Abstract
Contamination is a critical issue in high-throughput metagenomic studies, yet progress towards a comprehensive solution has been limited. We present SourceTracker, a Bayesian approach to estimating the proportion of a novel community that comes from a set of source environments. We apply SourceTracker to new microbial surveys from neonatal intensive care units (NICUs), offices, and molecular biology laboratories, and provide a database of known contaminants for future testing.
Advances in sequencing technology and informatics, including the MIxS (Minimum Information about any (x) Sequence) metadata standards, are producing an exponential increase in data acquisition and integration. These advances are revolutionizing our understanding of the roles microbes play in health and disease, biogeochemical cycling, etc. Although considerable attention has been paid to reducing sources of error from PCR1 and sequencing2, sample contamination has been relatively unstudied. Preparing contaminant-free DNA is challenging, and the sensitivity of PCR and whole-genome amplification methods means that even trace contamination can become a serious issue3. Ideally, computational methods could identify both the source and quantity of contamination, and could help prevent future instances. Furthermore, accurately estimating the proportion of contamination from a given source environment would have far-reaching applications in source tracking for forensics, pollution, public health, etc.
We have developed SourceTracker, a Bayesian approach to identifying sources and proportions of contamination in marker-gene and functional metagenomics studies. Our approach models contamination as the mixture of entire source communities into a sink community, where the mixing proportions are unknown. Previous approaches to microbial source tracking (MST) have focused on detection of fecal contamination in water4–6, limited to detection of predetermined indicator species and custom-tailored biomarkers from source communities. One notable exception7 uses community structure to measure similarity between sink samples and potential source environments. Other prior work uses data-driven identification of indicator species, but lacks a probabilistic framework8. SourceTracker’s distinguishing features are its direct estimation of source proportions, and its Bayesian modeling of uncertainty about known and unknown source environments.
We also present barcoded pyrosequencing datasets of bacterial 16S ribosomal RNA gene sequences covering surface contamination in office buildings, hospitals, and research labs, and reagents used for metagenomics studies (Supplementary Table 1; data collection described in Online Methods). Using SourceTracker, we compared these data to published datasets from environments likely to be sources of indoor contaminants, namely human skin, oral cavities, and feces9, and temperate soils10 (Supplementary Table 2). We treated these natural environments as sources contributing organisms to the indoor sink environments through natural migration (as with office samples) or inadvertent contamination (as with no-template PCR controls) (schematic in Supplementary Fig. 1).
Although qualitative assessment of source and sink similarities can be performed by visualizing UniFrac distances11 (Supplementary Fig. 2), or taxon relative abundance (Supplementary Fig. 3), they cannot tell us the proportion of each sink sample (e.g., a cotton swab) comprising taxa from a known source environment (e.g., soil). The problem would be trivial if source and sink environments shared no taxa, but usually some taxa are shared. Source tracking methods must therefore leverage potentially useful information contained in the abundance of species with low or moderate source environment endemicity.
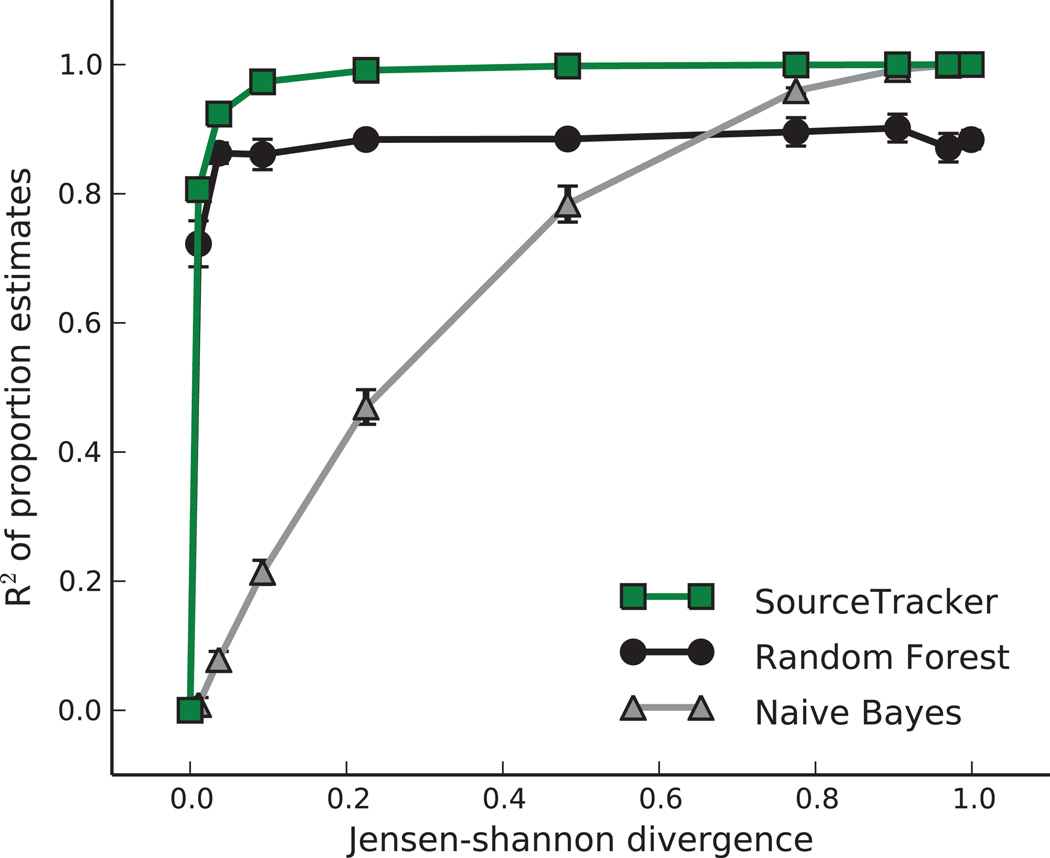
Previous work uses probabilistic indicator species for naïve Bayes estimation6. Although naïve Bayes actually estimates the probability that each source generated the entire sink sample, these probabilities can sometimes act as proxies for the proportions of the sink contributed by each source. We compared the accuracies of naïve Bayes and SourceTracker as we varied the distributions of taxa in two simulated source environments from perfectly identical to perfectly non-overlapping (Fig. 1). Naïve Bayes was accurate when disambiguation is easy, but inaccurate elsewhere. SourceTracker performed well even when disambiguation is difficult (R2 ≥ .8, Jensen-Shannon divergence ≥ 0.05; Fig. 1). We also evaluated the accuracy of the random forests (RF) classifier used in previous source-tracking work7. Like naïve Bayes, RF estimates the probability that the entire test sample came from a single source, but these probabilities are often reasonable estimates of the mixing proportions for source tracking purposes. RF generally performed better than naïve Bayes, but worse than SourceTracker. SourceTracker outperforms these methods because it allows uncertainty in the source and sink distributions, and because it explicitly models a sink sample as a mixture of sources.
Figure 1. Comparison of SourceTracker and alternative models.
Three models were used to estimate the proportions of two source environments in a set of simulated samples, as the degree of overlap between the environments was varied from a Jensen-Shannon divergence (JSD) of 0 (completely identical, and thus impossible to disambiguate), to a JSD of 1 (completely non-overlapping, and thus trivial to disambiguate). The coefficients of determination (R2) of the estimated proportions are plotted. Each point represents the mean R2 for three trials of 100 samples each; error bars show s.e.m. (n = 3).
The Bayesian approach requires consideration of all possible assignments of the test sample sequences to the different source environments, but direct exploration is intractable. Fortunately, we can explore this joint distribution using Gibbs sampling, a technique widely used in the exploration of complex posterior distributions in applications like topic modeling12. Community-wide source tracking is analogous to inferring the mixing proportions of conversation topics in a test document, except that the source environment distributions over taxa (topic distributions over words) are known from the training data, and each test sample may contain taxa from an unknown, uncharacterized source. The application of Gibbs sampling to topic modeling has been discussed in detail previously13.
SourceTracker considers each sink sample x as a set of n sequences mapped to taxa, where each sequence can be assigned to any one of the source environments v ∈ {1..V}, including an Unknown source. These assignments are treated as hidden variables, denoted zi=1..n ∈ {1..V}. To perform Gibbs sampling, we initialize z with random source environment assignments, and then iteratively re-assign each sequence based on the conditional distribution:
where mtv is the number of training sequences from taxon t in environment v, nv is the number of test sequences currently assigned to environment v, and ¬i excludes the ith sequence. The first fraction gives the posterior distribution over taxa in the source environment; the second gives the posterior distribution over source environments in the test sample. Both are Dirichlet distributions, and Gibbs sampling allows us to integrate over their uncertainty. The Dirichlet parameters, α and β, act as imaginary prior counts that smooth the distributions for low-coverage source and sink samples, respectively. They also allow Unknown source assignments to accumulate when part of a sink sample is unlike any of the known sources. By inferring source proportions for multiple sink samples simultaneously, we can allow them to share an Unknown source. We could also include several Unknown sources. Full details and an overview of Gibbs sampling are provided in our Online Methods.
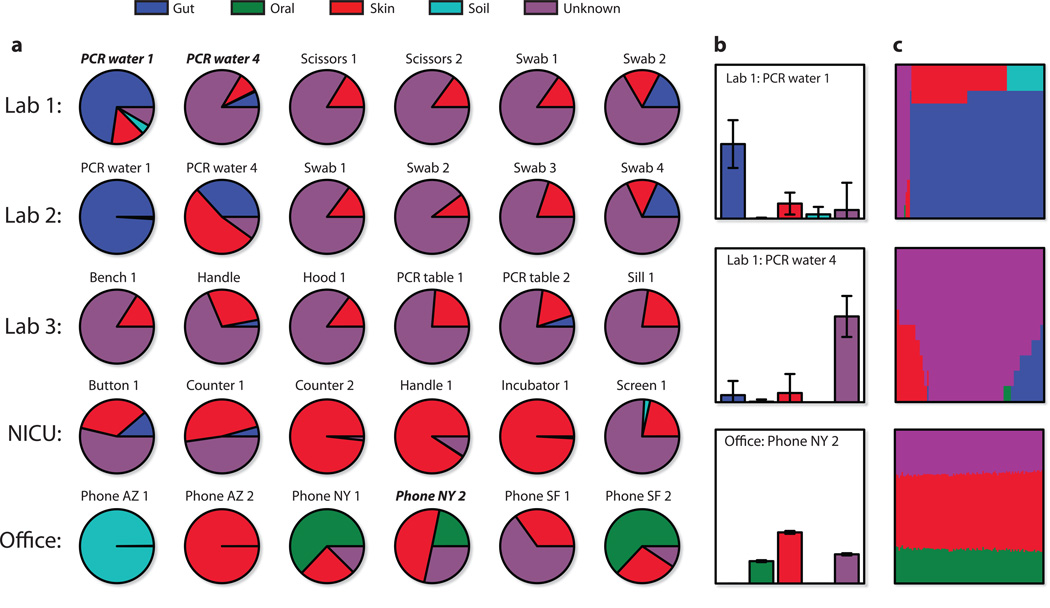
For each of our indoor sink environments, we used SourceTracker to estimate the proportion of bacteria from Gut, Oral, Skin, Soil, and Unknown (i.e., one or more sources absent from the training data) (Fig. 2 and Supplementary Figs. 4 and 5). In general, wet-lab surface communities tended to be composed mainly of bacteria from Skin and Unknown, with the exception of PCR water, which was generally more similar to Gut. NICU and office communities were dominated by Skin bacteria, except for two Arizona samples dominated by Soil bacteria and several telephone samples dominated by Oral bacteria. From these results we can also determine the most common contaminating taxa (Fig. 3).
Figure 2. SourceTracker proportion estimates for a subset of sink samples.
Source environment proportions were estimated using SourceTracker and 45 training samples from each source environment. (a) Pie charts of the mean proportions for 100 draws from Gibbs sampling. (b) Bar charts for three samples including standard deviations of the proportion estimates. (c) Direct visualization of 100 Gibbs draws for the samples in (b); each column shows the mixture from one draw, with columns sorted by the most prevalent source. The first sample, Lab 1: PCR water 1, shows several possible mixtures: all Unknown; Gut and Skin (most common); and Gut and Soil. The second sample shows poor disambiguation between Gut, Skin, and Unknown. Most mixtures were stable like the third sample; the first two were chosen for demonstrative purposes.
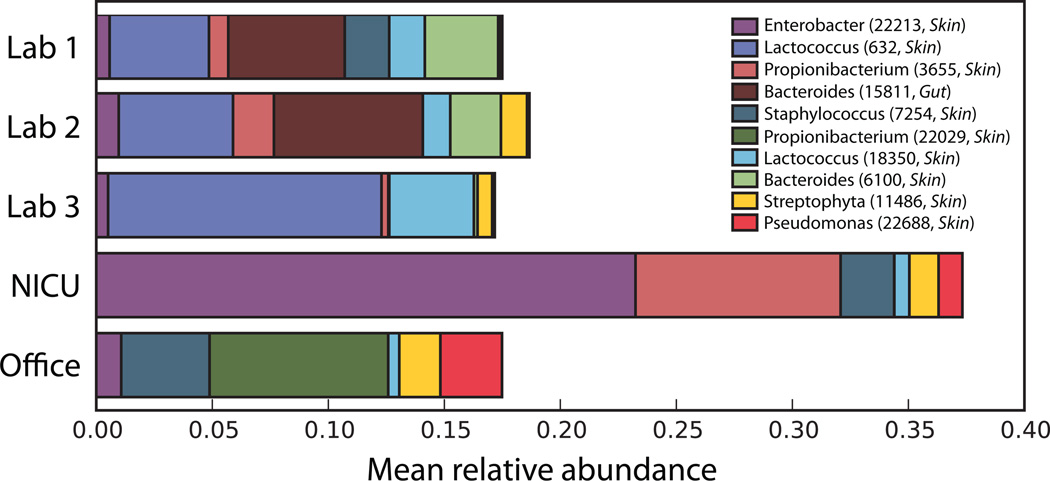
Figure 3. Relative abundance of common contaminating operational taxonomic units (OTUs).
For all sink sequences assigned to a known source environment (Gut, Oral, Skin, or Soil) by SourceTracker, these ten OTUs had the highest average relative abundance across sink environments. Note that the OTU classified as Enterobacter, a lineage commonly seen in the gut, was more prevalent in the Skin training samples than the Gut training samples.
For low-coverage sink samples, or when source environments lack a “core” set of taxa, SourceTracker will report high variability in the proportion estimates (Fig. 2). In some data sets, variation within each source environment (the “non-core” taxa) might be accounted for by using phylogenetic information, by automatically identifying distinct niches within the broader source environment, by modeling postmixture population dynamics, or by modeling potential biases inherent in the DNA extraction procedures used; these are important directions for future work. SourceTracker also assumes that an environment cannot be both a source and a sink, and we recommend research into bi-directional models.
SourceTracker can also be used to detect low-level contamination, with sensitivity adjusted by the prior parameter β. For simulations with 1% and 5% contamination, SourceTracker achieved nearly perfect specificity for a wide range of sensitivities, demonstrating that it is not restricted to low-biomass sink environments where contamination rates are likely to be higher (area under the receiver operating characteristic curve = .971 for 1%, .989 for 5%; Supplementary Fig. 6).
Based on our results, simple analytical steps can be suggested for tracking sources and assessing contamination in newly acquired data sets. Although source-tracking estimates are limited by the comprehensiveness of the source environments used for training, large-scale projects such as the Earth Microbiome Project will dramatically expand the availability of such resources. SourceTracker is applicable not only to source tracking and forensic analysis in a wide variety of microbial community surveys (e.g., “where did this biofilm come from?”), but also to shotgun metagenomics and other population genetics data. We have made our implementation of SourceTracker available as an R package (http://sourcetracker.sf.net), and we advocate automated tests of deposited data to screen samples that may be contaminated prior to deposition.
ONLINE METHODS
Data collection
We collected the Office samples from surfaces in 54 offices in three different office buildings (18 per building) located in New York, NY; San Francisco, CA; and Tucson, AZ, respectively (Hewitt, K.M., Gerba, C.P., Maxwell, S.L. & S.T.K., unpub. data). In each office, we sampled the same two surfaces, phone and chair, by swabbing approximately 13 cm2 with dual tip sterile cotton swabs (BBL CultureSwab™, catalog # 220135). Phone and chairs had already been determined by culture-based methods to be the most contaminated surfaces in these offices (unpub. data). We also collected samples from surfaces in two different large Level three Neonatal Intensive Care Units (NICUs) in San Diego, CA using the same methods. After sampling, we stored swabs in sterile-labeled tubes, placed them on ice and shipped them overnight, or drove them directly to the lab for DNA extraction.
For the Lab 1 and Lab 2 data sets, we cut sterile nylon-flocked swabs (Copan) and swabs of sterile scissors into MoBio 0.7 mm garnet bead tubes (Mo Bio Laboratories) using autoclaved and flamed scissors in a biosafety cabinet, placed them at −80°C within 1 hour, and stored them for <1 week prior to DNA extraction.
For the Lab 3 data set, we used sterile nylon-flocked swabs (Copan) to sample indoor surfaces including desktops, lab benches, windowsills, a keyboard, and a door handle over a three-month period from January-March 2010 in Philadelphia, PA. We cut swabs into MoBio 0.7 mm garnet bead tubes (Mo Bio Laboratories) using autoclaved and flamed scissors in a biosafety cabinet, placed them at −80°C within 1 hour, and stored them for <1 week prior to DNA extraction.
DNA extraction, PCR, and pyrosequencing
For the Office and NICU samples, we removed the cotton from the swab using a flame-sterilized razor blade and deposited the cotton threads into a lysozyme reaction mixture. The reaction mixture had a total volume of 200 µl and included the following final concentration: 20 M Tris, 2 mM EDTA (pH 8.0), 1.2% P40 detergent, 20 mg ml-1 lysozyme, and 0.2 µm filtered sterile water (Sigma Chemical Co.). We incubated the samples in a 37°C water bath for thirty minutes. Next, we added Proteinase K (DNeasy Tissue Kit, Qiagen Corporation) and AL Buffer (DNeasy Tissue Kit, Qiagen Corporation) to the tubes and gently mixed them. We incubated the samples in a 70°C water bath for 10 min. We subjected all samples to purification using the DNeasy Tissue Kit. Following extraction, we quantified the DNA using a NanoDrop ND-1000 Spectrophotometer (NanoDropTechnologies). PCR barcoded primers and conditions were previously described14. PCR purification, dilutions and pyrosequencing (FLX) were all conducted by the core facility at the University of South Carolina (Environmental Genomics Core Facility).
For the Lab 1 and Lab 2 data sets, we extracted genomic DNA from swabs using the QIAamp DNA Stool Minikit (Qiagen) with the following modifications. We added 1500 ul of ASL buffer and 5mM DTT to the nylon tips of frozen swabs. We beadbeat tubes with BioSpec Products Inc. Minibeadbeater-16 for 1 min. and incubated at 95 °C for 10 min. We performed the remaining steps as per manufacturer protocol. We performed PCR amplification of 16S rRNA genes using the V1V2 primers and conditions described in Wu et al.15 in duplicate. We quantified purified amplicons using Quant-iT PicoGreen kit (Invitrogen) and pooled them in equimolar ratios. We also performed PCR on molecular biology grade water (Sigma) and included it in the pool. We carried out pyrosequencing using primer A and the Titanium amplicon kit on a 454 Life Sciences Genome Sequencer FLX instrument (Roche).
For the Lab 3 data set, we extracted genomic DNA from swabs using the same extraction kit and technique as Lab 1 and 2 above. We performed PCR amplification of 16S rRNA genes using the V1V2 primers and conditions described in Wu et al., 2010. We quantified purified amplicons using Quant-iT PicoGreen kit (Invitrogen) and pooled them in equimolar ratios. We also performed PCR on molecular biology grade water (Sigma) and included it in the pool. We carried out pyrosequencing using primer A and the Titanium amplicon kit on a 454 Life Sciences Genome Sequencer FLX instrument (Roche).
We provide the DNA barcodes and primers for all samples collected in a supplementary table (Supplementary Table 1).
Combined preprocessing of contamination data sets
We processed the DNA sequence data for all source and sink samples in combination using the QIIME pipeline16. In order to avoid bias, we selected subsets of the same size (45 samples) from each of the four source environments (Supplementary Table 2). We sequenced samples in multiplex using error-correcting nucleotide barcodes, and we used QIIME to demultiplex the samples and perform quality filtering. We then used flowgram clustering17 to remove sequencing noise. We clustered similar sequences (≥ 97% similarity) into OTUs with uclust18, and assigned taxonomic identity to each OTU using the Ribosomal Database Project’s taxonomy assignment tool19. We aligned representative sequences from each OTU against the greengenes reference ‘core set’ of 16S rRNA gene sequences (http://greengenes.lbl.gov). We then removed likely chimeric PCR products using Chimera Slayer20. We used the remaining aligned sequences to construct a phylogeny relating the sequences, via FastTree21.
Identification and removal of Chimeras
As noted above, we removed likely chimeric PCR products using Chimera Slayer20. Note that we first aligned representative sequences from each OTU to the greengenes core set. Any OTU not aligning to the greengenes core set at > 75% identity to the nearest BLAST hit in the core set was discarded. These discarded sequences may contain chimeras, as well as other artifacts. However, once completed we also used Chimera Slayer to screen the resulting sequences for chimeras. The number of chimeras removed were: 58 sequences from Lab 1 samples (4%), 105 from Lab 2 (4%), 4208 from Lab 3 (5%), 422 from Office (0.3%), and 1365 from NICU (0.6%).
Principal Coordinates Plots
After randomly selecting 500 sequence reads per sample and dropping low-coverage samples to control for sequencing effort, we used UniFrac11 to measure the phylogenetic dissimilarity of all samples and performed Principal Coordinates Analysis (PCoA) on the matrix of unweighted UniFrac distances using QIIME16.
Gibbs sampling overview
To begin the Gibbs sampling procedure we assign each sequence to a random source environment. We assume that these assignments are correct (even though they are random), and tally the current proportions of the source environments in the test sample. We then remove one sequence from the tallies and re-select its source environment assignment, where the probability of selecting each source is proportional to the probability of observing that sequence’s taxon in that source, multiplied by the current estimate of the probability of observing that source in the test sample. After the re-assignment, we update the tally for the selected source environment, and repeat the process on another randomly selected sequence. After we have re-assigned all of the sequences many times in this manner, each set of assignments we observe is a representative draw from the distribution over all possible sequence-source assignments. To estimate the variability of this distribution, we can repeat the procedure as many times as we like, and we can report summary statistics for the mixing proportions or even visualize their distributions directly (Fig. 2c).
Dirichlet prior parameters
A larger value of β causes a smoother posterior distribution over environments in the sink sample. This is valuable when we want to avoid overfitting in sink samples with few sequences. By assigning different relative values of β to each environment, we can also incorporate prior knowledge about the expected distribution of source environments in our sink samples. α represents a prior count of each taxon in each source environment. This allows taxa that are unlikely under the known source environment distributions to accumulate in an Unknown environment during the sampling procedure. In order to simplify the choice of values for α and β, we treat them as prior counts relative to the number of sequences in the test sample, rather than absolute prior counts. For all inferences performed in this paper, we set both α and β to 0.0001. We use a separate and larger value of α (0.1) for the prior counts of each taxon in the Unknown environment, in order to prevent that environment from overfitting each individual test sample. If we had a prior belief that some of the test samples shared the same Unknown environment, we could perform inference on them jointly, and reduce this separate α value accordingly.
As is typical in Gibbs sampling, we first performed a set of “burn-in” passes (25 passes) through the entire set of sequences in a data sample before drawing a mixture sample from the joint posterior. We also re-started the entire sampling process with new random hidden variable values 100 times, thereby collecting a total of 100 samples from the posterior distribution for each sample. Each iteration on a sink sample with V source environments requires O(V2n) operations. Before running Gibbs sampling, we rarefied all samples to an artificial sequence depth of 1,000. We kept any samples whose original sequence depth was less than 1,000 at that lower depth.
Simulations
For the comparison of SourceTracker to naïve Bayes and Random Forests22 (Fig. 1), we simulated two source environments with varying degrees of overlap in their distribution over taxa by defining a single uniform Dirichlet prior over 100 taxa with varying concentration levels, and drawing two multinomial distributions from it. By varying the concentration parameter, we were able to control the degree of overlap between the two multinomials. The simulation procedure (Supplementary Fig. 7) was repeated three times.
For the application of SourceTracker with Gibbs sampling to the detection task, we used all of the Gut and Skin training samples to estimate the multinomial distribution over taxa in each environment. To generate “contaminated” samples, we drew 100 simulated samples from each environment at sequencing depth 1,000 and mixed them together with 1% (or 5%) Skin and 99% (or 95%) Gut. We also generated 100 pure Gut samples at depth 1,000. We then ran SourceTracker as described above to estimate the proportion of Skin taxa in the simulated Gut samples. We used a contamination threshold of one-half of the contamination rate, and varied the Dirichlet parameter β to adjust the sensitivity of the model (higher β means higher sensitivity). For each value of β, with its corresponding level of sensitivity, we measured the specificity of the contamination predictions made by SourceTracker, and plotted the series of values as receiver operating characteristic curves (Supplementary Fig. 6a—b).
Supplementary Material
Acknowledgements
We wish to acknowledge funding from National Institutes of Health (R01HG4872, R01HG4866, U01HL098957, P01DK78669); the Crohn’s and Colitis Foundation of America; and the Howard Hughes Medical Institute. We additionally wish to acknowledge Bharath Prithiviraj for helpful insight into previous related work, and the editor and reviewers for their valuable suggestions.
Footnotes
Author Contributions
D.K. designed the algorithm, software, and computational experiments; D.K., R.K., and S.T.K. wrote the manuscript; J.K., E.S.C., J.Z., M.C.M., R.G.C., and F.D.B. contributed to the manuscript; J.K. and M.C.M. contributed to algorithm design; J.K. processed the data after sequencing; E.S.C. collected the data; R.G.C. and F.D.B. organized and supervised the data collection; R.G.C., F.D.B., R.K., and S.T.K. supervised the project.
References
- 1.Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. Appl. Environ. Microbiol. 2005;71(12):8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quince C, et al. Nat. Methods. 2009;6(9):639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 3.Tanner MA, Goebel BM, Dojka MA, Pace NR. Appl. Environ. Microbiol. 1998;64(8):3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson JM, Santo Domingo JW, Reasoner DJ. Environ Sci Technol. 2002;36(24):5279–5288. doi: 10.1021/es026000b. [DOI] [PubMed] [Google Scholar]
- 5.Wu CH, et al. PloS one. 2010;5(6):e11285. doi: 10.1371/journal.pone.0011285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg J, Price B, Ware A. Water Res. 2010;44(8):2629–2637. doi: 10.1016/j.watres.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Smith A, Sterba-Boatwright B, Mott J. Water Res. 2010;44(14):4067–4076. doi: 10.1016/j.watres.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Dufrêne M, Legendre P. Ecol Monogr. 1997;67(3):345–366. [Google Scholar]
- 9.Costello EK, et al. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauber CL, Hamady M, Knight R, Fierer N. Appl. Environ. Microbiol. 2009;75(15):5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone C, Knight R. Appl. Environ. Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blei DM, Ng AY, Jordan MI. The Journal of Machine Learning. 2003 [Google Scholar]
- 13.Griffiths TL, Steyvers M. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5228–5235. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fierer N, Hamady M, Lauber CL, Knight R. Proc. Natl. Acad. Sci. U.S.A. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu GD, et al. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, et al. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeder J, Knight R. Nat. Methods. 2010;7(9):668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Garrity GM, Tiedje JM, Cole JR. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas BJ, et al. Genome Res. 2011 [Google Scholar]
- 21.Price MN, Dehal PS, Arkin AP. Mol. Biol. Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breiman L. Machine Learning. 2001;45(1):5–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.