Abstract
The t(4;8)(p16;p23) translocation, in either the balanced form or the unbalanced form, has been reported several times. Taking into consideration the fact that this translocation may be undetected in routine cytogenetics, we find that it may be the most frequent translocation after t(11q;22q), which is the most common reciprocal translocation in humans. Case subjects with der(4) have the Wolf-Hirschhorn syndrome, whereas case subjects with der(8) show a milder spectrum of dysmorphic features. Two pairs of the many olfactory receptor (OR)–gene clusters are located close to each other, on both 4p16 and 8p23. Previously, we demonstrated that an inversion polymorphism of the OR region at 8p23 plays a crucial role in the generation of chromosomal imbalances through unusual meiotic exchanges. These findings prompted us to investigate whether OR-related inversion polymorphisms at 4p16 and 8p23 might also be involved in the origin of the t(4;8)(p16;p23) translocation. In seven case subjects (five of whom both represented de novo cases and were of maternal origin), including individuals with unbalanced and balanced translocations, we demonstrated that the breakpoints fell within the 4p and 8p OR-gene clusters. FISH experiments with appropriate bacterial-artificial-chromosome probes detected heterozygous submicroscopic inversions of both 4p and 8p regions in all the five mothers of the de novo case subjects. Heterozygous inversions on 4p16 and 8p23 were detected in 12.5% and 26% of control subjects, respectively, whereas 2.5% of them were scored as doubly heterozygous. These novel data emphasize the importance of segmental duplications and large-scale genomic polymorphisms in the evolution and pathology of the human genome.
Introduction
Recent evidence has shown that the presence of a heterozygous submicroscopic parental inversion can mediate other chromosomal rearrangements and that these inversions are relatively common in the population (Jobling et al. 1998; Giglio et al. 2001). The meiotic mechanisms responsible do not substantially differ from those observed in classical inversions. In 1998, it was shown (by Jobling et al. [1998]) that the Xp/Yp translocation, accounting for XX male case subjects and XY female case subjects, occurred preferentially on Y chromosomes bearing a submicroscopic inversion of Yp. Previously, this inversion had been considered to be a neutral polymorphism, and these data had been considered to be peculiar to the sex chromosomes in that, owing to their common origin, they share several homologous regions and normally pair at pseudoautosomal region 1. However, the importance of these observations has been underlined by the demonstration (Giglio et al. 2001) that some recurrent 8p rearrangements occur as a consequence of an inversion polymorphism—mediated by two olfactory receptor (OR)–gene clusters—that is present in the parent transmitting the disease-related chromosome. Since OR-gene clusters exist at 4p16, as well as 8p23.1 (see the Database of Human Olfactory Receptor Genes Web site), we supposed that they might again be implicated in the genesis of the recurrent t(4;8)(p16;p23) translocation. This translocation has been reported in no fewer than 14 single or familial case subjects in either the balanced form or the unbalanced form (Wieczorek et al. 2000). Since it involves regions of very similar size and banding pattern, it may be undetected at the 400-band resolution that has been used, over the years, for much routine cytogenetic analysis. Thus, it is possible that this translocation is the most frequent one after the t(11q;22q) translocation, which is the most common reciprocal translocation in humans (Kurahashi et al. 2000). Case subjects with der(4) have the Wolf-Hirschhorn syndrome (WHS [MIM 194190]), whereas case subjects with der(8) have a less severe dysmorphic/mental retardation syndrome (Tranebjaerg et al. 1984). Our hypothesis that the translocation was mediated by OR-gene clusters, was reinforced by the finding that, in five patients with a der(4)t(4;8)(p16;p23), the 4p breakpoints were found to be at either ∼5 Mb or ∼14 Mb from the telomere of 4p (Wieczoreck et al. 2000). We reasoned that these two regions were compatible with the location of the 4p OR-gene clusters. We further hypothesized that heterozygous submicroscopic inversions of both 4p and 8p could prevent correct synapsis of the inverted regions at meiosis. As a result, both tetrads might assume a configuration that could result in an illegitimate but “homologous” crossover between the OR-gene clusters present on both 4p and 8p. In other words, the double heterozygous inversion would make two nonhomologous chromosomes available to recombine with each other.
Subjects and Methods
Subjects
We analyzed six case subjects (designated as “1”–“6” in tables 1 and 2) affected by WHS associated with der(4)t(4;8)(p16;p23) and their parents. We also analyzed one case subject (designated as “7” in tables 1 and 2) who had t(4;8)(p16;p23) and was the father of a case subject with WHS not included in this study. Case subjects 1 and 4 are those who were designated as “2” and “3,” respectively, by Wieczorek et al. (2000); case subject 5 was designated as “2” by Tonnies et al. (2001). A control population composed of 40 Ph.D. students was also analyzed (all blood samples were collected after informed consent was obtained).
Table 1.
Results of the FISH Experiments with Probes from 4p16 in the der(4)t(4;8)[Note]
|
Results for Case Subject |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Rearrangement | 797/01, der(4), de novo | TO, der(4), de novo | 1217/01, der(4), de novo | 796/01, der(4), de novo | 0B225, der(4), de novo | AnRag, der(4)mat | Bian, t(4;8) |
| Phenotype | WHS | WHS | WHS | WHS | WHS | WHS | Father of a case subject with WHS |
| Probe:a | |||||||
| 808b21 | − | − | − | ||||
| 520m5 | − | − | − | ||||
| 693B19 | − | − | −b | ||||
| 399e10 | − | − | − | − | −b | ||
| 529e10 | − | − | − | − | − | − | −b |
| 747h12 | + | + | + | + | |||
| 324i10 | − | + | + | ||||
| 265o12 | + | + | + | ||||
| 368b4 | + | + | + | + | + | ||
| 323f5 | + | + | |||||
| 732l22 | − | + | + | + | + | ||
| 274b16 | − | + | + | ||||
| 210l21 | − | − | + | + | + | + | |
| 69d13 | + | + | |||||
| 164f16 | + | ||||||
| 52i22 | + | + | |||||
| 190l6 | − | + | |||||
| 367j11 | − | ||||||
| 565i3 | − | + | + | + | |||
| 117j13 | − | + | |||||
| 448g15 | − | + | + | ||||
| 690d17 | + | + | |||||
| 751l19 | + | + | + | + | + | + | + |
| 264e23 | + | + | + | ||||
| 423d16 | + | + | + | ||||
| 363n10 | + | + | + | ||||
| 3m2 | + | + | + | + | + | + | + |
| 26l2 | + | + | + | ||||
| 124a18 | + | + | + | + | |||
Note.— Case subjects 1–6 are unbalanced, having 46,XX or XY,der(4)t(4;8)(p16;p23). Case subject 7 is balanced, having 46,XY,t(4;8)t(4;8)(p16;p23). Boxed regions identify BACs within the 4p OR-gene clusters.
Presence of a signal is indicated by a plus sign (+); absence of a signal is indicated by a minus sign (−). Signals within this region are also present on other OR-gene clusters.
Signals on the der(8).
Table 2.
Results of the FISH Experiments with Probes from 8p23 in the der(4)[Note]
|
Results for Case Subject |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Rearrangement | 797/01, der(4) de novo | TO, der(4), de novo | 1217/01, der(4), de novo | 796/01, der(4), de novo | 0B225, der(4), de novo | AnRag, der(4)mat | Bian, t(4;8) |
| Phenotype | WHS | WHS | WHS | WHS | WHS | WHS | Father of a case subject with WHS |
| Probe:a | |||||||
| 29g18 | + | + | + | + | + | ||
| 143g5 | + | + | + | + | + | + | |
| 42i21 | + | + | + | + | + | + | + |
| 173o4 | − | − | − | − | − | − | −b |
| 23o1 | − | − | − | −b | |||
| 77p24 | − | − | −b | ||||
Note.— Case subjects 1–6 are unbalanced, having either 46,XX or XY,der(4)t(4;8)(p16;p23). Case subject 7 is balanced, having 46,XY,t(4;8)t(4;8)(p16;p23). The boxed region identifies BACs within the 8p REPD.
Presence of a signal is indicated by a plus sign (+); absence of a signal is indicated by a minus sign (−). Signals within this region are also present on other OR-gene clusters.
Signals on the der(8).
General Procedure
The chromosome 8 translocation breakpoint had been refined by FISH, through use of clones of the 8p contigs already built in our previous study on inv dup(8p) (Giglio et al. 2001). Three OR-gene clusters are present on 8p (see the Database of Human Olfactory Receptor Genes Web site), and we previously reported the characterization of the central and proximal clusters containing 8p central and proximal repeated sequences (REPs), denoted as “8p REPD” and “8p REPP,” respectively.
Physical maps around the two OR-gene clusters at 4p are not well characterized, probably because of difficulties inherent in their duplicon-rich nature (Bailey et al. 2001). By in silico data mining, BAC-library screening, and FISH analysis of the resulting clones on the translocated chromosomes, we constructed a more precise physical map around the two OR-gene clusters at 4p. We named the distal and proximal REPs “4p REPD” and “4p REPP,” respectively.
Construction of the 4p16 Contigs and Map of the Region
FISH with BACs from RP11 library on metaphases from case subjects 1–7 defined a 12-Mb region that contained the translocation breakpoints. Data from the National Center for Biotechnology Information (NCBI) Entrez Genome View, the Celera Publication Site, and the Washington University in Saint Louis (WUSTL) Genome Sequencing Center showed that this region contained misassembly, misassignment, and/or decreased sequence coverage. Genome “holes” appeared, with sizes ranging between 100 and 200 kb. Thus, we estimated that they could be filled by one or two BACs. To build a unique contig, we analyzed the single BACs in the NCBI BLAST database. PCR products generated by primers designed on the end sequences of nonoverlapping BACs were used to screen high-density filters of the entire RP11/1 and RP11/2 libraries (BACPAC Resources). The resulting BACs were then used to confirm their mapping at 4p16, and their ends were sequenced and “blasted,” to test if they were filling the holes between contigs. Combining the FISH data on case subjects 1–7 with the mapping data from NCBI Entrez Genome View, Celera Publication Site, and WUSTL Genome Sequencing Center has permitted the location and orientation of BACs that have not yet been anchored.
Clone Isolation
BAC ends were amplified with specific primers. The software package Primer 3 was used to create primers from the sequence information. The amplification mix consisted of a 2-μM concentration of each primer, 200 μM deoxyribonucleotide triphosphates, 1× Mg2+ buffer (PerkinElmer), 1.25 U of Taq DNA polymerase (PerkinElmer), and 50 ng of genomic DNA. PCR conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 40 s, 68°C for 40 s, and 72°C for 40 s; and 72°C for 5 min.
FISH Analysis
FISH analysis was performed on both metaphase and interphase chromosomes from peripheral and/or lymphoblastoid cell lines from the six case subjects with der(4) (i.e., case subjects 1–6), the t(4;8)(p16;p23) translocation–bearing father of a case subject with der(4) (i.e., case subject 7), and 40 control subjects. We also analyzed the parent who transmitted the disease-related chromosome of the five de novo case subjects. All probes used were from RP11 and GenomeSystemInc libraries. Probe and slide preparation, DNA hybridization, and analysis were performed using conventional methods. At least 20 cells per case subject were analyzed by direct microscopic visualization and digital-imaging analysis.
Microsatellite Analysis
The DNA from case subjects 2, 3, and 5 and their parents was extracted from blood in EDTA by conventional methods. Primers for locus D4S2935 were from Applied Biosystems, and the 8p-specific amplimers and assay conditions have been described elsewhere (Giglio et al. 2001).
Results
4p OR Contigs
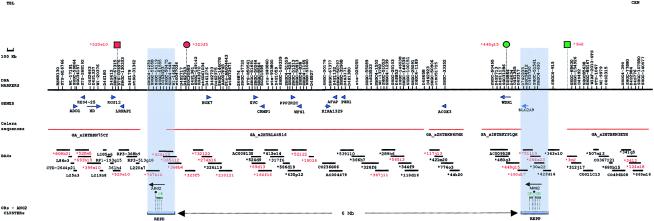
The two 4p 0R contigs are shown in figure 1. The two 8p OR contigs have been illustrated elsewhere (fig. 1 in Giglio et al. 2001). The OR-gene clusters at 4p and 8p are composed of blocks of nearly identical sequences (>95% identity), each spanning ∼400 kb. They contain genes, pseudogenes, and fragments of the OR and angiopoietin genes (fig. 1 of the present paper and fig. 1 in Giglio et al. 2001). Unfortunately, none of the holes that are present in the different maps (NCBI Entrez Genome View, Celera Publication Site, and WUSTL Genome Sequencing Center) could be completely filled, indicating that these holes were bigger than had been predicted by the genome databases.
Figure 1.
OR contigs at 4p. BACs marked with an asterisk (*) are from the RP11 library. BACs shown in red were used in FISH experiments on metaphases and interphases from case subjects 1–7. REPD and REPP indicate the two duplicons containing the OR- and angiopoietin-gene clusters. Each of them is ∼400 kb. In both REPD and REPP, all the genes or pseudogenes have the 3′ oriented toward the 4p telomere. The distance between the two REPs is ∼6 Mb. Blue arrows on the BACs indicate the genes they contain; their orientation is that provided by the NCBI Entrez Genome View and the Celera Publication Site databases. The markers in the region are indicated above the continuous black line. The breakpoint of the der(4) in case subjects 1 and 2 fell in REPP; the breakpoint of the der(4) in case subjects 3–7 fell in the 4p REPD. Red and green circles indicate the BACs at the edge of the inverted region (fig. 4a). Red and green squares indicate BACs used, in FISH, to demonstrate that they were not inverted, thus demonstrating that the inverted region is included between the two REPs.
The Translocation Breakpoints
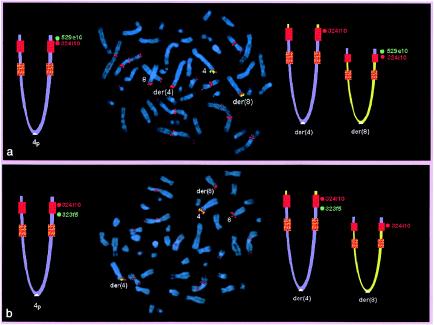
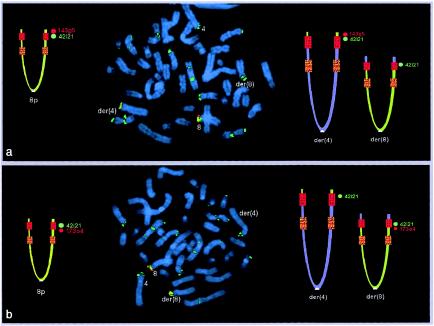
In case subjects 1 and 2, the 4p breakpoints were identified as falling in the 4p REPP (table 1). In fact, the first BAC present in the der(4) was RP11-690d17, which is the most distal clone partially belonging to REPP; all the more distal ones were absent. BAC RP11-3m2, proximal to REPP, was present in the der(4). In case subjects 3–7, the 4p breakpoint coincided with the 4p REPD (table 1 and fig. 2). In fact, all probes distal to REPD—including BAC RP11-529e10, the probe most proximal to REPD—were absent in the der(4) from case subjects 3–6 and were translocated to the der(8) in case subject 7. The 8p breakpoint was always detected within the 8p REPD (table 2 and fig. 3): BACs GS29g18 and GS143g5, distal but adjacent to the 8p REPD, were translocated to the der(4), whereas GS173o4, the probe most proximal to 8p REPD, was still present in the der(8).
Figure 2.
Metaphase FISH on case subject 7 (with the 46,XY,t(4;8)(p16;p23.1) translocation), showing the der(4) breakpoint. In the ideograms, the normal 4p is on the left, and the two derivative chromosomes are on the right; the red and orange squares indicate the REPD and the REPP, respectively. Red signals correspond to the distal 4p-REPD BAC RP11-324i10; these signals are present in several OR-gene cluster regions. a, Probe RP11-529e10 (green signals), which maps distal to the 4p REPD. This clone is translocated to the der(8). b, Probe RP11-323f5 (green signals), which is one of the more distal clones in the region included between the two 4p REPs. This clone remains on the der(4). These data demonstrate that the 4p breakpoint in case subject 7 is within the REPD (see also fig. 1).
Figure 3.
Metaphase FISH in case subject 7 (with the 46,XY,t(4;8)(p16;p23.1) translocation), showing the der(8) breakpoint. In the ideograms, the normal 8p is on the left, and the two derivative chromosomes are on the right; the red and orange squares indicate the REPD and the REPP, respectively. Green signals correspond to GS-42i21, which maps within the 8p REPD; these signals are present in several OR-gene cluster regions. a, Probe GS-143g5 (red signals), which is distal but adjacent to the 8p REPD. This clone is transposed to the der(4). b, Probe GS-173o4 (red signals), which is the most distal clone within the region included between the two 8p REPs. This clone remains on the der(8). These data demonstrate that the 8p breakpoint is within the REPD (see also fig. 1 in Eichler 2001).
Parents Transmitting the Disease-Related Chromosome
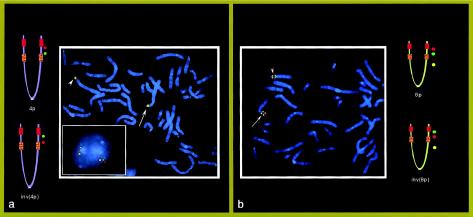
In all five de novo case subjects with der(4), the disease-related chromosome was of maternal origin (for case subjects 1 and 4, see Wieczoreck et al. 2000; data not shown for case subjects 2, 3, and 5). To assess the maternal organization of these regions, we investigated the maternal chromosomes by FISH with appropriate 4p and 8p BACs. Interestingly, all five mothers were found to be heterozygous for both 4p and 8p inversions (fig. 4). Clones RP11-323f5 and RP11-448g15, included between the two 4p REPs, were inverted in one maternal chromosome 4, whereas clones GS-173o4 and GS-257o3, included between the two 8p REPs (Giglio et al. 2001), showed that one maternal chromosome 8 was inverted. Clones RP11-529e10 and RP11-3m2, external to the 4p REPs, and clones GS143g5 and 2244f17 (FBAC-4434 library), external to the 8p REPs, were not inverted in either maternal chromosomes 4 or 8 (data not shown). These data demonstrated that the two pairs of 4p and 8p OR-gene clusters delimited the inversion boundaries.
Figure 4.
FISH in the mother of case subject 3 (with 46,XX). a, Normal and inverted 4p. At left, the ideograms show that clones RP11-323f5 (red signals) and RP11-448g15 (green signals), included between the two 4p REPs, are inverted in the chromosome 4 (arrowhead). In the square, a nucleus shows the same inversion; yellow signals refer to the control probe RP11-520m5, which is distal to the 4p REPD (see fig. 1), and red and green signals refer to RP11-323f5 and RP11-190l6, respectively (these two clones are at a distance of ∼2 Mb and thus are more appropriate for interphase FISH). b, Normal and inverted 8p. At right, the ideograms show that clones GS-173o4 (red signals) and GS-257o3 (green signals), included between the two 8p REPs, are inverted in one chromosome 8 (arrowhead). RP11-563o19 (yellow signals), at 8p12, was used as a control probe.
Analysis of a Control Population
To better understand the biological significance of our findings, we investigated normal control subjects. Five of 40 (12.5%) were found to be heterozygous for the inversion at 4p, and 13 of 50 (26%) were found to be heterozygous at 8p. Consistent with Hardy-Weinberg equilibrium, among 40 control subjects, one doubly heterozygous individual (2.5%) was found. These data suggest a highly significant association (P<10-6, by Fisher's exact test) between the polymorphic heterozygous submicroscopic inversion and the translocation event.
Discussion
The extensive segmental duplication of the human genome has recently become a focus of scientific attention for its evolutionary implications (Johnson et al. 2001; Crosier et al. 2002; Samonte and Eichler 2002) and for its role in the mediation of genomic disorders (Eichler 2001; Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002). In fact, an increasing number of human diseases—those now called “genomic disorders”—are recognized as resulting from recurrent rearrangements that involve unstable genomic regions (Ji et al. 2000; Emanuel and Shaikh 2001; Stankiewich and Lupski 2002). Such genomic abnormalities have been shown to result from nonallelic homologous recombination between specific low-copy repeats (“duplicons” or “segmental duplications”). Misalignment and recombination between homologous segmental duplications lying on the same chromosome has been demonstrated to lead to deletions, duplications, supernumerary chromosomes, and benign and pathological inversions. Although these events had been considered as stochastic, we have recently discovered that some 8p imbalances, previously considered as occurring “de novo,” are, on the contrary, the precise consequence of heterozygous inversion polymorphisms generated by the OR-gene duplicons at 8p23 through unusual meiotic configurations (Giglio et al. 2001). Similar findings at the Williams-Beuren 7q11.23 deletion region (Osborne et al. 2001) support the idea that heterozygosity for these inversions plays a role in the causing of susceptibility to unequal recombination. We have now demonstrated, for the first time to our knowledge, that double-heterozygous inversion polymorphisms that are present in nonhomologous chromosomes (generated again by OR-gene clusters) may also mediate interchromosomal rearrangements. A relatively small number of submicroscopic inversions, all flanked by low-copy repeats, have been reported (Lakich et al. 1993; Bondeson et al. 1995; Small et al. 1997; Jobling et al. 1998; Saunier et al. 2000; Giglio et al. 2001; Osborne et al. 2001), and most of them have been ascertained in clinical cases. Their occurrence, however, may be underestimated, since 5%–10% of the human genome is now thought to consist of interspersed, highly homologous duplications that have arisen over the past 35 million years of human genomic evolution (Eichler 2001). It is becoming clear that neighboring duplicons can produce, in addition to duplications and deletions, inversion polymorphisms that, in turn, can cause predisposition to other chromosomal imbalances.
The concept of “predisposition” to chromosomal rearrangements implies that inversion heterozygotes may be at risk of having more than one child with a de novo chromosomal rearrangement. Although two siblings with the Williams-Beuren deletion had been described elsewhere (Kara-Mostefa et al. 1999), a comprehensive survey of the numerous cases of inv dup(8p) andder(4)t(4;8)(p16;p23) showed that all of them represented isolated cases. In addition, the facts that heterozygous 8p polymorphisms occur in ∼1 in 4 control subjects (26%) and that case subjects with inv dup(8p) have an estimated frequency of 1 in 20,000 (Floridia et al. 1996) suggest that the population frequency of these heterozygous inversions is high, relative to the frequency of the rearrangements to which they predispose. The theoretical expectation of recurrence and the empirical data may, however, be reconciled by taking into account the fact that cases coming to clinical attention are only those that are compatible with relatively normal embryonic development. Our studies on 8p imbalances have indicated that the prerequisite for the inv dup(8p) formation is the occurrence of a dicentric 8qter-cen-8p::8p-cen-8qter chromosome, followed by a breakage at anaphase II (Giglio et al. 2001). The absence of a breakage would result in a fetus that bears an almost complete trisomy 8 and that thus would be unlikely to be viable. Moreover, we have shown that heterozygous inversion carriers can generate not only inv dup(8p) chromosomes but also their reciprocal product (i.e., small acentric marker chromosomes). It is likely that these markers are lost, resulting in a fetus with normal chromosomes, unless a neocentromere is immediately formed. The last occurrence seems to be a very rare event (Warburton 2001). Thus, the fact that case subjects with de novo chromosomal rearrangements are usually unique in their family does not preclude our hypothesis that submicroscopic genomic polymorphisms predispose to unbalanced chromosomal rearrangements.
Acknowledgments
O.Z. is grateful to M. Fraccaro for his lifelong encouragement and helpful discussion. This work was supported by cofin00-MIUR and cofin01-MIUR (to S.G., M.V., M.R., G.Gr., and O.Z.), by Italian Telethon Foundation (grants GP0247Y01 [to O.Z.] and C.50 [to M.R.]), by Associazione Italiana Ricerca sul Cancro (grant to M.R.), and by Ministero della Salute (grant to O.Z. and A.R.). V.C. is supported by a Telethon fellowship. We would like to thank G. Gillessen-Kaesbach, F. Majewski, and P. Meinecke for sending blood samples of the mothers of patients with WHS.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- BACPAC Resources, http://www.chori.org/bacpac/
- BLAST, http://www.ncbi.nih.gov/BLAST/
- Celera Publication Site, http://publication.celera.com/cds/login.cfm
- Database of Human Olfactory Receptor Genes, A, http://bioinformatics.weizmann.ac.il/HORDE/humanGenes/
- Entrez Genome View, http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/hum_srch?chr=hum_chr.inf&query (for Homo sapiens genome view, build 29)
- Genome Sequencing Center, http://www.genome.wustl.edu/gsc/ (for specific sequence, see http://genome.wustl.edu:8021/pub/gsc1/fpc_files/freeze_2001_08_06/MAP/4map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WHS [MIM 194190])
References
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson ML, Dahl N, Malmgren H, Kleijer WJ, Tonnesen T, Carlberg BM, Pettersson U (1995) Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum Mol Genet 4:615–621 [DOI] [PubMed] [Google Scholar]
- Crosier M, Viggiano L, Guy J, Misceo D, Stones R, Wei W, Hearn T, Ventura M, Archidiacono N, Rocchi M, Jackson MS (2002) Human paralogues of KIAA0187 were created through independent pericentromeric-directed and chromosome-specific duplication mechanisms. Genome Res 12:67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17:661–669 [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, Rossi E, Gimelli G, Croci G, Franchi F, Gilgenkrantz S, Grammatico P, Dalpra L, Wood S, Danesino C, Zuffardi O (1996) The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am J Hum Genet 58:785–796 [PMC free article] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, Weber JL, Ledbetter DH, Zuffardi O (2001) Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 68:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD (2000) Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10:597–610 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Williams GA, Schiebel GA, Pandya GA, McElreavey GA, Salas GA, Rappold GA, Affara NA, Tyler-Smith C (1998) A selective difference between human Y-chromosomal DNA haplotypes. Curr Biol 8:1391–1394 [DOI] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE (2001) Positive selection of a gene family during the emergence of humans and African apes. Nature 413:514–519 [DOI] [PubMed] [Google Scholar]
- Kara-Mostefa A, Raoul O, Lyonnet S, Amiel J, Munnich A, Vekemans M, Magnier S, Ossareh B, Bonnefont JP (1999) Recurrent Williams-Beuren syndrome in a sibship suggestive of maternal germ-line mosaicism. Am J Hum Genet 64:1475–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Emanuel BS (2000) Alu-mediated PCR artifacts and the constitutional t(11;22) breakpoint. Hum Mol Genet 9:2727–2732 [DOI] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J (1993) Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 5:236–244 [DOI] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nat Rev Genet 3:65–72 [DOI] [PubMed] [Google Scholar]
- Saunier S, Calado J, Benessy F, Silbermann F, Heilig R, Weissenbach J, Antignac C (2000) Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am J Hum Genet 66:778–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K, Iber J, Warren ST (1997) Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet 16:96–99 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski J (2002) genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- Tonnies H, Stumm M, Neumann L, Volleth M, Grumpelt U, Musebeck J, Annuss G, Neitzel H (2001) Two further cases of WHS with unbalanced de novo translocation t(4;8) characterised by CGH and FISH. J Med Genet 38:E21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranebjaerg L, Petersen A, Hove K, Rehder H, Mikkelsen M (1984) Clinical and cytogenetic studies in a large (4;8) translocation family with pre- and postnatal Wolf syndrome. Ann Genet 27:224–229 [PubMed] [Google Scholar]
- Warburton PE (2001) Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet 17:243–247 [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, Albrecht B, Meinecke P, Riess O, Gillessen-Kaesbach G (2000) Unexpected high frequency of de novo unbalanced translocations in patients with Wolf-Hirschhorn syndrome (WHS). J Med Genet 37:798–804 [DOI] [PMC free article] [PubMed] [Google Scholar]