Abstract
Rationale and Objectives
The clinical gold standard for breast cancer diagnosis relies on invasive biopsies followed by tissue fixation for subsequent histopathological examination. This process renders the specimens to be much less suitable for biochemical or metabolic analysis. Our previous metabolic imaging data in tumor xenograft models showed that the mitochondrial redox state is a sensitive indicator that can distinguish between normal and tumor tissue. In this study, we investigated whether the same redox imaging technique can be applied to core biopsy samples of human breast cancer and whether the mitochondrial redox state may serve as a novel metabolic biomarker that may be used to distinguish between normal and malignant breast tissue in the clinic. Our long-term objective was to identify novel metabolic imaging biomarkers for breast cancer diagnosis.
Materials and Methods
Both normal and cancerous tissue specimens were collected from the cancer-bearing breasts of three patients shortly after surgical resection. Core biopsies and tissue blocks were obtained from tumor and normal adjacent breast tissue, respectively. All specimens were snap-frozen with liquid nitrogen, embedded in chilled mounting medium with flavin adenine dinucleotide and reduced nicotinamide adenine dinucleotide reference standards adjacently placed, and scanned using the Chance redox scanner (ie, cryogenic nicotinamide adenine dinucleotide/oxidized flavoprotein fluorescence imager).
Results
Our preliminary data showed cancerous tissues had up to 10-fold higher oxidized flavoprotein signals and had elevated oxidized redox state compared to the normal tissues from the same patient. A high degree of tumor tissue heterogeneity in the redox indices was observed.
Conclusions
Our finding suggests that the identified redox imaging indices could differentiate between cancer and noncancer breast tissues without subjecting tissues to fixatives. We propose that this novel redox scanning procedure may assist in tissue diagnosis in freshly procured biopsy samples before tissue fixation.
Keywords: Mitochondria, oxidized flavoprotein (Fp), NADH, redox state, metabolic biomarker, the Chance redox scanner
Biochemical analysis of cancer core biopsies has little diagnostic value in the clinic so far. In this study, we demonstrated a novel approach using the intrinsic biochemical properties of breast tissues as measured by the Chance redox scanner (named after Dr. Britton Chance) (1–3) to distinguish between normal and cancerous breast tissues using clinical biopsy samples. The Chance redox scanner is a low-temperature ex vivo fluorescence imaging system that uses a raster-scanning fiberoptic probe coupled with a pair of rotating filter wheels to select proper excitation and emission light wavelengths to measure the endogenous fluorescence signals of reduced nicotinamide adenine dinucleotide (NADH) and oxidized flavoprotein (Fp) including flavin adenine dinucleotide (FAD) in tissue. With a tissue sample mounted in a liquid nitrogen-filled chamber and a miller to remove top tissues at different depth (z axis), the scanner can image the three-dimensional distribution of NADH and Fp with a high spatial resolution down to 50 × 50 × 20 µm3. It requires only a small tissue sample (1 mm in two dimensions) for measurement of tissue mitochondrial redox states.
Redox scanning can become a diagnostic tool. NADH and reduced FAD are reducing equivalents used for the electron transport chain to generate ATP in mitochondria. It has been shown that the redox ratio [Fp/NADH or the normalized form, Fp/(Fp + NADH)] was a good representation of the tissue redox state and a sensitive indicator of mitochondrial metabolic state (1,4). It was discovered previously that the Fp redox ratio [ie, Fp/(Fp + NADH)] correlated with the degree of tumor invasiveness in mouse xenografts of human melanoma and breast cancer cell lines (3,5–7). In another study, the premalignant pancreatic tissue in the PTEN-null transgenic mouse model was shown to be more oxidized and heterogeneous in the mitochondrial redox state than the normal one (8,9). Redox scanning was also used to image the redox ratio variations in human dysplastic cervical tissues compared to normal controls (10).
Although the redox state of tissues may serve as a promising cancer biomarker, redox scanning has not been fully evaluated as a diagnostic tool for breast cancer. The current gold standard to establish malignant versus benign breast tissue diagnosis requires an invasive biopsy followed by tissue fixation for subsequent histological examination. The tissue fixation renders the tissue less suitable for additional biochemical and metabolic analyses such as redox scanning. Redox scanning requires viable tissue with a preserved metabolic state, whereas histological evaluation often requires tissue fixation resulting in nonviable tissues. These two seemingly incompatible requirements are now met by our redox imaging protocol using snap-frozen biopsy samples, which may be further processed for histological evaluation.
We report the preliminary results on imaging the redox state of the breast biopsies from patients to discriminate between cancer and noncancer tissues. A portion of the data were previously reported in a conference proceeding (11). Our results indicate that this novel redox scanning procedure on clinical specimens may assist in cancer diagnosis in ex vivo tissues.
METHODS
Tissue collection from patients was performed according to a protocol approved by the internal review board of the University of Pennsylvania. Patient 1 was a 40-year-old woman with a 2.3-cm poorly differentiated, grade 3, and stage II invasive ductal carcinoma (IDC), with triple negative (TRN) status for estrogen/progesterone receptors (ER/PR) and human epidermal growth factor receptor 2 (HER2), negative nodes. Patient 2 was a 41-year-old woman with a poorly differentiated, grade 3, and stage II 3.2-cm IDC with BRCA1 mutation, TRN, and negative nodes. Patient 3 was a 83-year-old woman with a 5.5-cm invasive lobular carcinoma (ILC) with ER greater than 90%, PR 30%, HER2-negative, and positive nodes (2/12). This patient had locally advanced stage III breast cancer and had been receiving anastrozole (arimidex) for about 8 months.
Both normal and cancerous tissues were collected from the affected breasts. Shortly after completion of surgical resection to remove the breast cancer, two to four biopsy specimens were collected using a 20-gauge core biopsy needle (~1 × 1 × 5 mm3) from the central portion of the palpable tumor within the surgical specimen. A thin tissue block from normal adjacent breast tissue at the periphery of the surgical specimen was also obtained right after the core biopsy samples were taken. All tissue specimens were then immediately stored in liquid nitrogen until redox scanning was performed.
Breast tissues collected from patient 1 were placed on saline-moistened paper at room temperature and then wrapped with aluminum foil and dipped into liquid nitrogen. The estimated time interval between tumor resection from the body and the specimen snap-freezing was 5 to 10 minutes. The core biopsy specimens from various locations in the surgically removed tumor from patient 1 were dipped into liquid nitrogen temporally in the following order: a, b, d with a time interval of about 30 to 60 seconds between consecutive specimens. The breast tissues from patient 2 were placed on saline-moistened paper surrounded by ice for 5 minutes after the biopsy. The estimated time interval between tumor resection from the body and the specimen snap-freezing was 10 to 15 minutes. Both cancerous and normal tissues were given as a thin tissue block of about 0.3 × 2 × 2 cm3 for samples from patient 3. The estimated time interval between tumor removal from the patient and snap-freezing was about 5 minutes.
Small portions of the frozen core biopsies were directly embedded in a glycerol/ethanol/water mounting buffer according to the procedure described previously (7,8,12,13). The tissue pieces sliced off from the snap-frozen tissue blocks were embedded in the mounting buffer in such a way that the starting layer for redox scanning was the surface exposed to the air before snap-freeze. NADH and FAD reference standards (NADH 863 µM and FAD 500 µM in Tris pH~7 buffer for all samples except for the control sample of patient 2:NADH 1150 µM and FAD 750 µM) were placed adjacently to the tissue specimen. Cold mounting buffer slush was added on top of the tissues to secure them.
The embedded samples were first carefully milled by the miller equipped with the redox scanner under liquid nitrogen to expose the tissue surface. The exposed surface was then raster-scanned two-dimensionally with a fiberoptic probe coupled to a pair of time-sharing four-channel filter wheels that include an Fp channel (excitation 430 ± 25 nm, emission 525 ± 32 nm) and an NADH channel (excitation 360 ± 13 nm, emission 430 ± 25 nm) (7,12). The typical scanning time for an image matrix of 64 × 64 and step size of 100 µm is about 30 minutes. Two to four sections spacing between 40 and 120 µm (depth range 0 to 240 µm) were scanned for each core biopsy sample. Two sections with spacing of 200 µm were scanned for each tissue block sample. The Fp and NADH fluorescence signals were collected with a photomultiplier tube, and the data acquisition was controlled with a computer.
A customized MATLAB program was used to construct all images of NADH and Fp and the redox ratios. A region of interest (ROI) was carefully drawn along the tissue boundary to mask out the background, and a signal-to-noise-ratio threshold of 3 was used to remove low-signal pixels within the ROI. Both the NADH and Fp images are displayed as concentrations in the unit of µmol/L that is calibrated to the fluorescence from the corresponding standards. The Fp redox ratio images are displayed as the concentration-based ratio Fp/(Fp + NADH) in the range of 0 to 1.
Values of the redox indices (Fp, NADH, and the Fp redox ratio) were first averaged for the ROI in each individual section, then averaged across sections of a specimen, and then averaged across tissue specimens from the same patient. The reported group means were then obtained by averaging those means across patients. The reported standard deviations are the variations in the final distribution of means across patients. The statistical significance between the cancerous and normal tissues was first tested by the paired Student t test comparing the mean values of the redox indices of the cancerous tissue to those of the normal tissue for each patient and then tested by the univariate analysis of general linear method (IBM SPSS statistics version 20; http://www-01.ibm.com/software/analytics/spss/) with tissue depth as a covariate. The univariate analysis results were further confirmed by the multivariate analysis of the general linear method using the same software on the three redox indices together. For multiple tissue samples from patient 1, single-factor analysis of variance was performed to compare the differences among the mean values of three core biopsies from different locations. For each patient, we also calculated the ratio of Fptumor to Fpnormal (ie, the mean Fp value of the cancerous tissue divided by that of the normal tissue).
RESULTS AND DISCUSSION
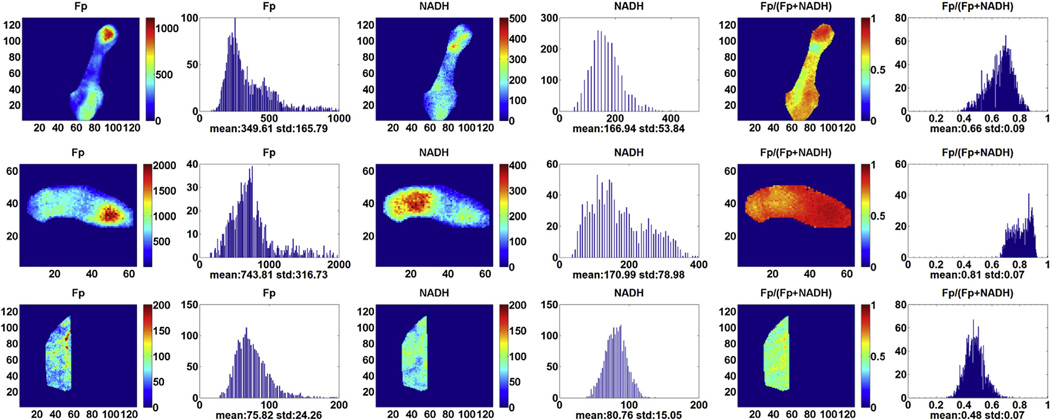
The typical redox images of breast tumor tissue and normal breast tissue are displayed in Figure 1. The top two rows show the redox images of the tumor tissue from patient 1 at different locations. The bottom row contains the redox images of the normal tissue from the same patient. Tissue heterogeneity can be clearly seen in all of the images. We quantified the redox indices using the global averaging method and tabulated the results in Table 1. The first and most notable finding is the high values of oxidized Fps, suggesting that the electron transfer chain to oxygen was operating efficiently before snap-freezing or cells were undergoing starvation or apoptosis. Fp nominal concentration in cancerous tissue (652 ± 93 µmol/L) is significantly higher than that in normal tissue (118 ± 89 µmol/L) with P = .027. There is no significant difference in NADH between cancerous and normal tissues.
Figure 1.
Typical redox images of breast tumor tissue at different locations of patient 1 (top two rows, image matrix 128 × 128 and 64 × 64, resolution 40 µm) and normal breast tissue of the same patient (bottom row, image matrix 128 × 128, resolution 100 µm). The redox ratio ranges between 0 and 1; the flavoproteins (Fp) or reduced nicotinamide adenine dinucleotide (NADH) images are in the unit of µmol/L in reference to the corresponding standards. The x axes of the corresponding histograms represent the redox ratio or concentration. The y axes represent the number of pixels in the tissue region of interest having a specific value of redox ratio or concentration.
TABLE 1.
Values of the Redox Indices of the Tumor and Normal (Control) Breast Tissues from Three Patients Analyzed by the Global Averaging Method
| Type | Fp, µmol/L |
NADH, µmol/L |
Fp Redox Ratio |
|---|---|---|---|
| Control (n = 3) | 118 ± 89 | 138 ± 45 | 0.40 ± 0.09 |
| Tumor (n = 3) | 652 ± 93 | 382 ± 283 | 0.61 ± 0.10 |
| P (paired t test) | .027 | .25 | .071 |
Fp, flavoproteins; NADH, reduced nicotinamide adenine dinucleotide.
The next noticeable difference is the Fp redox ratio between tumor and normal control tissue bearing a marginal significance. Because the global averaging method largely ignores tissue heterogeneity along tissue depth, we further analyzed the data using the univariate model of the general linear method with tissue depth as a covariate to test the statistical significance. The result showed that the Fp redox ratio of tumor tissue becomes significantly different from the normal control (P < .001) (Table 2). The statistical significance of the result was further confirmed by the multivariate analysis. Higher Fp redox ratio indicated that tumor tissue was more oxidized than normal tissue, which is consistent with our previous studies on mouse xenografts (5–7).
TABLE 2.
Values of the Redox Indices of the Tumor and Normal (Control) Breast Tissues from Three Patients Analyzed by the General Linear Method Univariate Analysis
| Type | Fp, µmol/L |
NADH, µmol/L |
Fp Redox Ratio |
|---|---|---|---|
| Control (6 sections) | 118 ± 85 | 138 ± 42 | 0.40 ± 0.08 |
| Tumor (14 sections) | 620 ± 428 | 251 ± 210 | 0.67 ± 0.12 |
| P | .015 | .20 | 1.72E–04 |
Fp, flavoproteins; NADH, reduced nicotinamide adenine dinucleotide.
The ratios of the mean Fp values between tumor and normal tissues (Fptumor/Fpnormal) for the three patients are 9.2, 11.1, and 2.7 for patient 1, 2, and 3, respectively. The average of Fptumor/Fpnormal from the three patients is 7.7 ± 4.4, varying largely among patients, likely due to the differences in clinical characteristics and treatment history of each patient. Patients 1 and 2 both were diagnosed with IDC that did not express ER, PR, or HER2-neu receptors (ie, TRN) and did not receive neoadjuvant therapy. Both tumors had much higher ratios of Fptumor/Fpnormal (almost 10-fold) than that of patient 3, who was diagnosed with a locally advanced ILC that was ER positive and had been treated with an aromatase inhibitor, anastrozole, for 8 months before her surgery. Whether the ratio Fptumor/Fpnormal may correlate with clinical characteristics of breast cancer such as breast cancer subtype or other prognostic factors warrants further investigation using a larger sample size.
Despite our limited sample size and heterogeneous tumor characteristics, we observed that the Fp level was strikingly higher in cancerous tissues (up to 10-fold higher) and cannot be fully accounted for by the slightly higher oxygen diffusion coefficient in tumor tissue than that in normal tissue (14–18). The tissue-penetration distance of oxygen via diffusion ranges from 800 to 1100 µm for a time window of 5 to 10 minutes assuming a diffusion constant D of 10−5 cm2 /s and the formula of distance = for 1D diffusion along the z axis, where t is the diffusion time. The reported redox scanning data were from the top layers of both the tissue blocks within about 800 µm and the core biopsy samples within about 500 µm. Thus, it is reasonable to assume that the reoxygenation due to oxygen diffusion should be similar for both normal and cancerous tissue samples.
Furthermore, for each patient the normal tissue was harvested immediately after the procurement of cancer specimens. It is reasonable to assume that both the cancerous and normal tissues from the same patient underwent a similar temporal process before snap-freezing. The ratio Fptumor/Fpnormal from the same patient’s breast thus may cancel out or largely reduce the influence of the differences in the sample’s lag time before snap-freezing among the three patients. Therefore, the ratio Fptumor/Fpnormal presumably reflects some intrinsic biological characteristics of each individual breast tumor.
It is well known that cancer tissue is often heterogeneous in cellular composition, function, and metabolism. Apart from the intrasample heterogeneity as shown by the redox images in Figure 1, we also identified intersample heterogeneity among several samples obtained from patient 1. Table 3 summarizes the redox indices of tumor core biopsies obtained by core needle at different locations from patient 1. The highly significant difference in the redox indices is readily recognized. The increase of all the redox indices (NADH and Fp concentrations and the Fp redox ratio) corresponds with the temporal order of the sample snap-freezing a → b → d. But this cannot be explained completely by the increased reoxygenation due to delayed snap-freeze and longer air exposure, which should result in increasing Fp and decreasing NADH. Therefore, the redox signal heterogeneity observed may partially reflect the intrinsic tumor heterogeneity in metabolism.
TABLE 3.
The Redox Indices of the Cancerous Core Biopsies Taken from Different Locations of Patient 1
| Tissue Location | Fp Redox Ratio |
NADH, µmol/L |
Fp, µmol/L |
| a (2 sections) | 0.54 ± 0.01 | 45 ± 13 | 57 ± 20 |
| b (4 sections) | 0.69 ± 0.03 | 139 ± 28 | 346 ± 38 |
| d (5 sections) | 0.82 ± 0.03 | 221 ± 38 | 1115 ± 344 |
| P* | .00001 | .0004 | .0017 |
Fp, flavoproteins; NADH, reduced nicotinamide adenine dinucleotide.
Single-factor analysis of variance was used to show the statistical difference between locations.
Further investigation is warranted to evaluate the contribution of the intrinsic biological difference among tissues, the air-exposure difference or other factors in sample handling procedures in affecting the marked differences in the redox indices between the normal and cancerous tissues among the three patients and within the same patient (as noted in patient 1). We will standardize the sample collection procedure to minimize its possible contribution to the redox indices in the future and perform redox scanning on more samples to confirm the results presented here.
CONCLUSIONS
The mitochondrial redox states of human breast cancer tissue samples measured fluorometrically with the Chance redox scanner showed marked differences between the cancerous and normal tissues. The breast cancer tissue exhibited significantly higher signals of oxidized flavoproteins and more oxidized redox state than the normal tissues. The redox scanning results clearly indicated that the redox state in tumor tissues is heterogeneous. We propose that this novel redox scanning procedure may be used as a metabolic and complementary cancer diagnosis tool in freshly procured tissues such as those obtained at the time of percutaneous core needle biopsy for diagnostic purposes in the clinical setting. Redox imaging of breast biopsies may open up new avenues to explore the diagnostic and prognostic value of the redox state of cancer tissues.
ACKNOWLEDGMENTS
This article is dedicated to the memory of Dr. Britton Chance, who devoted himself to the research of this study with excitement and sleepless nights and his profound insights into science as well as his great attention to detail until the last moment of his life. This work was supported by Susan G. Komen Foundation grant KG081069 (L.Z.L.), National Institutes of Health (NIH) grant R01CA155348 (L.Z.L.), National Cancer Institute Cancer Center Support Grant 2-P30-CA-016520-35 (J.T.), Linda and Paul Richardson Breast Cancer Research Funds (J.T.), Center of Magnetic Resonance and Optical Imaging (CMROI) – an NIH supported research resource grant P41RR02305 (R.R.), and Small Animal Imaging Program (SAIR) grant 2U24-CA083105 (J.G. and L.C.). We are grateful for Dr H. Zhao’s help on the statistical analysis of our data and the help of Ms Min Feng in collecting some of the biopsy samples.
REFERENCES
- 1.Chance B, Schoener B, Oshino R, et al. Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem. 1979;254(11):4764–4771. [PubMed] [Google Scholar]
- 2.Quistorff B, Haselgrove JC, Chance B. High spatial resolution readout of 3-D metabolic organ structure: an automated, low-temperature redox ratio-scanning instrument. Anal Biochem. 1985;148(2):389–400. doi: 10.1016/0003-2697(85)90244-1. [DOI] [PubMed] [Google Scholar]
- 3.Li LZ, Xu HN, Ranji M, et al. Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J Innov Optical Health Sci. 2009;2:325–341. doi: 10.1142/S1793545809000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozawa K, Chance B, Tanaka A, et al. Linear correlation between acetoa-cetate beta-hydroxybutyrate in arterial blood and oxidized flavoprotein reduced pyridine-nucleotide in freeze-trapped human liver-tissue. Biochim Biophys Acta. 1992;1138(4):350–352. doi: 10.1016/0925-4439(92)90014-e. [DOI] [PubMed] [Google Scholar]
- 5.Li LZ, Zhou R, Zhong T, et al. Predicting melanoma metastatic potential by optical and magnetic resonance imaging. Adv Exp Med Biol. 2007;599:67–78. doi: 10.1007/978-0-387-71764-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li LZ, Zhou R, Xu HN, et al. Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proc Natl Acad Sci U S A. 2009;106(16):6608–6613. doi: 10.1073/pnas.0901807106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu HN, Nioka S, Glickson JD, et al. Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J Biomed Optics. 2010;15(3):036010. doi: 10.1117/1.3431714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu HN, Nioka S, Chance B, et al. Heterogeneity of mitochondrial redox state in premalignant pancreas in a PTEN null transgenic mouse model. Adv Exp Med Biol. 2011;201:207–213. doi: 10.1007/978-1-4419-7756-4_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu HN, Nioka S, Chance B, et al. Imaging heterogeneity in the mitochondrial redox state of premalignant pancreas in the pancreas-specific PTEN-null transgenic mouse model. Biomarker Res. 2013;1:6. doi: 10.1186/2050-7771-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramanujam N, Richards-Kortum R, Thomsen S, et al. Low temperature fluorescence imaging of freeze-trapped human cervical tissues. Opt Express. 2001;8(6):335–343. doi: 10.1364/oe.8.000335. [DOI] [PubMed] [Google Scholar]
- 11.Xu HN, Tchou J, Chance B, et al. Imaging the redox states of human breast cancer core biopsies. Adv Exp Med Biol. 2013;765:343–349. doi: 10.1007/978-1-4614-4989-8_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu HN, Wu B, Nioka S, et al. Quantitative redox scanning of tissue samples using a calibration procedure. J Innov Opt Health Sci. 2009;2:375–385. doi: 10.1142/S1793545809000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu HN, Wu B, Nioka S, et al. Calibration of redox scanning for tissue samples. Proc SPIE. 2009;7174:71742F. doi: 10.1142/S1793545809000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans NTS, Naylor PFD, Quinton TH. The diffusion coefficient of oxygen in respiring kidney and tumour tissue. Respir Physiol. 1981;43(3):179. doi: 10.1016/0034-5687(81)90100-6. [DOI] [PubMed] [Google Scholar]
- 15.Groebe K, Vaupel P. Evaluation of oxygen diffusion distances in human breast cancer xenografts using tumor-specific in vivo data: role of various mechanisms in the development of tumor hypoxia. Int J Radiat Oncol Biol Physics. 1988;15(3):691. doi: 10.1016/0360-3016(88)90313-6. [DOI] [PubMed] [Google Scholar]
- 16.Krogh A. The rate of diffusion of gases through animal tissues, with some remarks on the coefficient of invasion. J Physiol. 1919;52(6):391–408. doi: 10.1113/jphysiol.1919.sp001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macdougall JDB, McCabe M. Diffusion coefficient of oxygen through tissues. Nature. 1967;215(5106):1173. doi: 10.1038/2151173a0. [DOI] [PubMed] [Google Scholar]
- 18.Vaupel P, Mayer A, Briest S, et al. Hypoxia in breast cancer: role of blood flow, oxygen diffusion distances, and anemia in the development of oxygen depletion. Adv Exp Med Biol. 2005;566:333. doi: 10.1007/0-387-26206-7_44. [DOI] [PubMed] [Google Scholar]