Abstract
The molecular basis of the skipping of constitutive exons in many messenger RNAs is not fully understood. A well-studied example is exon 9 of the human cystic fibrosis transmembrane conductance regulator gene (CFTR), in which an abbreviated polypyrimidine tract between the branch point A and the 3′ splice site is associated with increased exon skipping and disease. However, many exons, both in CFTR and in other genes and have short polypyrimidine tracts in their 3′ splice sites, yet they are not skipped. Inspection of the 5′ splice sites immediately up- and downstream of exon 9 revealed deviations from consensus sequence, so we hypothesized that this exon may be inherently vulnerable to skipping. To test this idea, we constructed a CFTR minigene and replicated exon 9 skipping associated with the length of the polypyrimidine tract upstream of exon 9. We then mutated the flanking 5′ splice sites and determined the effect on exon skipping. Conversion of the upstream 5′ splice site to consensus by replacing a pyrimidine at position +3 with a purine resulted in increased exon skipping. In contrast, conversion of the downstream 5′ splice site to consensus by insertion of an adenine at position +4 resulted in a substantial reduction in exon 9 skipping, regardless of whether the upstream 5′ splice site was consensus or not. These results suggested that the native downstream 5′ splice site plays an important role in CFTR exon 9 skipping, a hypothesis that was supported by data from sheep and mouse genomes. Although CFTR exon 9 in sheep is preceded by a long polypyrimidine tract (Y14), it skips exon 9 in vivo and has a nonconsensus downstream 5′ splice site identical to that in humans. On the other hand, CFTR exon 9 in mice is preceded by a short polypyrimidine tract (Y5) but is not skipped in vivo. Its downstream 5′ splice site differs from that in humans by a 2-nt insertion, which, when introduced into the human CFTR minigene, abolished exon 9 skipping. Taken together, these observations place renewed emphasis on deviations at 5′ splice sites in nucleotides other than the invariant GT, particularly when such changes are found in conjunction with other altered splicing sequences, such as a shortened polypyrimidine tract. Thus, careful inspection of entire 5′ splice sites may identify constitutive exons that are vulnerable to skipping.
Introduction
Alternative splicing, defined as regulated and productive variation in patterns of splice-site utilization, increases the diversity of mRNAs and proteins expressed in the cell (Black 2000; Graveley 2001). Consequences for mRNA commonly include skipping or inclusion of one or more exons, substitution of alternate exons, and deletion or incorporation of a short block of extra coding sequence adjacent to a constitutive exon. Although much alternative splicing is regulated and purposeful, constitutive exons can also occasionally be skipped because of suboptimal splice sites. Exon 9 of the cystic fibrosis transmembrane conductance regulator gene (CFTR [MIM 602421]) is missing in a fraction of CFTR transcripts in nearly all individuals (Chu et al. 1991, 1992, 1993). CFTR mRNAs missing exon 9 encoded a protein that was misfolded and nonfunctional (Delaney et al. 1993; Strong et al. 1993), suggesting that skipping of this exon may be a case of nonproductive, rather than purposeful, alternative splicing. The degree of CFTR exon 9 skipping is inversely correlated with the length of a polymorphic polythymidine tract upstream of the exon. Transcripts derived from genes that carry five thymidines (5T) at this locus have high levels of exon 9 skipping, whereas those with seven or nine thymidines (7T and 9T, respectively) have successively lower levels of skipping (Chu et al. 1993). The 5T variant can have a profound effect on splicing, as is illustrated by the absence of exon 9 in as many as 95% of CFTR transcripts in respiratory epithelia of 5T homozygotes (Chu et al. 1992). Approximately 10% of individuals worldwide carry the 5T variant (Kiesewetter et al. 1993), which is of clinical importance because it is associated with congenital bilateral absence of the vas deferens (CBAVD [MIM 277180]), a form of male infertility (Kiesewetter et al. 1993; Osborne et al. 1994; Chillón et al. 1995; Jarvi et al. 1995; Zielenski et al. 1995). Penetrance of the 5T allele for the CBAVD phenotype is variable and has been estimated to be 0.6 (Zielenski et al. 1995). Interestingly, rare 5T homozygotes have been reported with a cystic fibrosis phenotype (CF [MIM 219700]) (Noone et al. 2000), suggesting that 5T is a partially penetrant disease-causing allele for CF as well.
Several sequence elements in addition to the polythymidine tract appear to contribute to exon 9 skipping. It has been proposed that longer alleles of a polymorphic tract of TG repeats immediately upstream of the polythymidine tract exacerbate exon 9 skipping (Cuppens et al. 1998; Niksic et al. 1999). Indeed, a protein that binds this TG tract has been identified and shown to modulate exon 9 splicing efficiency (Buratti and Baralle 2001; Buratti et al. 2001). A 6-bp splicing enhancer and a 10-bp silencer within exon 9, as well as a silencer of unknown length in intron 9, have also been identified (Pagani et al. 2000). Overexpression of a variety of cellular and viral splicing factors has been shown to increase exon 9 skipping in transcripts derived from minigene constructs (Nissim-Rafinia et al. 2000; Pagani et al. 2000). The overexpression of one particular viral factor, ORF3, actually decreases exon 9 skipping, although the mechanism responsible for this effect is unknown (Nissim-Rafinia et al. 2000). These studies support a model in which splicing of exon 9 is subject to a number of sequence elements within and near the exon and to their interactions with trans-acting factors in the splicing machinery.
A fundamental group of cis-acting sequences that are known to influence splicing—and that heretofore have not been thoroughly studied with respect to CFTR exon 9—are the 5′ splice sites flanking the exon. Inspection of the 5′ splice sites upstream and downstream of CFTR exon 9 suggested that they may be particularly relevant in this case, since neither conforms to consensus. Using a minigene construct, we show that mutating each of the 5′ splice sites to conform to consensus has opposite effects upon exon 9 splicing. Interestingly, exon skipping caused by the short (5T) upstream polythymidine tract can be eliminated by conforming the downstream 5′ splice site to consensus. A cross-species comparison of these splice-site utilizations and CFTR exon 9 splicing efficiency in vivo supported conclusions derived from the minigene studies. Taken together, these observations point to the importance in exon inclusion of the entire 5′ splice-site sequence.
Material and Methods
Construction of Minigene
Genomic DNA from control samples carrying 5T and 9T alleles were amplified using the following oligonucleotides: ex8f, 5′-GACTCTAGAACATTGGAATATAACTTAACG-3′; ex8r3, 5′-GACGCGGCCGCGTGAGTGATCCTCC-3′; ex9f, 5′-CAGGCGGCCGCTGATAATGGGCAAATATC-3′; ex9r, 5′-CAGCCGCGGCTAGAGTAAATTATCAGTGG-3′; ex10f, 5′-CAGCCGCGGCACACTGTATTGTAATTGTC-3′; ex10r, 5′-CAGCTGCAGTGCCAGGCATAATCCAGG-3′. PCR was performed on a Perkin Elmer 480 thermal cycler, using an initial denaturation of 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and by an extension of 72°C for 10 min. Conditions were 20 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 1 U Taq polymerase (all from Life Technologies), and 200 μM each dNTP (Pharmacia) per 50 μl reaction. PCR fragments were subcloned into vector pCR2.1 with TA cloning kit (Invitrogen) and were sequenced to ensure authenticity. The above oligonucleotides incorporated unique restriction-enzyme sites that enabled in-frame fusion of the products into an ornithine-amino-transferase (OAT) cDNA construct in the pGEM4 vector (Stratagene) (OAT vector kindly provided by the laboratories of H. Dietz and D. Valle). Vector and PCR fragments were joined in a four-way ligation at 12°C overnight (T4 ligase [Life Technologies]) and transformed into MAX Efficiency DH5α cells (Life Technologies), and the resulting clones were checked for fragment order, orientation, and authenticity, by use of restriction digestion and sequencing. The OAT-CFTR fusion minigene was removed from pGEM4 by EcoRI digestion and was subcloned into mammalian expression vector pBK-RSV (Stratagene). Two variants were initially made, one containing (TG)10T9 and the other (TG)12T5.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed on the (TG)12T5 minigene construct by use of the Transformer kit (Clontech), according to the manufacturer’s instructions. Selection oligonucleotides in multiply mutated constructs alternated between a unique BsaI site and a unique BsmBI site: Bsa-Bsm+, 5′-CCACCGAGACGCCATTGGGGC-3′, Bsm-Bsa+, 5′-TACCCCACCGAGACCCCATTGGGGCCAATACGC-3′. The following mutagenic oligonucleotides were used: +3C-2, 5′-CAGCCTTCTGGGAGGAGGTDAGAATTTTTAAAAAATTGTTTGCTC-3′, where D≠C; int9donr, 5′-CAGGCAAGGTRAGTTCTTTTGTTCTTCAC-3′, where R=A or G; and int9d-CT, 5′-GGAGCAGGCAAGGTACTGTTCTTTTGTTCTTCAC-3′. Mutated minigenes were verified by sequencing, grown to bulk and isolated with a Maxiprep kit (Qiagen).
Cell Culture, Transfections, and Isolation of RNA
Human embryonic kidney 293 cells (HEK293 [American Type Culture Collection]) were grown in Eagle's minimal essential medium supplemented with 10% fetal bovine serum (both from Biofluids) and 1% penicillin/streptomycin (Life Technologies), in 5% CO2 at 37°C in T25 flasks (Falcon). Cells were trypsinized (Biofluids) and transferred to six-well plates (Falcon), were allowed to adhere for >24 h, and were then transfected with 3 μg plasmid and 10 μl Lipofectin in OptiMEM (both from Life Technologies), according to the manufacturer’s instructions. Transfections were terminated at 8 h with serum-containing growth media, and cells were allowed to continue growing until 24 h after transfection, at which time RNA was harvested with RNAzol B (TelTest), according to the manufacturer’s instructions, and was quantified with at least three readings on GeneQuant RNA/DNA Calculator (Pharmacia).
RT-PCR and Analysis by Capillary Electrophoresis
cDNA was synthesized from 5 μg RNA, using oligo-dT (Roche) and SuperScript II reverse transcriptase (Life Technologies), according to the manufacturer’s instructions. cDNA was synthesized for 50 min at 42°C, then shifted to 70°C for 15 min to inactivate the RT. One microliter of cDNA was amplified under the following conditions: 20 mM Tris-HCl, 50 mM KCl, 1 mM MgCl2, 200 μM each dNTP, 1 mM dithiothreitol, 1 U Taq polymerase per 50-μl reaction (Life Technologies), by use of an initial denaturation at 95°C for 5 min, followed by 24, 27, 30, or 33 cycles (separate tubes run in parallel) of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and by an extension of 72°C for 7 min. The primer pair used for RT-PCR was designed to specifically detect only transcripts derived from our minigene: OAT-specific forward primer E4S, 5′-GTGCTGTCAACCAAGGGC-3′, and CFTR-specific ex10r labeled with 5′-fluorescein phosphoramidite (Glen Research). PCR products were sized and quantified by use of capillary electrophoresis on ABI Prism 310 (Applied Biosystems), using peak-area measurements.
Data Management and Statistical Analysis
Means, standard deviations, and graphs were generated with Microsoft Excel. Figures were composed using Microsoft PowerPoint. Statistical analyses were performed using the JMP statistical package, version 3.2.2 (SAS Institute). For each pair and for comparisons between groups, analysis of variance was performed. P<.05 was considered significant. Values were corrected for multiple comparisons, using the Bonferroni method.
Isolation of Total RNA from Ovine Tissues
Tissues were snap-frozen in liquid nitrogen in cryotubes (Nunc) and were stored in liquid nitrogen. Tissue was homogenized in glass homogenizers in 4 M guanidinium isothiocyanate (Fluka). Total RNA was isolated from cesium chloride (BDH Laboratory Supplies) gradients (specific density 1.7 g/cm3) and was resuspended in 0.1 mM EDTA and 10 mM Tris (pH 7.4).
RT-PCR for Ovine Tissues
CFTR cDNA synthesis was primed with 3′ primer B2L 5′-GGAAGGCAGCCTATGTGAGA-3′ under oil at 65°C for 10 min. cDNA was generated by use of the RT Superscript (Life Technologies) at 42°C for 1 h. PCR was then performed with 5′ primer B2R 5′-AGCCATCAATTTACAGACAC-3′ and B2L, under the following conditions: 1 cycle at 94°C for 5 min, 30 cycles of 94°C for 1 min and 60°C for 1 min, and extension at 72°C for 3 min, followed by a final cycle of extension at 72°C for 5 min.
Southern Blot Analysis of Ovine RT-PCR Products
RT-PCR products were separated on a 1.5% agarose gel in Tris-borate EDTA. RT-PCR products were transferred to Hybond-N+ membranes (Amersham), using standard methods, and were probed with randomly primed α32P-labeled ovine CFTR B fragment (U20418:1019–1914) (Megaprime DNA labeling kit; Amersham), as described elsewhere (Tebbutt et al. 1995). Quantification of exon 9+ and exon 9− products was determined by phosphorimaging (Amersham).
Results
CFTR Minigene Replicates In Vivo Exon Skipping
To investigate cis-acting sequences that affect CFTR exon 9 splicing, we created a minigene composed of exon 9, flanking intron sequence, and portions of exons 8 and 10 (fig. 1A). The minigene was fused in-frame to an ornithine aminotransferase cDNA construct driven by a Rous sarcoma virus promoter. To determine whether the length of the polythymidine tract affected the splicing of our minigene construct, we created one 9T and one 5T. Because it has been suggested that the distance of the branch point A from exon 9 plays a role in splicing efficiency (Cuppens et al. 1998), the number of TG repeats preceding the polypyrimidine tract in the 3′ splice site of intron 8 was adjusted (to 12 TGs in the 5T construct and 10 TGs in the 9T construct) such that the distance from the branch point A to exon 9 in each construct was constant. The minigenes were transfected separately into HEK293 cells, and fluorescent RT-PCR and capillary electrophoresis were performed to determine the relative proportions of exon 9+ and exon 9− transcripts derived from each minigene (fig. 1B). Accurate determination of the ratio of 9− to 9+ transcripts, rather than their absolute quantitation, was necessary in this study. We therefore chose to compare the peak area of fluorescence generated by each amplicon, since this represented the number of DNA molecules newly synthesized by RT-PCR. This technique has been used by a number of investigators, because it avoids the imprecision introduced by hybridization-based techniques (Teng et al. 1997; Cuppens et al. 1998; Nissim-Rafinia et al. 2000).
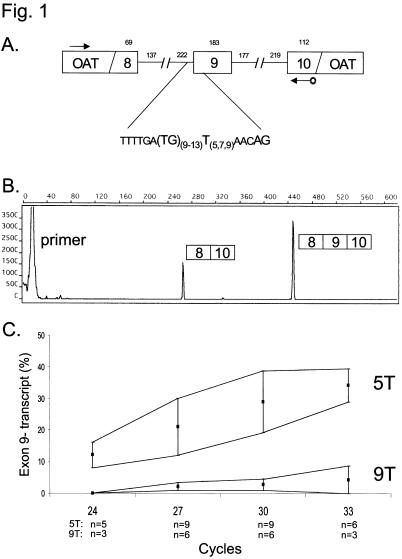
Figure 1.
Exon 9 splicing of a CFTR minigene construct. Exon 9 splicing is dependent on the length of the polypyrimidine tract in intron 8. A, Diagram of the CFTR minigene. CFTR exons 8, 9, and 10 are fused in-frame to an ornithine aminotransferase (OAT) cDNA construct. Both exon 9+ and exon 9− transcripts are in-frame. Most of the 6.5-kb intron 8 and 10.6-kb intron 9 have been deleted. Note two polymorphic loci immediately preceding exon 9. Arrows indicate RT-PCR primers; 3′ primer is tagged with the fluorescent label 6-FAM. B, Capillary electrophoresis trace of a typical RT-PCR, showing the relative proportions of product derived from transcripts lacking and including exon 9. C, Results from 9T and 5T variants, showing range of exon skipping observed at 24, 27, 30, and 33 cycles (mean ± SD).
To confirm that PCR was in log phase (so that measurements reflected the concentration of starting template rather than a distortion due to saturation of the PCR), we compared the fluorescent signals after 24, 27, 30, and 33 cycles of PCR. There was a minor increase in the proportion of exon 9− transcripts at greater cycle numbers, probably because of preferential amplification of the shorter 9− fragment (fig. 1C). A small fraction of transcripts from the 9T minigene were missing exon 9 (mean [SD] 2.8% [1.8] exon 9− transcripts at 30 cycles), whereas a substantial proportion of transcripts derived from the 5T minigene were missing exon 9 (mean [SD] 29.0% [9.8] exon 9− transcripts at 30 cycles) (fig. 1C). Variability in the splicing efficiency of the 5T construct was due to differences in passage number of the HEK293 cells. The difference between the 9T and 5T constructs was statistically significant for all cycle numbers studied (P<.001 at 27, 30, and 33 cycles; P<.01 at 24 cycles). Thus, the minigene replicated exon 9 skipping associated with polypyrimidine tract length seen in vivo (Chu et al. 1993; Mak et al. 1997; Teng et al. 1997). Since we wished to determine whether sequence alterations increased or decreased exon 9 skipping, we analyzed RT-PCR products after 30 cycles of amplification from variants of the 5T minigene for subsequent experiments.
Exon 9 Skipping Increases When the Upstream 5′ Splice Site Conforms to Consensus
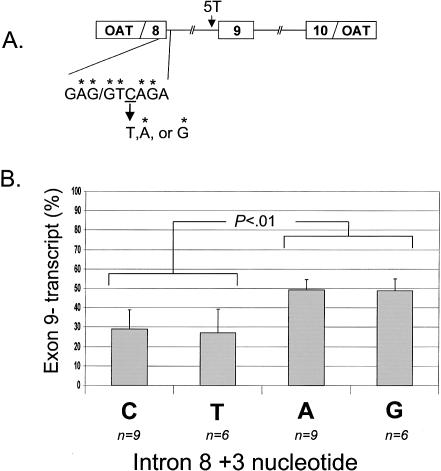
Inspection of the 5′ splice site upstream of CFTR exon 9 revealed a nonconsensus pyrimidine at the +3 position (fig. 2A), giving the site a Shapiro-Senapathy score (an indicator of splice-site strength) of 84.3 (of 100) and an information content of 5.2 bits, according to an information-theory–based algorithm (Shapiro and Senapathy 1987; Rogan et al. 1998). Because only 1 in 40 splice sites contains a cytosine at the +3 position (Clark and Thanaraj 2002), the coincidence of finding this unusual splice-site feature adjacent to exon 9 raised the possibility that it contributes to exon 9 skipping. We hypothesized that mutation of the native +3 cytosine to a consensus adenine would improve splicing of exon 8 to exon 9 and would reduce exon skipping. Such a change resulted in a splice-site score of 94.3 and an information content of 10.0 bits. However, conversion of the site to consensus resulted in higher, not lower, levels of transcripts missing exon 9 (fig. 2B). Subsequent substitution of the +3 nucleotide with guanine or thymidine revealed the following pattern. Nonconsensus pyrimidines at this location have similar splice-site scores, and they spliced with roughly equal efficiency (mean [SD] for C=29.0% [9.8] exon 9− transcripts, and mean [SD] for T=27.1% [12.1]) The presence of either consensus purine resulted in higher splice-site scores but also in higher rates of exon skipping (mean [SD] for A=49.2% [5.5] exon 9− transcripts, and mean [SD] for G=48.8% [6.1]). The difference was significant (P<.01 by ANOVA). Thus, creation of a consensus upstream 5′ splice site did not, in fact, increase splicing of exon 8 to exon 9 but increased splicing of exon 8 to exon 10 instead. This raised a question about the ability of the downstream 5′ splice site to define exon 9 and to facilitate efficient splicing of exon 9 to exon 10. Therefore, we next turned our attention to this site.
Figure 2.
Native nonconsensus nucleotide at +3 position of intron 8 5′ splice site. The nucleotide reduces skipping of exon 9. A, Diagram of minigene showing the endogenous cytosine at +3 position and the substitutions made. Asterisks (*) indicate nucleotides that conform to consensus (RAG/GTRAG; R = purine). B, Substitution of consensus purines in the +3 position. The substitution results in a significant increase in exon skipping, compared with nonconsensus pyrimidines.
Exon 9 Skipping Decreases Significantly When the Downstream 5′ Splice Site Conforms to Consensus
Inspection of the 5′ splice site downstream of CFTR exon 9 revealed that it also deviated from consensus, and corresponded to a splice-site score of 74.8 and information content of 5.9 bits. The site could be made to conform to consensus by the insertion of an adenine at position +4 (fig. 3A), resulting in a score of 100.0 and in 12.0 bits of information. The creation of this change in the minigene resulted in a substantial decrease in exon 9 skipping among exon 9− transcripts (mean [SD] 1.6% [2.5] vs. 29.0% [9.8]) (fig. 3B). We concluded that the newly altered downstream 5′ splice site increased the efficiency with which exon 9 spliced to exon 10, thus decreasing splicing of exon 8 to exon 10 and resulting in a reduced proportion of transcripts missing exon 9.
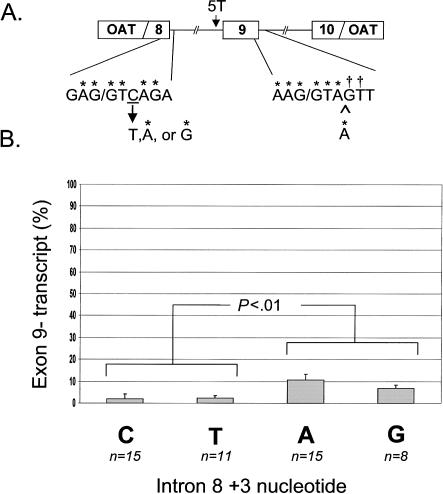
Figure 3.
Insertion of adenine at position +4 of the downstream 5′ splice site. The insertion reduced exon skipping. A, Consensus splice site created by insertion of an adenine at +4 of intron 9. Daggers (†) indicate nucleotides that conform to consensus after the insertion. B, Effect of combined mutations at both 5′ splice sites. Experiments from figure 2 were repeated, with the added difference of an adenine insertion in the intron 9 5′ splice site. Statistical significance was calculated by ANOVA with Bonferroni correction.
Taken together, these observations suggested that exon 9 splicing is subject to a balance between the 5′ splice sites up- and downstream of the exon. To determine the relative contribution of each of the sites to exon 9 skipping, we evaluated splicing efficiency when the +3 nucleotide in the upstream site was altered in a minigene with a consensus downstream site (effectively combining the two previous experiments). When a nonconsensus T was introduced to the upstream site in this new construct, splicing was as efficient as with the exon 9− native nonconsensus C (mean [SD] 2.4% [2.7] vs. 1.6% [2.5] (fig. 3B). Interestingly, use of a consensus A or G resulted in a slight increase in skipping, although splicing was still far more efficient than with the native intron 9 5′ splice site (mean [SD] with A 10.7% [2.6] vs. 49.2% [5.5], P<.001; mean [SD] with G 6.8% [1.5] vs. 48.9% [6.1], P<.001) (fig. 3B). Minigenes that contained both a consensus upstream and a consensus downstream 5′ splice site generated transcripts that used cryptic splice sites in addition to transcripts that simply skipped exon 9. The pattern of aberrant transcripts, which accounted for 10%–13% of the total product, was reproducible. Thus, a consensus 5′ splice site downstream of exon 9 reduces exon skipping, whereas a consensus 5′ splice site upstream increases exon skipping associated with the abbreviated polypyrimidine tract.
Cross-Species Comparison of Splice Sequences and Exon 9 Skipping
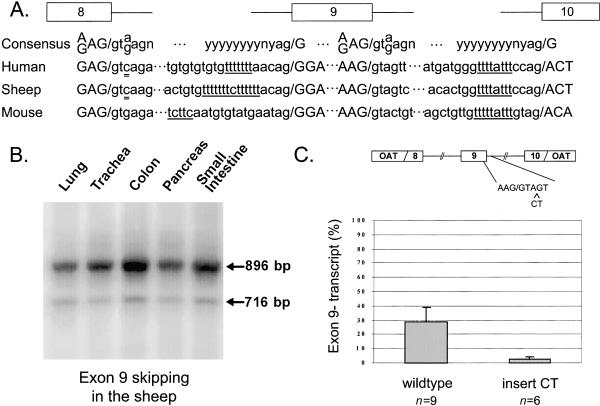
To substantiate hypotheses derived from the minigene studies, we performed a comparison of intron 8 and intron 9 splice-site sequences in humans, mice, and sheep, and we studied exon 9 splicing in sheep (results of human and mouse studies had already been reported elsewhere [Chu et al. 1993; Delaney et al. 1993]). Sheep have upstream and downstream 5′ splice-site sequences very similar to those of humans (score of 80.1, with 2.0 bits upstream, and 69.0, with 4.1 bits downstream). Importantly, however, the polypyrimidine tract in the intron 8 3′ splice site is extensive and is not accompanied by an adjacent poly-TG tract, as it is in humans (fig. 4A). These observations suggested that the exon skipping associated with a brief polythymidine tract and long TG variants in humans should not be present in sheep. Nevertheless, RT-PCR analysis of sheep CFTR transcripts from lung, trachea, colon, pancreas, and small intestine demonstrated that exon 9 is skipped in all sheep tissues studied (fig. 4B), at levels comparable to those of human 9T variants (see legend to fig. 4).
Figure 4.
Cross-species comparison of CFTR exon 9 splice sequences and splicing data. A, Sequence comparison of 5′ and 3′ splice sites of introns 8 and 9 in humans, sheep, and mice. Consensus sequences for 5′ sites are shown at top; y indicates a pyrimidine, and n indicates any nucleotide. Underlines indicate polypyrimidine tracts, double underlines indicate nonconsensus pyrimidines. B, RT-PCR of sheep CFTR RNA, using primers B2R and B2L from several tissues. Proportion of exon 9− transcripts: lung 14.6%, trachea 7%, and pancreas 9.3%. The 9+ product intensities for small intestine and colon were outside the linear range so proportions of 9− could not be calculated. Sequencing revealed that the 896-bp fragment includes exon 9 and the 716-bp fragment is missing exon 9. C, Insertion of a CT dinucleotide into the endogenous human intron 9 5′ splice site replicates mouse intron 9 5′ splice site and results in efficient splicing of exon 9 in the minigene.
On the other hand, mouse sequence at the 5′ splice site upstream of exon 9 is different from those of sheep and humans and conforms to consensus (91.1, 9.1 bits); in addition, the only discernible polypyrimidine tract in the mouse intron 8 3′ splice site is brief, at a distance of 16 bases from the intron/exon junction, and is nonuniform (fig. 4A). Yet exon 9 is not skipped in CFTR transcripts from murine epithelia (Delaney et al. 1993). Interestingly, the mouse downstream 5′ splice site also differs from human and sheep but does not conform to consensus (69.5, 3.8 bits). According to the conclusions drawn from the minigene studies, however, it should serve as an adequate splice site and should efficiently splice exon 9 to exon 10 and reduce skipping. Indeed, replication of the mouse intron 9 5′ splice site by insertion of a CT dinucleotide in the human minigene resulted in a substantial reduction of exon 9 skipping (2.5% [1.9] exon 9−). These data are consistent with the hypothesis that the exact composition of both 5′ splice sites are significant determinants of exon 9 splicing efficiency.
Discussion
Sequence elements that influence splicing efficiency have been identified within and near exon 9, indicating that splicing of this exon is a complex affair involving multiple competing signals (Cuppens et al. 1998; Niksic et al. 1999; Nissim-Rafinia et al. 2000; Pagani et al. 2000; Buratti and Baralle 2001; Buratti et al. 2001). It has been shown that the length of the polypyrimidine tract in intron 8 of the CFTR gene is a primary determinant of the efficiency of exon 9 splicing (Chu et al. 1991, 1992, 1993). However, even the most efficient variant of this tract, with 9T, is associated with some degree of exon 9 skipping (Chu et al. 1993). Our observation that the 5′ splice site upstream of exon 9 deviates from consensus uniquely among 5′ splice sites of the CFTR gene led us to propose that this exon is inherently vulnerable to skipping and that shorter alleles of the polypyrimidine tract exacerbate this skipping. To test this concept, we created a minigene containing only two introns, so the number and type of splicing events that could occur were limited. However, only immediately flanking intron sequence was used, leaving the possibility that critical splicing signals deep within the introns would be missing. For example, others have identified a splicing repressor in intron 9 that may involve intron sequences not present in our minigene (Pagani et al. 2000). Despite these limitations, exon 9 skipping correlated with polypyrimidine tract length in the minigene, as it does in vivo. In addition, we used a cross-species comparison of splice-site sequences and in vivo splicing data to explore hypotheses generated by the minigene studies. Remarkably, despite an extensive polypyrimidine tract, the sheep constitutively skipped exon 9 in vivo. This result demonstrates that sheep exon 9 is vulnerable to skipping, even with an optimal polypyrimidine tract, and supports the view that sequences elsewhere are responsible for exon skipping. On the other hand, exon skipping was absent in the mouse when two strong splice sites and a very brief polypyrimidine tract were present. Thus, comparison of CFTR exon 9 splicing in the sheep and mouse revealed that exon 9 splicing correlated with splice-site composition and not with polypyrimidine tract length. These observations are consistent with the concept that pervasive exon 9 skipping in humans is due to the unusual composition of the flanking 5′ splice sites.
Mutations at splice-site nucleotides other than the invariant GT-AG can and do significantly alter splicing, often resulting in disease (Rogan et al. 1998; Ketterling et al. 1999; Ohno et al. 1999; von Kodolitschet al. 1999; Bolz et al. 2001; Tzetis et al. 2001; Clark and Thanaraj 2002). Documented mutations in 5′ splice sites are twice as abundant as those in 3′ splice sites (Clark and Thanaraj 2002). Although mutations per se have not been documented to alter exon 9 skipping, the above-referenced works point to the profound effects that subtle changes of +3 to +6 nucleotides in 5′ splice sites can have on splicing. Sequence analysis predicted the 5′ splice site upstream of exon 9 to be of low efficiency (computed according to known algorithms [Shapiro and Senapathy 1987; Rogan et al. 1998]). Thus, we expected that mutation of this site to match the consensus sequence would improve the efficiency of the site and would increase exon 9 inclusion. It was therefore surprising that the opposite result was obtained. We considered the following explanations. First, the specific changes we introduced to the upstream site may have reduced, rather than increased, its use because of the sequence context of the region surrounding the exon 8–intron 8 junction. For example, it has been shown that introns tend to favor a G at the +3 position, rather than an A, in GC-rich regions of the genome (Clark and Thanaraj 2002). However, the nucleotide content (∼36% GC) of this region of CFTR is similar to that of the gene as a whole. A second possibility is that the +3C in the upstream site alters splicing efficiency in our minigene system but would not cause the same splicing changes in vivo. We believe that the occurrence of this unusual splice-site feature upstream of a frequently skipped exon that significantly increases that exon’s inclusion (at least in our minigene) seems unlikely to have arisen by chance. Keeping in mind the speculative nature of extrapolating experimental results to in vivo phenomena or evolutionary history, we believe it is possible that the unusual nucleotide in the upstream site was a mutation that was retained because of its tendency to include an otherwise frequently skipped exon. Answering this question will require a more detailed cross-species analysis.
A third possibility requires a consideration of exon definition. Exon definition proposes that the splice sites immediately flanking an exon influence one another, through protein-bridging interactions, and that a change in one can alter binding at the other, thereby changing the efficiency with which the exon is recognized (Berget 1995). Indeed, a 5′ splice site that conforms to consensus is likely to bind U1snRNP more strongly and is capable of enhancing the binding of the essential splicing factor U2AF65 to a weak polypyrimidine tract at the upstream 3′ splice site (Hoffman and Grabowski 1992). It is therefore plausible that the increased inclusion of exon 9 that we observed when the downstream 5′ splice site matched consensus was due to enhanced exon definition. Finally, changes made to the upstream 5′ splice site may have increased its utility as a splice donor, as we predicted; however, in the context of a weak 3′ site preceding exon 9 (because of the 5T allele), it spliced with greater efficiency to exon 10 instead. This would imply that the two 5′ splice sites compete for splicing to exon 10 and that the degree to which exon 9 is included or skipped is a result of the balance achieved by these (and other) splicing signals. There is ample evidence in the literature that the efficiency with which an exon is included is, in part, a result of the balance of neighboring splice sites (Dirksen et al. 1995; Zandberg et al. 1995; Chen and Helfman 1999). Our observation that creation of a consensus upstream 5′ splice site resulted in higher levels of exon skipping, even after the downstream 5′ splice site was mutated to conform to consensus, supports the balance concept. If the changes in the downstream 5′ splice site improved exon definition, then improvement in the upstream 5′ site should have increased, rather than decreased, exon 8 to exon 9 splicing. Thus, we favor a model of exon 9 splicing in which a precarious balance is struck between the upstream and downstream 5′ sites for splicing to exon 10. Variation in other sequence elements (such as the polypyrimidine tract and the various regulatory elements identified by Pagani et al.) tip this balance in favor of exon inclusion or exclusion.
If all the sequence elements thus far identified as affecting exon 9 splicing efficiency are utilized in vivo, then the splicing of exon 9 appears to be a complex event. Whether this complexity arose as a compensation for unfavorable splice-site composition or to facilitate regulation of CFTR transcripts is a subject for future study. Our results emphasize the importance that splice-site nucleotides other than GT-AG can play in making a constitutive, functionally important exon susceptible to being skipped. Finally, these data suggest that thorough inspection of intron/exon junctions in other genes for the presence of unusual splice sites may help to identify other exons vulnerable to skipping.
Acknowledgments
We thank Drs. Hal Dietz and Iain McIntosh (Johns Hopkins University), for key insights and helpful discussions, and Rita McWilliams and Joshua Groman, for statistical analysis. We also thank Drs. Nathalie Mouchel and Scott Tebbutt for ovine sequence analysis. This work was funded by Vaincre La Mucoviscidose, the Cystic Fibrosis Trust (United Kingdom), and grants from the United States National Institutes of Health. F.C.B.-C. is in receipt of a Medical Research Council postgraduate studentship.
Electronic–Database Information
Accession numbers and the URL for data herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CFTR [MIM 602421], CF [MIM 219700], and CBAVD [MIM 277180])
References
- Berget SM (1995) Exon recognition in vertebrate splicing. J Biol Chem 270:2411–2414 [DOI] [PubMed] [Google Scholar]
- Black DL (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103:367–370 [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedo Cabrera M, Vila MC, Molina OP, Gal A, Kubisch C (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27:108–112 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor tdp-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276:36337–36343 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20:1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Helfman DM (1999) Donor site competition is involved in the regulation of alternative splicing of the rat β-tropomyosin pre-mRNA. RNA 5:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillón M, Casals T, Mercier B, Bassas L, Lissens W, Silber S, Romey M-C, Ruiz-Romero J, Verlingue C, Claustres M, Nunes V, Férec C, Estivill X (1995) Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N Engl J Med 332:1475–1480 [DOI] [PubMed] [Google Scholar]
- Chu CS, Trapnell BC, Curristin SM, Cutting GR, Crystal RG (1992) Extensive post-translational deletion of the coding sequences for part of nucleotide-binding fold 1 in respiratory epithelial mRNA transcripts of the cystic fibrosis transmembrane conductance regulator gene is not associated with the clinical manifestations of cystic fibrosis. J Clin Invest 90:785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1993) Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat Genet 3:151–156 [DOI] [PubMed] [Google Scholar]
- Chu CS, Trapnell BC, Murtagh JJ Jr, Moss J, Dalemans W, Jallat S, Mercenier A, Pavirani A, Lecocq JP, Cutting GR (1991) Variable deletion of exon 9 coding sequences in cystic fibrosis transmembrane conductance regulator gene mRNA transcripts in normal bronchial epithelium. EMBO J 10:1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark F, Thanaraj TA (2002) Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet 11:451–464 [DOI] [PubMed] [Google Scholar]
- Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, Jorissen M, Droogmans G, Reynaert I, Goossens M, Nilius B, Cassiman JJ (1998) Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes: The polymorphic (TG)m locus explains the partial penetrance of the 5T polymorphism as a disease mutation. J Clin Invest 101:487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney SJ, Rich DP, Thomson SA, Hargrave MR, Lovelock PK, Welsh MJ, Wainwright BJ (1993) Cystic fibrosis transmembrane conductance regulator splice variants are not conserved and fail to produce chloride channels. Nat Genet 4:426–430 [DOI] [PubMed] [Google Scholar]
- Dirksen WP, Sun Q, Rottman FM (1995) Multiple splicing signals control alternative intron retention of bovine growth hormone pre-mRNA. J Biol Chem 270:5346–5352 [DOI] [PubMed] [Google Scholar]
- Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17:100–107 [DOI] [PubMed] [Google Scholar]
- Hoffman BE, Grabowski PJ (1992) U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev 6:2554–2568 [DOI] [PubMed] [Google Scholar]
- Jarvi K, Zielenski J, Wilschanski M, Durie P, Buckspan M, Tullis E, Markiewicz D, Tsui LC (1995) Cystic fibrosis transmembrane conductance regulator and obstructive azoospermia. Lancet 345:1578 [DOI] [PubMed] [Google Scholar]
- Ketterling RP, Drost JB, Scaringe WA, Liao DZ, Liu JZ, Kasper CK, Sommer SS (1999) Reported in vivo splice-site mutations in the factor IX gene: severity of splicing defects and a hypothesis for predicting deleterious splice donor mutations. Hum Mutat 13:221–231 [DOI] [PubMed] [Google Scholar]
- Kiesewetter S, Macek M Jr, Davis C, Curristin SM, Chu C-S, Graham C, Shrimpton AE, Cashman SM, Tsui LC, Mickle J, Amos J, Highsmith WE Jr, Shuber A, Witt DR, Crystal RG, Cutting GR (1993) A mutation in the cystic fibrosis transmembrane conductance regulator gene produces different phenotypes depending on chromosomal background. Nat Genet 5:274–278 [DOI] [PubMed] [Google Scholar]
- Mak V, Jarvi KA, Zielenski J, Durie P, Tsui LC (1997) Higher proportion of intact exon 9 CFTR mRNA in nasal epithelium compared with vas deferens. Hum Mol Genet 6:2099–2107 [DOI] [PubMed] [Google Scholar]
- Niksic M, Romano M, Buratti E, Pagani F, Baralle FE (1999) Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum Mol Genet 8:2339–2349 [DOI] [PubMed] [Google Scholar]
- Nissim-Rafinia M, Chiba-Falek O, Sharon G, Boss A, Kerem B (2000) Cellular and viral splicing factors can modify the splicing pattern of CFTR transcripts carrying splicing mutations. Hum Mol Genet 9:1771–1778 [DOI] [PubMed] [Google Scholar]
- Noone PG, Pue CA, Zhou Z, Friedman KJ, Wakeling EL, Ganeshananthan M, Simon RH, Silverman LM, Knowles MR (2000) Lung disease associated with the IVS8 5T allele of the CFTR gene. Am J Respir Crit Care Med 162:1919–1924 [DOI] [PubMed] [Google Scholar]
- Ohno K, Brengman JM, Felice KJ, Cornblath DR, Engel AG (1999) Congenital end-plate acetylcholinesterase deficiency caused by a nonsense mutation and an A→G splice-donor-site mutation at position +3 of the collagenlike-tail-subunit gene (COLQ): how does G at position +3 result in aberrant splicing? Am J Hum Genet 65:635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Alton EWFW, Tsui LC (1994) CFTR intron 9 [sic] poly-T tract length in men with congenital bilateral absence of the vas deferens. Ped Pulm Suppl 10:214 [Google Scholar]
- Pagani F, Buratti E, Stuani C, Romano M, Zuccato E, Niksic M, Giglio L, Faraguna D, Baralle FE (2000) Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J Biol Chem 275:21041–21047 [DOI] [PubMed] [Google Scholar]
- Rogan PK, Faux BM, Schneider TD (1998) Information analysis of human splice site mutations. Hum Mutat 12:153–171 [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong TV, Wilkinson DJ, Mansoura MK, Devor DC, Henze K, Yang Y, Wilson JM, Cohn JA, Dawson DC, Frizzell RA (1993) Expression of an abundantly alternatively spliced form of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is not associated with a cAMP-activated chloride conductance. Hum Mol Genet 2:225–230 [DOI] [PubMed] [Google Scholar]
- Tebbutt SJ, Wardle CJ, Hill DF, Harris A (1995) Molecular analysis of the ovine cystic fibrosis transmembrane conductance regulator gene. Proc Natl Acad Sci USA 92:2293–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng H, Jorissen M, Van Poppel H, Legius E, Cassiman JJ, Cuppens H (1997) Increased proportion of exon 9 alternatively spliced CFTR transcripts in vas deferens compared with nasal epithelial cells. Hum Mol Genet 6:85–90 [DOI] [PubMed] [Google Scholar]
- Tzetis M, Efthymiadou A, Doudounakis S, Kanavakis E (2001) Qualitative and quantitative analysis of mRNA associated with four putative splicing mutations (621+3A→G, 2751+2T→A, 296+1G→C, 1717−9T→C−D565G) and one nonsense mutation (E822X) in the CFTR gene. Hum Genet 109:592–601 [DOI] [PubMed] [Google Scholar]
- von Kodolitsch Y, Pyeritz RE, Rogan PK (1999) Splice-site mutations in atherosclerosis candidate genes: relating individual information to phenotype. Circulation 100:693–699 [DOI] [PubMed] [Google Scholar]
- Zandberg H, Moen TC, Baas PD (1995) Cooperation of 5′ and 3′ processing sites as well as intron and exon sequences in calcitonin exon recognition. Nucleic Acids Res 23:248–255 [PMC free article] [PubMed] [Google Scholar]
- Zielenski J, Patrizio P, Corey M, Handelin B, Markiewicz D, Asch R, Tsui LC (1995) CFTR gene variant for patients with congenital absence of vas deferens. Am J Hum Genet 57:958–960 [PMC free article] [PubMed] [Google Scholar]