, 2790, 3308 |
| 7549A→G (S2517G) | 53 | EC23/EC24a | 1/214 | 1056 |
| 8230G→A (G2744S) | 57 | EC26 | 1/214 | 1054 |
| 8497C→G (R2833G) | 58 | EC26 | 1/214 | 980 |
| 9524G→A (R3175H) | 67 | CYTO | 1/214 | 3051 |
| Splicing variants: | ||||
| IVS4+1G→A | 4 | EC1 | 13/214 | 1185, 1197, 1285, 1288, 1289, 1291, 1321, 1324 |
| 1450G→Cc (A484P) | 14 | EC5 | 2/214 | 1517 |
| IVS20+1G→A | 20 | EC7 | 3/214 | 1066, 2059, 3036 |
| 3105A→Cc (T1035T) | 25 | EC10 | 1/214 | 2059 |
| 4488G→Cc (Q1496H)b | 35 | EC14 | 1/214 | 1444 |
| IVS45−9G→Ab | 46 | EC19 | 7/214 | 1071, 3036, 3150, PKSRB, PKSR13a |
| 7872G→Ac (E2624E) | 54 | EC25 | 1/214 | 2797 |
| Nonsense: | ||||
| 172C→T (Q58X) | 3 | EC1 | 1/214 | 1291 |
| 4504C→T (R1502X) | 36 | EC14 | 5/214 | 1184, 1197, 1198, 1288 |
| 6307G→T (E2103X) | 47 | EC20 | 1/214 | 3150 |
| Deletions: | ||||
| 193delC | 3 | EC1 | 1/214 | 216 |
| 1087delG | 10 | EC4 | 2/214 | 2738 |
| 1112delT | 10 | EC4 | 2/214 | 359, 792 |
| 6155delC | 46 | EC19 | 1/214 | 1130 |
| 6968delC | 49 | EC22 | 1/214 | 51 |
| Insertions: | ||||
| 3840insATGA | 31 | EC12 | 1/214 | 216 |
| 9626insC | 68 | CYTO | 1/214 | 897 |
Table 3.
Presumed Nonpathogenic Variants in CDH23
| Intronic Variants | Exonic Variants | ||
| Nucleotide Change | Exon-SpecificPCR Product | Nucleotide(Codon) Change | Exon |
| IVS-121C→T | 1 | 7C→T (R3C) | 1 |
| IVS-88A→C | 1 | 198G→A (V66V) | 3 |
| c.-1C→T | 1 | 366T→C (V122V) | 5 |
| IVS1+12C→T | 1 | 1038G→A (P346P) | 10 |
| IVS5+13G→A | 5 | 1053C→T (S351S) | 10 |
| IVS5+26A→G | 5 | 1469G→C (G490A) | 14 |
| IVS6+41C→A | 6 | 1487G→A (S496N) | 14 |
| IVS6+64C→T | 6 | 2316T→C (N772N) | 21 |
| IVS8+101G→A | 8 | 2388T→C (D796D) | 21 |
| IVS9−24G→A | 10 | 3009T→C (S1003S) | 25 |
| IVS10+13A→G | 10 | 3231T→G (P1077P) | 27 |
| IVS11−107A→T | 12 | 3664G→A (A1222T) | 30 |
| IVS11−89C→T | 12 | 4051G→A (D1351N) | 31 |
| IVS14−12G→A | 15 | 4299T→A (P1433P) | 34 |
| IVS15−47T→C | 16 | 4310G→A (R1437Q) | 34 |
| IVS16+79A→G | 16 | 4341T→C (D1447D) | 34 |
| IVS16−220C→T | 17 | 4723G→A (A1575T) | 37 |
| IVS16−46T→C | 17 | 4858G→A (V1620M) | 38 |
| IVS18−40G→T | 19 | 5023G→A (V1675I) | 38 |
| IVS18−21C→G | 19 | 5100C→T (Y1700Y) | 39 |
| IVS19−19G→A | 20 | 5411G→A (R18804Q) | 41 |
| IVS20+135C→T | 20 | 5541C→T (N1847N) | 42 |
| IVS21+26T→C | 21 | 5660C→T (T1887I) | 42 |
| IVS23−17C→T | 24 | 5996C→G (T1999S) | 45 |
| IVS25−60C→T | 26 | 6130G→A (E2044K) | 46 |
| IVS29−12C→T | 30 | 6197G→A (R2066Q) | 46 |
| IVS31−183G→C | 32 | 6315C→T (V2105V) | 47 |
| IVS31−160C→T | 32 | 6847G→A (V2283I) | 49 |
| IVS31−83A→G | 32 | 6918G→A (L2306L) | 49 |
| IVS33−53T→C | 34 | 7073G→A (R2358Q) | 50 |
| IVS33−71C→T | 34 | 7139C→T (P2380L) | 50 |
| IVS35+32C→G | 35 | 7467C→T (R2489R) | 52 |
| IVS37−135C→T | 38 | 7572G→A (A2524A) | 53 |
| IVS37−79G→A | 38 | 7762G→C (E2588Q) | 54 |
| IVS37−49T→C | 38 | 8798T→A (V2933E) | 60 |
| IVS38−48G→C | 39 | 8860G→A (D2954N) | 60 |
| IVS39+44C→G | 39 | 8885A→G (N2962S) | 60 |
| IVS39+73C→T | 39 | 8895C→T (P2965P) | 60 |
| IVS41−44T→C | 42 | 8907C→T (R2969R) | 60 |
| IVS41−10A→G | 42 | 9015G→A (A3005A) | 61 |
| IVS48−81G→A | 49 | 9873G→A (T3291T) | 69 |
| IVS52+6G→A | 52 | ||
| IVS58+25G→A | 58 | ||
| IVS60−14C→A | 61 | ||
| IVS61+8G→A | 61 | ||
Nonsense mutations
Three mutations causing premature stop codons were identified in six probands with Usher syndrome type ID. Heterozygous nucleotide substitutions resulted in two premature stop codons—Q58X and E2103X, in exons 3 and 47, respectively. A 4504C→T transition, producing R1502X in exon 36, was identified exclusively in four Swedish families. Genealogical research revealed that all four families originated from or near the Västerbotten region in northern Sweden, suggesting a founder effect for R1502X.
Deletions/insertions
Seven mutations that created a frameshift leading to a subsequent premature stop codon were identified in seven probands with Usher syndrome. One mutation, 9626insC, is located in the cytoplasmic domain. Sequence analysis of family 216 identified a maternally inherited 4-base insertion of an ATGA between nucleotide position 3840 and nucleotide position 3841 in exon 31, resulting in a deduced TGA stop codon eight amino acids downstream. The insertion of these nucleotides also alters codon 1281, the same methionine codon that Bolz et al. (2001) previously had reported to be deleted in a patient who was compound heterozygous for Usher syndrome.
Splice-site mutations
Seven splice-site defects were detected, two of which accounted for the most frequent mutations in CDH23. An IVS4+1G→A mutation of the splice-donor site of exon 4 was detected in eight Swedish families. The frequency of IVS4+1G→A in the Swedish probands suggests that it may also be a Swedish founder CDH23 mutation. Haplotype conservation of the two Swedish mutations is currently under study.
The second most frequent CDH23 mutation identified in the present study is an IVS45−9G→A mutation of the splice-acceptor region preceding exon 46. Exon-trapping experiments done by von Brederlow et al. (2002) have revealed aberrant splicing creating a novel acceptor splice site 9 bp upstream of the 5′ end of the exon, consequently resulting in the insertion of 7 bp from the 3′ end of intron 45. Another mutation disrupting the conserved donor splicing sequence is an IVS20+1G→A transition found in one allele of three independent families.
The four remaining predicted splicing defects are nucleotide substitutions occurring at the first or last nucleotide of an exon. One of these mutations, Q1496H, a mutation that has been reported elsewhere, is heterozygous in one of the Spanish probands. Published exon-trapping experiments have shown that this mutation causes the use of a cryptic intronic splice site used downstream of the G→C nucleotide substitution (Bolz et al. 2001).
Missense mutations
A total of 19 different missense mutations were detected in our study; 12 were identified in families with RNSD, and the remaining 7 were identified in families with Usher syndrome type I. Each missense mutation identified in the probands with Usher syndrome was compound heterozygous with a presumed null allele, except for two cases: one family was homozygous for T1209A, and two families were homozygous for R1746Q; each presents an atypical Usher syndrome phenotype (Bolz et al. 2001; present study).
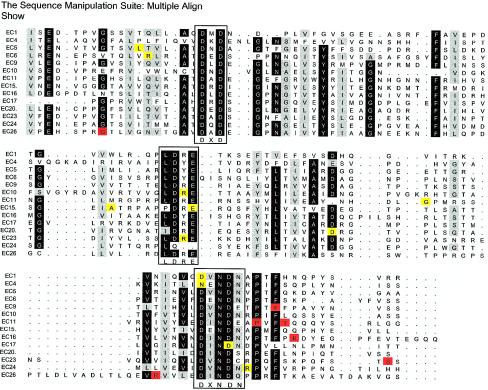
An American family with RNSD was compound heterozygous for two missense mutations in CDH23: D124G in exon 5 and R2465W in exon 52. Amino acid alignment of several cadherin 23 domains reveals that the D124G mutation disrupts a conserved DXNDN calcium-binding motif in the first EC domain. Likewise, the R2465W mutation disrupts the LDRE calcium-binding motif in the 23rd EC domain (fig. 1). Molecular modeling (Brouwer et al., unpublished data) of the R2465W mutation shows, furthermore, an interrupted intra- and interdomain salt bridge, which should strongly reduce the interaction between EC23 and EC24 and which therefore may change the tertiary structure required for proper cadherin 23 function.
Figure 1.
Alignment of cadherin 23 EC domains with missense mutations causing Usher syndrome type ID and recessive DFNB12. DNASTAR Lasergene and the program of The Sequence Manipulation Suite were used to align 14 EC domains. Corresponding EC domains are indicated to the left of the lineup. Dark-gray–shaded residues are >90% identical in all sequences; light-gray–shaded residues are 70% conserved in all aligned sequences. The LDRE, DXNDN, and DXD calcium-binding motifs are boxed. The sites of missense mutations identified in the present study (tables 1 and 2) are highlighted in yellow (in the case of DFNB12) and red (in the case of USHID).
A homozygous D2148N was identified in one Dutch family with RNSD; an American family was heterozygous for the mutation. Restriction digestion with BsiEI confirmed segregation of this DFNB12 variant in both families with RNSD (data not shown). Additional EC alignments of these mutations revealed the amino acids to be conserved (fig. 1), and molecular modeling also indicates impairment in calcium binding (Brouwer et al., unpublished data). The remaining eight DFNB12 mutations are listed in table 1.
Seven different missense mutations were identified among 11 typical and atypical families with Usher syndrome type I. Eight of these families were found to have missense mutations that were compound heterozygous with a null allele. Six of the seven missense mutations in probands with Usher syndrome type ID were identified in the EC domains; the seventh missense mutation was found in the cytoplasmic domain. All Usher syndrome missense mutations are listed in table 2. None of the CDH23 missense mutations found in patients with Usher syndrome interfered with calcium-binding sites, in contrast to what was seen for the missense mutations found in patients with DFNB12; however, two of the substitutions did disrupt conserved amino acids (fig. 1).
Clinical Analyses
RNSD is defined as hearing impairment with no other identified organ-system involvement. Typical Usher syndrome type I is profound deafness, RP verified by an extinguished ERG, and absent bilateral vestibular function that first manifests as delayed (i.e., at age >18 mo) age at ambulation. Clinical investigations of both of our sample groups demonstrate a remarkable range of phenotypic variation. Clinical summaries presented in tables 4 and 5 contain only those families for which relevant audiologic, vestibular, and ophthalmologic examinations and documentation could be obtained; no anamnestic information is included, with the exception of the developmental reports of ages at independent ambulation.
Table 4.
Clinical Summary of Eight RNSD Cases with CDH23 Mutations
| Ethnicitya and Family | Mutation | Sensorineural Hearing Lossb | Results of Ophthalmologic Examinationc | Visual-FieldTyped | Age atAmbulation(mo) | VestibularFunctione |
| African American: | ||||||
| 2640 | L480Q/G1186D | Moderate to severe, progressive | ||||
| Dutch: | ||||||
| 1194 | R582Q/? | Moderate to severe, progressive | ±ERG at age 11 years, ± FE | 1 | 10 | |
| Moderate to severe | ±ERG at age 9 years, ±FE | 1 | 10 | |||
| European:d | ||||||
| 1566 | E1595K/? | Severe to profound | ||||
| Severe to profound | 12 | |||||
| 1846 | R2465W/D124G | Severe to profound, progressive | +ERG at age 11 years, ±FE | 10 | ||
| Severe to profound, progressive | +ERG at age 9 years, ±FE | 11 | ||||
| 3055 | N452S/D1846N | Severe to profound, asymmetric | ||||
| 3063 | R1060W/? | Moderate to severe | ±ERG at age 20 years, ±FE | 1 | 12 | + |
| German: | ||||||
| 1668 | D2148N/R2608H | Moderate to severe, progressive | ±ERG at age 29 years,+FE | 1 | 11 | + |
| Moderate to severe/right, severe to profound/left, progressive, asymmetric | ±ERG at age 25 years,+FE | 1 | + | |||
| Pakistani: | ||||||
| PKDF030 | A1586P/A1586P | Profound | +ERG at age 18 years | + | ||
| Profound | +ERG at age 22 years | + |
“European” denotes an ancestry of mixed European origins. Clinical findings on family 3409 will be reported elsewhere.
Verified by pure-tone audiometry.
For ERG response, − = extinguished, ± = subnormal, and+ = normal; for FE results, − = typical RP, ± = subnormal RP-like findings, and + = normal. FE = fundus examination.
1 = Normal visual fields OU (type 1). Visual-field loss was measured by Goldmann perimeter. Rates of visual-field loss are as described by Grover et al. (1997).
+ = Normal vestibular function, as determined by video electro-nystagmography with caloric testing and/or examination in a rotary chair.
Table 5.
Clinical Summary of Families with Usher Type I and CDH23 Mutations
| Ethnicity and Familya | Mutation | Sensorineural Hearing Lossb | Results of Ophthalmologic Examinationc | Visual-Field Typed | Age atAmbulation(mo) | VestibularFunctione |
| Typical Usher syndrome type I: | ||||||
| German: | ||||||
| 3150 | IVS45−9G→A/E2103X | Profound | 18 | − | ||
| Mexican, Native American: | ||||||
| 2738 | 1087delG/1087delG | Profound | −ERG at 17 years, −FE | 3 | 21 | − |
| Pakistani: | ||||||
| PKRB | IVS45−9G→A/IVS45−9G→A | Profound | −ERG at 35 years | − | ||
| Profound | −ERG at 20 years | − | ||||
| Spanish: | ||||||
| 1078 | P1206R/? | Profound | −ERG at 19 years, −FE | 4 | 18 | |
| Profound | −ERG at 11 years, −FE | 4 | 24 | |||
| 1444 | Q1496H/? | Profound | −ERG at 30 years, −FE | 4 | 18 | − |
| Profound | −ERG at 28 years, −FE | 4 | 22 | − | ||
| Swedish: | ||||||
| 1185 | IVS4+1G→A/IVS4+1G→A | Profound | −ERG at 10 years | 3 | 20 | − |
| Profound | 5 at 80 years | |||||
| 1324 | IVS4+1G→A/IVS4+1G→A | Profound | 4 | 24 | − | |
| Profound | – ERG at 21 years | 4 | 24 | − | ||
| 1291 | Q58X/IVS4+1G→A | Profound | 5 at 75 | |||
| 1197 | IVS4+1G→A/R1502X | Profound | 4 | 24 | ||
| Profound | −ERG at 15 years | 4 | 18 | |||
| 1198 | R1502X | Profound | 2 at 47 years | 36 | − | |
| 897 | 9626insC/? | Profound | −ERG at 18 years | 3 | 18 | − |
| Atypical Usher syndrome type I: | ||||||
| Dutch: | ||||||
| 1066 | IVS20+1G→A/? | Profound | −ERG at 5 years, −FE | 4 | 12 | |
| Profound | 4 | 18 | ||||
| 1071 | IVS45−9G→A/? | Profound | −ERG, −FE | 4 | 36 | |
| Severe to profound | −ERG at 23 years, −FE | 4 | ||||
| European: | ||||||
| 3308 | R1746Q/R1746Q | Profound | +ERG at 3.5 years | 19 | ± | |
| 2059 | IVS20+1G→A/T1035T | Severe, progressive, asymmetric | ±ERG at 12 years, ±FE | 14 | ||
| Moderate to severe, progressive | −ERG at 9 years, ±FE | 12 | ||||
| 216 | 193delC/3840insATGA | Profound | ±ERG at 20 years, −FE | 4 | 18 | − |
| Profound | 18 | − | ||||
| 792 | 1112delT/R1746Q | Profound | −ERG at 44 years, −FE | 4 | 26 | |
| Profound | −ERG at 38 years, −FE | 4 | 15 | |||
| R1746Q/WTf | No hearing loss | −ERG at 73 years, ±FE | 4 | |||
| 359 | 1112delT/? | Profound | ±ERG at 21 years, −FE | 24 | − | |
| Profound | ±ERG at 19 years, −FE | 4 | 18 | − | ||
| 1130 | 6155delC/? | Profound | −ERG at 14 | 4 | 10 | − |
| Profound | −ERG at 16 | 4 | 13 | |||
| 51 | 6968delC/? | Profound, progressive | −ERG at 12 years, −FE | 3 | 22 | |
| German: | ||||||
| 2790 | R1746Q/? | Severe to profound, asymmetric | ±ERG at 29 years, −FE | 4 | 13 | |
| Irish: | ||||||
| 809 | R1746Q/R1746Q | Profound | ±ERG at 27 years, ±FE | |||
| Profound | −ERG at 16 years, −FE | “Narrowed” | ||||
| Sephardic Jewish: | ||||||
| 1054g | G2744S/? | Severe | −ERG at 21 years, −FE | Severely reduced | 22 | |
| No hearing loss | ±ERG at 17 years, +FE | Moderately reduced | 36 | |||
| Spanish: | ||||||
| 3036 | IVS20+1G→A/IVS45−9G→A | Profound | −ERG at 16 years, −FE | 4 | 16 | ± |
| Profound | −ERG at 12 years, −FE | 4 | 16 | ± | ||
| Swedish: | ||||||
| 1184 | R1502X/R1502X | Profound | −ERG at 21 years | 3 | 14 | |
| Profound | −ERG at 23 years | 3 | 15 | + | ||
| 1288 | IVS4+1G→A/R1502X | Profound | 3 | 20 | + | |
| Profound | −ERG at 12 years | 4 | 24 | |||
| 980 | R2833G/? | Moderate to profound, progressive | 3 | 12 | + |
Families 220, 1056, 1285, 1289, 1321, 3051, and PKSR13a have been omitted, since adequate clinical information could not be obtained on them. “European” denotes an ancestry of mixed European origins. Clinical findings on family 3409 will be reported elsewhere.
Verified by pure-tone audiometry.
For ERG response, − = extinguished, ± = subnormal, and + = normal; for FE results, − = typical RP, ± = subnormal RP-like findings, and + = normal. FE = fundus examination.
1 = Normal visual fields (type 1); 2 = presence of a partial or complete ring scotoma (type 2); 3 = concentric central-field loss with a remaining peripheral island (type 3); 4 = marked concentric loss (type 4); 5 = partial peripheral restriction (type 5) OU. Visual-field loss was clinically measured by Goldmann perimeter. Rates of visual-field loss are as described by Grover et al. (1997).
− = Absent, ± = mild, and + = normal, as determined by video electro-nystagmography with caloric testing and/or rotary-chair examination.
Father is heterozygous for mutation and presented with RP without hearing loss.
Clinical description has been summarized by Bitoun et al. (1991).
DFNB12 phenotype
Probands with DFNB12 and their affected siblings show a less severe phenotype, compared with the probands with Usher syndrome. During their later years, they present a hearing loss ranging from moderate (40–60 dB) to profound (>90 dB) deafness. In two families, hearing loss had progressed from a mild (<40 dB) loss during childhood. In table 4, six individuals with DFNB12 present with moderate to severe hearing loss, five individuals show severe to profound hearing loss, and one consanguineous family has profound hearing loss. The age at diagnosis of sensorineural hearing loss in affected members has been documented as being 3 mo–6 years, suggesting an onset that is not congenital in all cases. Intrafamilial variation in hearing is also evident, as demonstrated in family 1668, in which serial audiograms of two affected brothers show a progressive hearing loss, with onset at age 4 years in one and at age 6 years in the other; the hearing loss progressed from moderate to severe, bilaterally, in the older brother but progressed asymmetrically, from severe to profound, just in the left ear of the younger brother. All patients with DFNB12 had been able to walk at age ⩽12 mo, and subsequent examinations have shown normal vestibular function. However, ERG, visual-field analysis, and/or fundus examination results demonstrated a subjectively mild retinal attenuation and/or slightly subnormal, or low-amplitude, ERG response; however, there is no visual-field deficit, and only one of five families tested has night blindness. The retinal pathology can be subtle, as is the case in family 1194 (table 4): although ERG results for affected members were normal, one of two pediatric ophthalmologists noticed slightly narrowed retinal arterioles with underlying granular pigment epithelium; despite the fact that vision scores in dim light were below normal and that each individual complained of night-blindness difficulties, the subnormal, albeit mild, fundus-examination results were reported as falling within normal limits. Consequently, this family had been referred to the Boys Town National Research Hospital laboratory, with a diagnosis of RNSD.
Usher syndrome type I phenotype
A remarkably diverse phenotype was observed in the patients with Usher syndrome type I. Although the mildest phenotype was consistently seen in families with DFNB12, families with Usher syndrome that carry an array of mutations demonstrated considerable variation in hearing, vision, and balance (table 5).
The majority of patients with Usher syndrome who were tested had congenital profound deafness. Six individuals with Usher syndrome type I deviated from this phenotype. Two families with Usher syndrome had severe hearing loss, two families had severe to profound hearing loss, one family had one affected individual with moderate to severe hearing loss and had a second affected individual with moderate to profound hearing loss. In these six individuals, serial audiologic data showed progression and asymmetry in two cases.
Age at ambulation was 10–36 mo in patients with Usher syndrome. Balance ability of 19 individuals with Usher syndrome type I was determined by ENG, calorics and/or examination in a rotary chair. All patients with typical Usher syndrome type I show a late age at ambulation, 18–36 mo, and no vestibular reflexes. One atypical patient, who had been able to walk at age 19 mo, presented with mild vestibular dysfunction. Balance-ability testing of five atypical cases, who had been able to walk at age 10–15 mo, demonstrated normal vestibular function.
Of 37 patients with Usher syndrome type I on whom ERG tests and/or fundus examinations were performed, 26 showed extinguished, or flat, ERG amplitudes, 19 had typical RP manifestations in the fundus, and 8 had subnormal ERG responses. Subnormal RP-like findings in the retina were found in four patients. One proband had a normal ERG response; the fundus examination of another proband with subnormal ERG amplitudes indicated a normal retina.
A collection of visual-field tests were examined, and rates of progression were quantified and categorized according to a classification system described by Grover et al. (1997). For those cases in which visual fields could not be determined, verbal descriptions of the testing results are given in table 5. Of the 34 individuals for whom visual-field data were available, 20 show a marked concentric loss of ⩽10° in both eyes, or oculi unitas (OU) (type 4). Eight patients presented a concentric central-field loss with a temporal island, or scotoma OU (type 3). Partial peripheral restrictions OU (type 5) were identified in two Swedish individuals with Usher syndrome type I, at ages 75 and 80 years. Another Swedish proband, at age 47 years, had a midperipheral complete-ring scotoma OU (type 2) that had not progressed since age 19 years.
Clinical analyses of cases with the R1746Q missense mutation show a variable retinal phenotype similar to that described by Bolz et al. (2001). An American family, 3308, homozygous for the R1746Q mutation originally was referred to our laboratory through the RNSD research project. This 5-year-old boy presented with congenital profound deafness but had been able to walk at age 19 mo. Clinical investigation and examination in a rotary chair revealed a mildly hypoactive vestibular response. Although the results of an ERG at age 3.5 years were normal, follow-up examinations are being recommended, to determine whether a diagnosis of Usher syndrome type I is valid. An Irish family, 809, also homozygous for R1746Q, presented with mild late-onset RP. Ophthalmologic examinations of members of family 792, who were compound heterozygous for R1746Q and 1112delT, revealed a severe course of RP. Interestingly, late-onset RP was seen in the 73-year-old father, who was heterozygous for the R1746Q allele. Ophthalmologic examinations of the father revealed a flat ERG, a nonpigmentary and diffuse atrophy of the fundus, and visual fields of <5°. The father did not present with hearing loss. No other individuals in this family who were heterozygous for CDH23 reported visual problems or any other carrier effect; however, the father has been tested only for mutations in exon 40 of CDH23, and the possibility exists that (a) another CDH23 mutation exists or (b) mutations in another RP gene are responsible for his condition. Family 2790, heterozygous for R1746Q, presented atypical Usher features: the hearing loss is asymmetric and severe to profound, and results of ERG tests were subnormal at age 29 years, despite a documented 10° visual-field result. Fundus examination revealed typical RP, and, although vestibular testing was not completed, the affected individual in this family reportedly had been able to walk at age 13 mo.
The following clinical findings for families 980 and 2059 underscore the fact that the severity of the phenotypes can be highly variable and may overlap with Usher syndrome types II and III. Swedish family 980 was referred to our molecular-testing lab as having Usher syndrome type II. The 49-year-old singleton proband presented with a sloping moderate to profound hearing loss and normal developmental milestones and vestibular function. The serial visual data for this individual show a late onset and slow progression of RP. On the basis of the variable retinal phenotype associated with mutations of CDH23, these retinal findings prompted us to include this proband in our analysis, and, indeed, a heterozygous R2833G missense mutation was identified; however, the second mutation has not yet been identified. Family 2059 was originally referred to our laboratory as having RNSD. One of the two affected siblings had a severe asymmetrical hearing loss at age 14 years, and the other had a moderate to severe hearing loss at age 8 years; both showed progressive hearing loss. Subsequent testing for mutations in GJB2 was negative. Genotyping analysis was consistent with linkage to the 10q region, and therefore our testing included this family, as putatively having DFNB12. While the mutation screening was in progress, visual exams revealed extinguished and subnormal ERG responses, as well as abnormal fundus-examination findings, in each affected individual. Because of reported normal developmental milestones and normal vestibular function, the diagnosis was changed to Usher syndrome type II, but testing for mutations in CDH23 continued. Family 2059 simultaneously underwent preliminary analysis for mutations in USH2A, until two splice-site mutations, IVS20+1G→A and 3105A→C (T1035T), were identified in CDH23.
The remarkable phenotype of family 1054 has been reported elsewhere (Bitoun et al. 1991). Two affected children born of Sephardic Jewish first cousins have a heterozygous G2744S mutation and presented with retinal degeneration. A severe hearing loss and an extinguished ERG, typical of RP, were identified in the older proband; the younger brother has a subnormal ERG response, with normal fundus-examination findings and normal hearing. The lack of a homozygous CDH23 mutation in this consanguineous family, the identification of only one missense mutation, G2744S, and the additional clinical findings of developmental delay, dysmorphism, and severe ataxia suggest either a non–Usher syndrome diagnosis or the existence of another syndrome; therefore, until a second mutation is been identified, we are hesitant to claim that this family has Usher syndrome type ID.
Discussion
In 27 (56.3%) of 48 multiplex families with Usher syndrome and in 6 (30%) of 20 multiplex families with RNSD, we have identified CDH23 mutations that link to chromosome 10q21.1-q22.1, which includes the USH1D, DFNB12, USH1F, and DFNB23 loci (Chaib et al. 1996; Wayne et al. 1996, 1997; Hereditary Hearing Loss Homepage). Mutations were seen in six sporadic cases of Usher syndrome type I and in five sporadic cases of RNSD. Forty-one families with Usher syndrome type I were part of a previous study (Astuto et al. 2000) in which results of linkage and heterogeneity analyses suggested that the chromosome 10q–associated Usher syndrome type I subtype was the second most common Usher syndrome type I subtype. Our CDH23 mutation analysis has corroborated those findings. On the basis of the proportion of CDH23 mutations among families with RNSD, it should be noted that the sample is refined, since cases with mutations of GJB2, SLC26A4, or OTOF have been excluded. Extrapolating from an original population of 157 families with RNSD, we estimate that 5% of RNSD is caused by mutations in CDH23. Technical error or hidden mutations in gene regions that we did not explore—regions such as introns and regulatory regions—may account for the individuals who carry single CDH23 heterozygous mutations. The possibility also exists that these individuals are CDH23 carriers and that the causative mutation is at a different locus, such as USH1F.
The majority of CDH23 mutations occur within the CDH23-gene region encoding the EC domain. Mutations were identified in all domains except the signal sequence. Only one mutation in the cytoplasmic domain had been previously identified (Bork et al. 2001); the remainder of the mutations occur in the EC repeats. We have identified two additional mutations in the cytoplasmic domain—R3175H and 9626insC—both associated with an Usher syndrome type I phenotype.
The distribution and location of the missense mutations identified in the present study illustrate that the mutations do not cluster within one domain. However, comparison of the locations between phenotypic groups show that 6 of 12 DFNB12 missense mutations change the highly conserved peptide sequences LDRE, DXNDN, and DXD, which are responsible for calcium-binding and rigidification (fig. 1). Molecular modeling of five of these amino acid substitutions strongly suggests that there is a defect in Ca2+ ion binding (Brouwer et al., unpublished data). No Usher syndrome type ID missense mutations change the conserved calcium-binding sequences, and only two of six EC-domain mutations substitute a conserved amino acid (fig. 1).
Conclusions drawn from the present study and from other groups (Bolz et al. 2001; Bork et al. 2001; Brouwer et al., unpublished data) show that missense mutations produce a less severe phenotype, both in families with Usher syndrome type I and in families with RNSD, whereas nonsense, splice-site, and frameshift mutations produce a more severe Usher syndrome type I phenotype (table 6). In our mutation data, a reduced frequency of null mutations of CDH23 is seen as the phenotype becomes milder. Approximately 88%, 67%, and 0% of null mutations of CDH23 are found in typical Usher syndrome type I, in atypical Usher syndrome type I, and in probands with RNSD, respectively. However, the genotype does not always predict the clinical phenotype, which varies both within and between families carrying the same CDH23 mutations, implying the existence of other genetic and/or environmental factors that influence phenotype.
Table 6.
Summary of CDH23 Mutations Identified in Present Study and in Patients Who Elsewhere Have Been Reported as Being Affected with Either RNSD or Usher Type I
| Mutation | Domain | Phenotypea | Reference |
| Q58X | EC1 | Usher syndrome type I | Present study |
| 193delC | EC1 | Atypical Usher syndrome | Present study |
| IVS4+1G→A | EC1 | Usher syndrome type I, atypical Usher syndrome | Present study |
| D124G | EC1 | RNSD | Present study |
| 1087delG | EC4 | Usher syndrome type I | Present study |
| 1112delT | EC4 | Atypical Usher syndrome | Present study |
| N452S | EC4 | RNSD | Present study |
| L480Q | EC5 | RNSD | Present study |
| A484P | EC5 | Usher syndrome type Ib | Present study |
| R582Q | EC6 | RNSD | Present study |
| IVS20+1G→A | EC7 | Atypical Usher syndrome | Present study |
| D990N | EC9 | RNSD | Bork et al. (2001) |
| T1035T | EC10 | Atypical Usher syndrome | Present study |
| R1060W | EC10 | RNSD | Present study |
| G1186D | EC11 | RNSD | Present study |
| P1206R | EC11 | Usher syndrome type I | Present study |
| T1209A | EC11/EC12c | Usher syndrome type Ib | Present study |
| 3840insATGA | EC12 | Atypical Usher syndrome | Present study |
| ÄM1281 | EC12 | Usher syndrome type I | Bolz et al. (2001) |
| Q1294X | EC12 | Usher syndrome type I | Bork et al. (2001) |
| D1341N | EC13 | RNSD | Brouwer et al. (unpublished data) |
| Q1496H | EC14 | Usher syndrome type I | Bolz et al. (2001) |
| R1502X | EC14 | Usher syndrome type I, atypical Usher syndrome | Present study |
| A1586P | EC15 | RNSD | Present study |
| E1595K | EC15 | RNSD | Present study |
| R1746Q | EC16/EC17c | Atypical Usher syndrome | Bolz et al. (2001); present study |
| D1846N | EC17 | RNSD | Present study |
| T1904T | EC18 | Usher syndrome type I | von Brederlow et al. (2002) |
| IVS45+1G→A | EC19 | Usher syndrome type I | Bork et al. (2001) |
| IVS45−9G→A | EC19 | Usher syndrome type I, atypical Usher syndrome | von Brederlow et al. (2002); present study |
| D2045N | EC19 | RNSD | Bork et al. (2001) |
| 6155delC | EC19 | Atypical Usher syndrome | Present study |
| E2103X | EC20 | Usher syndrome type Ib | Present study |
| R2107X | EC20 | Usher syndrome type I | Bork et al. (2001) |
| D2148N | EC20 | RNSD | Brouwer et al. (unpublished data) |
| D2202N | EC21 | RNSD | Bork et al. (2001) |
| 6933delT | EC21 | Usher syndrome type I | von Brederlow et al. (2002) |
| 6968delC | EC22 | Atypical Usher syndrome | Present study |
| IVS51+5G→A | EC23 | Usher syndrome type I | Bolz et al. (2001); von Brederlow et al. (2002) |
| R2465W | EC23 | RNSD | Present study |
| S2517G | EC23/EC24c | Usher syndrome type Ib | Present study |
| R2608H | EC24 | RNSD | Present study |
| E2624E | EC25 | Usher syndrome type Ib | Present study |
| G2744S | EC26 | Atypical Usher | Present study |
| R2833G | EC26 | Atypical Usher syndrome | Present study |
| I2950N | EC27 | RNSD | Bork et al. (2001) |
| R2956C | EC27 | RNSD | Bork et al. (2001) |
| P3059T | EC27-TMd | RNSD | Bork et al. (2001) |
| IVS66+1G→A | CYTO | Atypical Usher syndrome | Bork et al. (2001) |
| R3175H | CYTO | Usher syndrome type Ib | Present study |
| 9626insC | CYTO | Usher syndrome type I | Present study |
The lack of a distinct Usher syndrome type ID phenotype may complicate mutation analysis of all patients diagnosed with Usher syndrome types I–III. In the present study, we have identified a CDH23 mutation in an individual diagnosed with Usher syndrome type II. We therefore plan to test a series of cases with an Usher syndrome type II phenotype, to determine the frequency of CDH23 mutations within this diagnostic category. Identification of progressive hearing loss and variable balance dysfunction in patients excluded from harboring identified Usher syndrome type III mutations are also candidates for CDH23 mutation screening. Efficient mutation testing will be difficult because of the complexity of the CDH23-gene structure. However, some mutations of CDH23 occur with high frequency in certain populations. For example, R1502X and the IVS4+1G→A should be the first two CDH23 mutations screened in patients with Usher syndrome who have a Swedish background.
The syndromic and nonsyndromic association found in CDH23 alleles is not exclusive to this gene. Two other Usher syndrome type I subtypes are allelic with DFNB loci: DFNB2 is associated with USH1B (Liu et al. 1997; Weil et al. 1997), and DFNB18 is associated with USH1C (Ahmed et al., in press). Understanding the variations of pathophysiology underlying mutations of CDH23, as well as the other Usher syndrome genes, is paramount if effective treatments are to be developed.
Acknowledgments
We would like to thank all the patients and families, for their participation in the Usher syndrome and recessive nonsyndromic-deafness research projects, and the physicians, including Drs. Martini and Iannoccone, for referring families to us. We would like to thank Dr. Rocky Young at Texas Tech University, for ERG testing of two patients with mutations in DFNB12, and Dr. Ed Cohn, for his advice and contribution. We also would like to recognize Mehdi Sadeghi, for helping to collect Swedish clinical information; Zubair Ahmed, for genotyping the Pakistani families; and Edward Wilcox, for his contributions. This work was supported in part by National Institutes of Health grant P01 DC01813, National Institute on Deafness and Other Communication Disorders grants R01 DC00677-07, and a Foundation Fighting Blindness grant (all to W.J.K.); National Institutes of Health grant P60 DC00982 (to L.M.A.); the University Grants Commission, Islamabad, Pakistan (support to S.K. and S.R.); National Institute on Deafness and Other Communication Disorders/National Institutes of Health intramural funds 1Z01 DC000035-05 and 1Z01 DC000039-05 (both to T.B.F.); a Research to Prevent Blindness Clinician-Scientist Award (to J.R.H.); and the Mgr Van Overbeek Foundation, De Drie Lichten, and De Gelderse Blinden Verneniging (support to C.W.R.J.C. and H.K.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project Splice Site Prediction by Neural Network, http://www.fruitfly.org/seq_tools/splice.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human CDH23 [accession number AF312024])
- Hereditary Hearing Loss Homepage, http://dnalab-www.uia.ac.be/dnalab/hhh/index.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for USH1D [MIM 601067], DFNB12 [MIM 601386], and CDH23 [MIM 605516])
- Sequence Manipulation Suite, The, http://www.ualberta.ca/~stothard/javascript/
References
- Ahmed ZA, Smith TN, Riazuddin S, Makishima T, Ghosh M, Bokhari S, Menon PSN, Deshmukh D, Griffith AJ, Riazuddin S, Friedman TB, Wilcox ER. Nonsyndromic recessive deafness DFNB18 and Usher syndrome type 1C are allelic mutations of USH1C. Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Riazuddin S, Wilcox ER (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 69:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, Schwartz S, Lee C, Morton CC, Mullins RF, Ramesh A, Van Camp G, Hagemen GS, Woychik RP, Smith RJ (2001) Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet 10:1709–1718 [DOI] [PubMed] [Google Scholar]
- Astuto LM, Weston MD, Carney CA, Hoover DM, Cremers CW, Wagenaar M, Moller C, Smith RJ, Pieke-Dahl S, Greenberg J, Ramesar R, Jacobson SG, Ayuso C, Heckenlively JR, Tamayo M, Gorin MB, Reardon W, Kimberling WJ (2000) Genetic heterogeneity of Usher syndrome: analysis of 151 families with Usher type I. Am J Hum Genet 67:1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O'Brien RE, Farndon PA, Sowden J, Liu XZ, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B (2000) A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet 26:56–60 [DOI] [PubMed] [Google Scholar]
- Bitoun P, Bandini D, Rigaudiere F (1991) A hereditary syndrome with retinopathy and ataxia or deafness in two consanguineous brothers. Ophthalmic Paediatr Genet 12:149–152 [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-S, Vila MC, Molina OP, Gal A, Kubisch C (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27:108–112 [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, et al (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet 68:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman JA, Vernon M, Shaver KA (1983) Usher syndrome: definition and estimate of prevalence from two high-risk populations. J Chronic Dis 36:595–603 [DOI] [PubMed] [Google Scholar]
- Chaib H, Place C, Salem N, Dode C, Chardenoux S, Weissenbach J, El Zir E, Loiselet J, Petit C (1996) Mapping of DFNB12, a gene for a non-syndromal autosomal recessive deafness, to chromosome 10q21-22. Hum Mol Genet 5:1061–1064 [DOI] [PubMed] [Google Scholar]
- Grondahl J (1987) Estimation of prognosis and prevalence of retinitis pigmentosa and Usher syndrome in Norway. Clin Genet 31:255–264 [DOI] [PubMed] [Google Scholar]
- Grover S, Fishman GA, Anderson RJ, Alexander KR, Derlacki DJ (1997) Rate of visual field loss in retinitis pigmentosa. Ophthalmology 104:460–465 [DOI] [PubMed] [Google Scholar]
- Kelley PM, Harris DJ, Comer BC, Askew JW, Fowler T, Smith SD, Kimberling WJ (1998) Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am J Hum Genet 62:792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberling WJ, Moller C (1995) Clinical and molecular genetics of Usher syndrome. J Am Acad Audiol 6:63–72 [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJTV, Steel KP, Brown SDM (1997) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 16:188–190 [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F (2000) Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol 299:551–572 [DOI] [PubMed] [Google Scholar]
- Nuutila A (1970) Dystrophia retinae pigmentosa—dysacusis syndrome (DRD): a study of the Usher or Hallgren syndrome. J Genet Hum 18:57–88 [PubMed] [Google Scholar]
- Tamayo ML, Bernal JE, Tamayo GE, Frias JL, Alvira G, Vergara O, Rodriguez V, Uribe JI, Silva JC (1991) Usher syndrome: results of a screening program in Colombia. Clin Genet 40:304–311 [DOI] [PubMed] [Google Scholar]
- Vernon M (1969) Usher's syndrome—deafness and progressive blindness: clinical cases, prevention, theory and literature survey. J Chronic Dis 22:133–151 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies usher syndrome type 1C. Nat Genet 26:51–55 [DOI] [PubMed] [Google Scholar]
- von Brederlow B, Bolz H, Janecke A, La O Cabrera A, Rudolph G, Lorenz B, Schwinger E, Gal A (2002) Identification and in vitro expression of novel CDH23 mutations in patients with Usher syndrome type 1D. Hum Mutat 19:268–273 [DOI] [PubMed] [Google Scholar]
- Wayne S, Der K, V, Schloss M, Polomeno R, Scott DA, Hejtmancik JF, Sheffield VC, Smith RJ (1996) Localization of the Usher syndrome type ID gene (Ush1D) to chromosome 10. Hum Mol Genet 5:1689–16 [DOI] [PubMed] [Google Scholar]
- Wayne S, Lowry RB, McLeod DR, Knaus R, Farr C, Smith RJH (1997) Localization of the Usher syndrome type IF (Ush1F) to chromosome 10. Am J Hum Genet Suppl 61:A300 [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 [DOI] [PubMed] [Google Scholar]
- Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 16:191–193 [DOI] [PubMed] [Google Scholar]