Abstract
The Ta (transcribed, subset a) subfamily of L1 LINEs (long interspersed elements) is characterized by a 3-bp ACA sequence in the 3′ untranslated region and contains ∼520 members in the human genome. Here, we have extracted 468 Ta L1Hs (L1 human specific) elements from the draft human genomic sequence and screened individual elements using polymerase-chain-reaction (PCR) assays to determine their phylogenetic origin and levels of human genomic diversity. One hundred twenty-four of the elements amenable to complete sequence analysis were full length (∼6 kb) and have apparently escaped any 5′ truncation. Forty-four of these full-length elements have two intact open reading frames and may be capable of retrotransposition. Sequence analysis of the Ta L1 elements showed a low level of nucleotide divergence with an estimated age of 1.99 million years, suggesting that expansion of the L1 Ta subfamily occurred after the divergence of humans and African apes. A total of 262 Ta L1 elements were screened with PCR-based assays to determine their phylogenetic origin and the level of human genomic variation associated with each element. All of the Ta L1 elements analyzed by PCR were absent from the orthologous positions in nonhuman primate genomes, except for a single element (L1HS72) that was also present in the common (Pan troglodytes) and pygmy (P. paniscus) chimpanzee genomes. Sequence analysis revealed that this single exception is the product of a gene conversion event involving an older preexisting L1 element. One hundred fifteen (45%) of the Ta L1 elements were polymorphic with respect to insertion presence or absence and will serve as identical-by-descent markers for the study of human evolution.
Introduction
Computational analysis of the draft sequence of the human genome indicates that repetitive sequences comprise 45%–50% of the human genome mass, 17% of which consists of ∼500,000 L1 LINEs (long interspersed elements) (Smit 1999; Prak and Kazazian 2000; Lander et al. 2001). L1 elements are restricted to mammals, having expanded as a repeated DNA sequence family over the past 100–150 million years (Smit et al. 1995). Full-length L1 elements are ∼6 kb long and amplify via an RNA intermediate in a process known as “retrotransposition.” L1 integration likely occurs by a mechanism termed “target-primed reverse transcription” (Luan et al. 1993; Kazazian and Moran 1998). This mechanism of mobilization provides two useful landmarks for the identification of L1Hs (L1 human specific) inserts: an endonuclease-related cleavage site (Jurka 1997; Cost and Boeke 1998; Cost et al. 2001) and direct repeats or target site duplications flanking newly integrated elements (Fanning and Singer 1987; Kazazian 2000).
L1 retrotransposons have had a significant impact on the human genome, through recombination (Fitch et al. 1991), alteration of gene expression (Yang et al. 1998; Rothbarth et al. 2001), and de novo insertions that disrupt ORFs and splice sites resulting in human disease (Kazazian et al. 1988; Kazazian 1998; Kazazian and Moran 1998). L1 elements are also able to transduce adjacent genomic sequences at their 3′ end, facilitating exon shuffling (Boeke and Pickeral 1999; Moran et al. 1999; Goodier et al. 2000). In addition, individual mobile elements may undergo post-integration gene conversion events in which short DNA sequences are exchanged by an undefined mechanism, thereby altering the levels of SNP associated with the individual L1 elements (Hardies et al. 1986). Thus, LINEs have exerted a significant influence on the architecture of the human genome.
Even though there are ∼500,000 L1 elements in the human genome, only a limited subset of L1 elements appear to be capable of retrotransposition (Moran et al. 1996; Sassaman et al. 1997). As a result of the limited amplification potential of this diverse gene family, a series of discrete subfamilies of L1 elements exists within the human genome (Deininger et al. 1992; Smit et al. 1995). Each of the L1 subfamilies appears to have amplified within the human genome at different times in primate evolution, making them different genetic ages (Deininger et al. 1992; Smit et al. 1995). The most recently integrated L1 elements within the human genome share a common 3-bp diagnostic sequence within the 3′ UTR, and they comprise almost all of the de novo disease-associated L1 elements within the human genome, as well as several elements that have been shown to be capable of retrotransposition in cell culture (Kazazian and Moran 1998; Boissinot et al. 2000; Sheen et al. 2000). This subfamily was first identified in human teratocarcinoma cells and has been collectively termed “Ta” (for transcribed, subset a) (Skowronski et al. 1988). Some members of the L1 Ta subfamily have inserted in the human genome so recently that they are polymorphic with respect to insertion presence/absence (Boissinot et al. 2000; Sheen et al. 2000). The L1 insertion polymorphisms are a useful source of identical-by-descent variation for the study of human population genetics (Boissinot et al. 2000; Santos et al. 2000; Sheen et al. 2000). Here, we report the analysis of the Ta subfamily of L1 elements from the draft sequence of the human genome.
Material and Methods
Cell Lines and DNA Samples
The cell lines used to isolate primate DNA samples were as follows: human (Homo sapiens) HeLa (American Type Culture Collection [ATCC] number CCL2), common chimpanzee (Pan troglodytes) Wes (ATCC number CRL1609), pygmy chimpanzee (P. paniscus) (Coriell Cell Repository number AG05253), gorilla (Gorilla gorilla) Lowland Gorilla (Coriell Cell Repository number AG05251B), green monkey (Cercopithecus aethiops) (ATCC number CCL70), and owl monkey (Aotus trivirgatus) (ATCC number CRL1556). Cell lines were maintained as directed by the source and DNA isolations were performed using Wizard genomic DNA purification (Promega). Human DNA samples from the European, African American, Asian or Alaskan native, and Egyptian population groups were isolated from peripheral blood lymphocytes (Ausabel et al. 1987), as described elsewhere (Stoneking et al. 1997).
Computational Analyses
The draft sequence of the human genome was screened using the Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990), available at the National Center for Biotechnology Information genomic BLAST Web site. A 19-bp oligonucleotide (5′-CCTAATGCTAGATGACACA-3′) that is diagnostic for the L1Hs Ta subfamily was used to query the human genome database with the following optional parameters: filter none and advanced options −e 0.01, −v 600, and −b 600. Copy-number estimates were determined from BLAST search results. Sequences that contained exact matches were subjected to additional analysis as outlined below.
A sequence region of 9,000–10,000 bp, including the match and 1,000–2,000 bp of flanking unique sequence, was annotated using RepeatMasker (version 7/16/00), from the University of Washington Genome Center, or Censor, from the Genetic Information Research Institute (Jurka et al. 1996). These programs annotate repeat-sequence content and were used to confirm the presence of L1Hs elements and regions of unique sequence flanking the elements. PCR primers flanking each L1 element were designed using Primer3 software, available from the Whitehead Institute for Biomedical Research, and were complementary to the unique sequence regions flanking each L1 element. The resultant primers were screened, by standard nucleotide-nucleotide BLAST (blastn), against the nonredundant (nr) and high-throughput (htgs) sequence databases, to ensure that they resided in unique DNA sequences. Primers that resided in repetitive sequence regions were discarded, and, if possible, new primers were then designed. A complete list of all the L1 elements that were identified using this approach and supplemental material from this manuscript are available from the Batzer Lab Web site, in the “Publications” section. Individual L1 DNA sequences were aligned using MegAlign, with the Clustal V algorithm and the default settings (DNAstar, version 5.0 for Windows), followed by manual refinement.
PCR Amplification
PCR amplification of 262 individual L1 elements was performed in 25-μl reactions that contained 50–100 ng of template DNA; 40 pmol of each oligonucleotide primer (table A1see table A1, available online only); 200 μM of deoxyribonucleoside triphosphates, in 50 mM KCl and 10 mM Tris-HCl (pH 8.4); 1.5 mM MgCl2; and 1.25 U of Taq DNA polymerase. Each sample was subjected to the following amplification conditions for 32 cycles: an initial denaturation at 94°C for 150 s, 1 min denaturation at 94°C, and 1 min at the annealing temperature (specific for each locus, as shown in table 1 and appendix A, available online onlyappendix A), followed by extension at 72°C for 10 min. For analysis, 20 μl of each sample was fractionated on a 2% agarose gel with 0.05 μg/ml ethidium bromide. PCR products were directly visualized using UV fluorescence. The human genomic diversity associated with each Ta L1 element was determined by the amplification of 20 individuals from each of four geographically distinct populations (African American, Asian or Alaskan native, European German, and Egyptian).
Table 1.
Summary of Ta L1 Element Computational and PCR Analysis[Note]
| Classification | No. ofElements |
| Successful PCR analysis | 262 |
| L1 elements inserted in other repeats | 137 |
| L1 elements located at the end of sequencing contigs | 69 |
| Total Ta L1 elements analyzed | 468 |
Note.— A full summary of GenBank accession numbers, PCR primers and conditions, and PCR amplicon sizes for these loci is shown in table A1table A1, available online only, and is also available at the Batzer Lab Web site.
Cloning and Sequence Analysis
L1 element–related PCR products were cloned using the Invitrogen TOPO TA Cloning Kit, according to the manufacturer's instructions, and were sequenced using an Applied Biosystems 3100 automated DNA sequencer, by the chain-termination method (Sanger et al. 1977). The DNA sequence for the common and pygmy chimpanzee orthologs of L1HS72 were assigned GenBank accession numbers AF489459 and AF489460, respectively. Additional diverse human sequences from L1HS72 were assigned GenBank accession numbers AF489450–AF489458. DNA sequences derived from L1 pre-integration sites were assigned GenBank accession numbers AF461364, AF461365, AF461368–AF461383, AF461386, and AF461387.
Results
L1 Ta Subfamily Copy Number and Age
To identify recently integrated Ta L1 elements from the human genome, we searched the draft sequence of the human genome (BLASTN database, version 2.2.1), using BLAST (Altschul et al. 1990) with an oligonucleotide that is complementary to a highly conserved sequence in the 3′ UTR of Ta L1 elements. This 19-bp query sequence (CCTAATGCTAGATGACACA) includes the Ta subfamily–specific diagnostic mutation ACA at its 3′ end at positions 5930–5932 relative to L1 retrotransposable element–1 (Dombroski et al. 1991). We identified 468 unique Ta L1 elements from 2.868×109 bp of available human draft sequence. Extrapolating this number to the actual size of the human genome (3.162×109 bp), we estimate that this subfamily contains ∼520 elements. Of the 468 elements retrieved, 69 resided at the end of sequence contigs and were not amenable to additional in vitro wet-bench analysis. Of the 399 remaining elements, 124 (31%) of the elements were essentially full length, and the remaining 275 were truncated to variable lengths. Alignment and sequence analysis of the full-length elements revealed that 44 contained two intact ORFs and therefore may be capable of retrotransposition. This estimate of putative retrotransposition-competent L1 elements is in good agreement with the initial analysis of the draft sequence of the human genome (Lander et al. 2001).
The ages of L1 elements can be determined by the level of sequence divergence from the subfamily consensus sequence by use of a neutral mutation rate for primate noncoding sequence of 0.15% per million years (Miyamoto et al. 1987). The mutation rate is known to be ∼10 times greater for CpG bases as compared to non-CpG bases, as a result of the spontaneous deamination of 5-methyl cytosine (Bird 1980). Thus, two age estimates that are based on CpG and non-CpG mutations can be calculated for the Ta subfamily of L1 elements. A total of 89,929 bp from the 3′ UTR of 459 Ta L1Hs elements were analyzed, and L1 elements characterized elsewhere were excluded from this analysis—along with nine elements that, according to the nucleotide present at position 6015 in the 3′ UTR of the elements, do not technically belong to the Ta subfamily (Ovchinnikov et al. 2001). Three hundred thirty-one total nucleotide substitutions were observed. Of these, 263 were classified as non-CpG mutations against the backdrop of 88,141 total non-CpG bases, thereby producing a non-CpG mutation density of 0.002984. Based on the non-CpG mutation density and a neutral rate of evolution (0.002984/0.0015), the average age of the Ta L1 elements was 1.99 million years. A total of 68 CpG mutations were found across these 459 L1 elements from 1,788 total CpG nucleotides, thereby yielding a CpG-mutation rate of 0.038031. With the expectation that the CpG mutation rate is ∼10-fold higher than the non-CpG mutation rate, the approximate age (obtained using the CpG mutation density) of the L1Hs Ta subfamily is 2.54 million years. These estimates are in good agreement with one another, as well as with previous estimates derived from an analysis of a small number of Ta L1 elements (Boissinot et al. 2000).
Nine of the 468 elements analyzed do not technically belong to the Ta subfamily of L1 elements, on the basis of a single-nucleotide substitution (L1HS19, -72, -274, -309, -318, -325, -390, -399, and -493) that is also considered diagnostic for the L1 Ta subfamily. Although they all have the 19-bp query sequence ending in ACA in the 3′ UTR at positions 5930–5932, they lack a G at position 6015 (Ovchinnikov et al. 2001) and instead contain an A at that position, which is a diagnostic feature found in older primate-specific L1PA10–L1PA2 subfamilies (Smit et al. 1995). Thus, these elements may be Ta L1 elements that have undergone fortuitous single-base substitutions of the ancestral nucleotide, may be Ta L1 elements that have undergone backward gene-conversion events, or may simply be older, “pre-Ta” L1 elements that were generated by a source gene (or source genes) that did not contain this diagnostic base. To determine the effect that the Ta versus non-Ta designation has on the calculated age estimate, we examined a total of 1,807 bp from the 3′ UTRs of these nine elements. There were 27 non-CpG mutations from a total of 1,771 non-CpG bases, thereby yielding a mutation density of 27/1,771, or 0.015246. Dividing by the neutral rate of evolution for primate noncoding sequence (0.015246/0.0015), we arrive at an estimated age of 10.16 million years. This is significantly older than the average age of 2.26 million years that was calculated from the larger data set (i.e., the data set of Ta L1 elements only). The CpG mutation density in the elements was also calculated. There were 2 CpG mutations from 36 CpG bases, thereby producing a CpG mutation density of 2/36, or 0.056. We divide this figure by the projected CpG mutation rate (0.056/0.015), arriving at an estimated age of 3.73 million years. This figure is lower than the non-CpG mutation rate, but it still suggests that these elements are at least twice as old as their true Ta counterparts. In addition, all but one of these Ta L1 elements (L1HS493) were monomorphic for the presence of the L1 element in the human population. Thus, the higher levels of nucleotide diversity and the absence of associated insertion polymorphism of eight of these L1 elements are consistent with their being older members of the L1 Ta subfamily, whereas L1HS493 may be the product of a gene-conversion event.
The nucleotide-sequence substitution patterns were further examined with respect to the levels of presence/absence of insertion polymorphism associated with each of the L1 elements (as outlined in detail below, in the “L1 Element–Associated Human Genomic Diversity” subsection). The 3′ UTRs of 139 fixed-present elements were analyzed for both CpG and non-CpG mutations and had an estimated average age of 2.45 million years. This calculation yields an age that is somewhat older than the average age that was predicted for the subfamily as a whole—a finding that was expected, since these elements are thought to have inserted during the early stages of L1Hs Ta expansion in the human genome, such that they have become fixed across diverse human populations. Similar calculations were repeated for the high-frequency, intermediate-frequency, and low-frequency L1 Ta insertion polymorphisms, with average ages of 2.24, 2.06, and 1.69 million years, respectively. Although the age differences across different insertion frequencies are not significantly different (P values >.05) when tested with a one-tailed t test, they do suggest a progressive decrease in the calculated age of each group, with corresponding decreases in insertion frequency. This is exactly what would be expected under a model in which newer elements arose more recently and have lower allele frequencies in the human population.
L1 Element–Associated Human Genomic Diversity
Of the 468 Ta L1Hs elements isolated in silico, 262 were further analyzed using a PCR-based assay and flanking unique sequence primers as described elsewhere (Sheen et al. 2000) (table 1; also see appendix A, available online only and appendix A). The remaining elements were not suitable for further analysis, for various reasons. Some (137) of the L1 elements were inserted into other repetitive regions of the genome such that flanking unique sequence PCR primers could not be designed. Sixty-nine additional elements resided at the end of sequencing contigs in GenBank, so the lack of flanking unique sequence information made PCR-primer design in this region impossible. Three elements—L1HS17, L1HS47, and L1HS63—produced inconclusive PCR results because of the amplification of paralogous genomic sequences as described elsewhere (Batzer et al. 1991). Another five elements produced nonspecific PCR results, and they were excluded from further analysis. Thirty-six of the Ta L1 elements mapped to chromosome X, and 10 mapped to chromosome Y (table 1; also see appendix A, available online only and appendix A). All of the Ta L1 elements from chromosomes X and Y were tested using human DNA samples in which the gender had been determined using a PCR-based assay that was described elsewhere (Eng et al. 1994). The human genomic diversity associated with the autosomal and sex-linked Ta L1 elements is summarized in table 2 and appendix A, available online onlyappendix A.
Table 2.
Summary of Ta L1 Element–Associated Human Genomic Diversity[Note]
| Classification | No. ofElements |
| Autosomal Ta L1 elements: | |
| HF | 36 |
| IF | 55 |
| LF | 15 |
| VLF/fixed absent | 3 |
| Fixed present | 129 |
| X-linked Ta L1 elements: | |
| HF | 1 |
| IF | 1 |
| LF | 4 |
| VLF/fixed absent | 0 |
| Fixed present | 8 |
| Y-linked Ta L1 elements: | |
| Polymorphic | 0 |
| Fixed present | 2 |
Note.— The L1 Ta insertion polymorphisms are classified according to allele frequency as high-frequency (HF) (present in more than 2/3 but not in all chromosomes tested), intermediate-frequency (IF) (present in more than 1/3 of chromosomes tested but in no more than 2/3 of the chromosomes), low-frequency (LF) (present in no more than 1/3 of the chromosomes tested), or very-low-frequency (VLF) (or “private”) insertion polymorphisms. A full summary of the genotypes for each locus, L1 allele-frequency data, and heterozygosity values is shown in tables A2 and A3tables A2 and A3, available online only, and is also available at the Batzer Lab Web site.
A high degree (45%) of insertion polymorphism was found in the 254 (i.e., 262-8) remaining elements that were subjected to the two-step PCR-based assay across 80 individuals from four geographically diverse human populations (table 2; also see appendix A, available online only and appendix A). One hundred thirty-nine of the Ta L1 elements were fixed present, meaning that every individual tested was homozygous (i.e., +/+) for the presence of the L1 repeat. These elements are likely to be slightly older than their polymorphic counterparts, having inserted into the human genome prior to the migration of humans from Africa. By contrast, 115 of the elements assayed by PCR were polymorphic, to some degree, in the populations that were surveyed. A survey of human genomic diversity associated with a severely truncated L1 element is shown in figure 1. A sample of the human genomic diversity associated with relatively long L1 insertion polymorphism is shown in figure 2. Thirty-seven of the Ta L1 elements were high-frequency insertion polymorphisms with an L1 allele frequency that was >0.67, so that most of the individuals were homozygous for the presence of the L1 element. Fifty-six of the polymorphic elements were intermediate frequency, with an L1 allele frequency >0.33 but <0.67 across the diverse human populations sampled. Nineteen of the 254 elements had insertion allele frequencies <0.33, and these were termed “low-frequency insertion polymorphisms.” These elements include some of the youngest members of the subfamily, having inserted into the human genome so recently that the element appears in the genomes of only a handful of individuals who were screened in our assay. Three Ta L1 elements—L1HS44, L1HS287, and L1HS373—appeared to be absent from the genomes of all the individuals tested, and one of these (L1HS373) is full length and has two functional ORFs, suggesting that it may be retrotransposition competent. Previous experiments with Alu elements have shown not only that these types of elements are indeed present within the genomic clone that was sequenced as part of the human genome project but also that they represent relatively rare, “private” mobile-element insertion polymorphisms (Carroll et al. 2001).
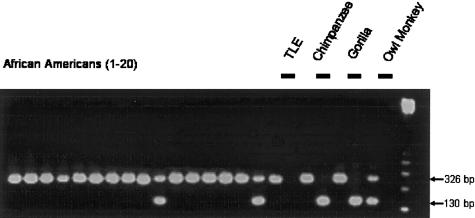
Figure 1.
Human diversity associated with a truncated Ta L1Hs element, as shown by an agarose gel chromatograph of the PCR products from a survey of the human genomic variation associated with L1HS7. Amplification of the pre-integration site of this locus generates a 130-bp PCR product; amplification of a filled site generates a 326-bp product (by use of flanking unique sequence primers). In this survey of human genomic variation, 20 individuals from each of four diverse populations were assayed for the presence or absence of the L1 element, with only the African American samples shown here; the control samples (gray lines) were TLE buffer (i.e., 10 mM Tris-HCl:0.1 mM EDTA), common chimpanzee, gorilla, and owl monkey DNA templates. Most of the individuals surveyed were homozygous for the presence of the L1 element; in addition, this particular L1 element was absent from the genomes of nonhuman primates.
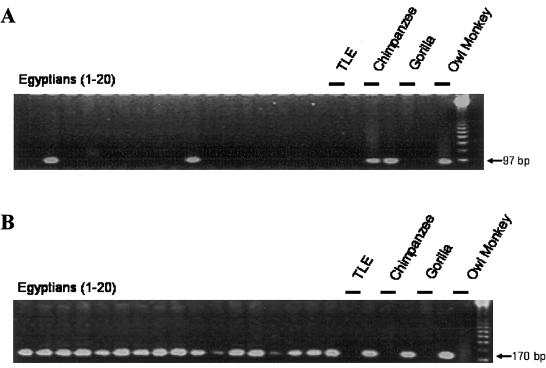
Figure 2.
Human diversity associated with a long L1Hs Ta insertion polymorphism, as shown by an agarose gel chromatograph of the PCR products from a survey of the human genomic variation associated with L1HS364. Because of the size (∼6,000 bp) of this L1 element, two separate PCRs are performed to genotype individual samples. In the first reaction, flanking unique sequence primers were used to genotype the empty alleles (A); amplification of empty alleles from this locus generates a 97-bp PCR product. In the second reaction, a Ta subfamily–specific internal primer termed “ACA” and the 3′ flanking unique sequence primer were used to genotype filled sites (B); the amplification of filled sites generates a 170-bp product. In this survey of human genomic variation, 20 individuals from each of four diverse populations were assayed for the presence or absence of the L1 element, with only the Egyptian samples shown here; the control samples (black lines) were TLE buffer, common chimpanzee, gorilla, and owl monkey DNA templates. This particular L1 insertion polymorphism is a high-frequency insertion polymorphism, and most of the individuals surveyed have L1 filled chromosomes.
Overall, the unbiased heterozygosity values across all of the L1 elements subjected to PCR analysis were similar across the four populations, with values of 0.265 in African Americans, 0.233 in Asians, 0.252 in European Germans (i.e., white Germans of European descent), and 0.250 in Egyptians (table 2; also see appendix A, available online only and appendix A). However, several of the polymorphic elements individually exhibited unbiased heterozygosity values that approached 0.5, the theoretical maximum for biallelic loci. A subset of 31 of the 115 L1 insertion polymorphisms are, to some degree, population specific, meaning that insertion frequencies differ by ⩾25% in one of the tester populations, relative to the other three populations that were surveyed. Detailed analysis of the human genomic variation associated with the polymorphic L1 elements will prove useful for the study of human population genetics.
To determine if the L1 insertion polymorphisms were in Hardy-Weinberg equilibrium (HWE), we performed a total of 460 χ2 tests for goodness of fit. A total of 77 deviations from Hardy-Weinberg expectations were observed in the comparisons. However, 73 of the deviations were the result of low expected numbers. The remaining four tests that deviated from HWE did not cluster by locus or population. A total of 23 deviations from HWE would be expected by chance alone at the 0.5% significance interval. In addition, we applied Fisher’s exact test to the data, using the Genetic Data Analysis program. The test yielded only 22 of 436 significant comparisons, which is approximately what would be expected on the basis of chance alone. By Fisher’s exact test, only 6 of the 436 comparisons were significant at the .01 level, and they did not cluster across all populations at any locus tested. Therefore, we conclude that these L1 insertion polymorphisms do not significantly depart from HWE.
Phylogenetic Origin
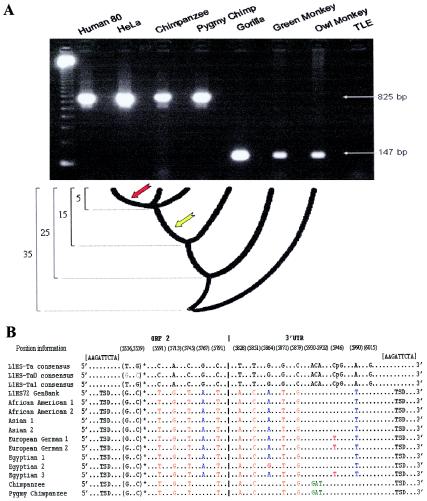
Almost all of the Ta L1 elements analyzed using PCR were located in the human genome and were absent from the orthologous positions within nonhuman primate genomes. Only a single truncated L1 element (L1HS72) produced unexpected results when subjected to the initial PCR by use of external flanking primers and nonhuman primate DNA as a template. The 825-bp amplicon that corresponded to the L1HS72 insertion was found in loci in all 80 human individuals tested, as well as in the orthologous loci from the common chimpanzee and pygmy chimpanzee genomes (fig. 3A). However, the gorilla, green monkey, and owl monkey only amplified the small PCR product corresponding to the empty allele or pre-integration site (fig. 3A). Subsequent PCRs by use of the internal subfamily-specific ACA primer and the 3′ flanking primer across the same DNA templates produced a characteristic L1 filled-site amplicon only in the human individuals and not in any of the nonhuman primate genomes (chimpanzee, gorilla, green monkey, and owl monkey). It appeared that we had potentially isolated a Ta L1 element that inserted into the genome before the divergence of humans from African apes, but the second PCR by use of the internal subfamily-specific ACA primer and the 3′ flanking primer again produced the expected product that corresponded to the presence of this Ta L1 element only in humans. These data suggest that there is a difference in the sequence structure of this L1 element in the human genome, as compared to the common and pygmy chimpanzee genomes, which contained putative Ta L1 filled alleles.
Figure 3.
L1HS72 gene conversion. A, Agarose gel chromatograph of the PCR products derived from the amplification of L1HS72 in a series of human and nonhuman primate genomes, with a schematic of the primate evolutionary tree over the past 35 million years shown below. The yellow notched arrow represents the approximate time period when the L1HS72 element first integrated, and the red notched arrow represents the approximate time period of the gene conversion event of the preexisting L1 element. The fragment-length marker is a 123-bp ladder. B, Sequence alignment generated by sequencing the L1HS72 amplicons from nine diverse humans. Sequences are compared relative to L1Hs Ta consensus sequence and the L1HS72 sequence obtained from GenBank with only the diagnostic bases shown and positions reported relative to L1 retrotransposable element–1 (Dombroski et al. 1991). The G and C at positions 5536 and 5539 are indicative of the Ta-0 subset, whereas the Ta-1 subset has T and G at these nucleotides (Boissinot et al. 2000). The G at position 6015 (in addition to the ACA at positions 5930–5932) is diagnostic for the L1Hs Ta subfamily (Ovchinnikov et al. 2001). The target-site duplication sequence (TSD) is shown in brackets. The mosaic elements seen in the human samples are believed to be the result of at least one gene conversion, some time after the divergence of humans from the great apes (approximately five million years ago), of a preexisting L1 element with a younger L1Hs element. In the representation of nucleotides, different colors are used to denote conserved sequences and sequence variations between samples: green denotes bases unique to the common and pygmy chimpanzee genomes; blue denotes nucleotides unique to the human samples; orange denotes shared bases conserved between the common chimpanzee, pygmy chimpanzee, and human samples; and red denotes SNPs, within L1HS72, in the human population.
Gene Conversion
To precisely define the sequence structure of the L1HS72 locus, we cloned and sequenced, for further analysis, the PCR amplicons from several human genomes, as well as those from the common chimpanzee and the pygmy chimpanzee (fig. 3B). Sequence analysis of the orthologous sites from the common and pygmy chimpanzee genomes revealed the presence of an older, primate-specific L1 element that had the greatest sequence identity to the L1PA3 subfamily (fig. 3B). Interestingly, this L1 element shared identical target-site duplications with that of the Ta L1 element that was present in the human samples that we studied. Both the human sequence and the chimpanzee sequence also contained many of the diagnostic mutations characteristic of an L1PA3 element. However, only the human L1 sequences contained the Ta diagnostic ACA mutation at positions 5930–5932 in the 3′ UTR. The common and pygmy chimpanzee sequences contained GAT at this position and an additional A mutation at diagnostic position 6015, both of which are characteristic of older L1PA elements (L1PA6–L1PA2). The most likely explanation for the presence of the L1Hs Ta ACA sequence in the human L1 element is a forward gene-conversion event that affected a preexisting older L1 element at this locus. To further investigate the putative gene conversion at this locus, we cloned and sequenced alleles derived from African American, Asian, European German, and Egyptian genomes. Although there was a limited sample size, all nine individuals who were sequenced contained the ACA sequence, and at least four samples (European Germans 1 and 2 and Egyptians 2 and 3) contained SNPs, three of which occur at a specific CpG dinucleotide (fig. 3B). Therefore, we conclude that gene-conversion events have altered the L1 Ta subfamily–specific diagnostic nucleotide positions at this locus within the human lineage.
To begin to examine the level of gene conversion across the entire Ta subfamily, we examined multiple-sequence alignments of the 459 Ta L1Hs elements. Close inspection of the multiple-sequence alignment revealed some highly variable sequence features that were unexpected among such a young L1 subfamily, in which we would expect low levels of nucleotide divergence. It appears that many of the single-base substitutions in Ta L1 elements are not completely random mutation events. In fact, it became clear that a substantial number of the elements possessed specific mutations that are diagnostic for older L1PA primate-specific elements in addition to the younger diagnostic mutations. These mosaic elements all possessed the 19-bp Ta L1 consensus sequence, but they also contained short tracts of sequence diagnostic for other L1 subfamilies.
There are two possible explanations for the presence of these mosaic elements. The first theory is that L1Hs Ta source genes, while acquiring the young diagnostic mutations of the L1Hs Ta subfamily, also retained many of the other diagnostic mutations of their older L1 subfamily progenitors. Over time, this gave rise to elements with combinations of young and old mutations, as proposed in the master-gene theory of LINE and short-interspersed-element (SINE) amplification (Deininger et al. 1992). The second theory is that some of these mosaic elements are products of gene-conversion events—that is, a nonreciprocal transfer of sequence between a pair of nonallelic genomic DNA sequences, such as interspersed repeats. The donor sequence is unchanged, and the recipient sequence gains some of the donor sequence; alternatively, a nonintegrated LINE cDNA may also serve as the donor sequence for the gene conversion. Gene conversion between SINEs and LINEs is a significant influence on the genomic landscape of young Alu elements, creating hybrid sequence mosaics of the various mobile-element subfamilies (Batzer et al. 1995; Kass et al. 1995; Roy et al. 2000; Roy-Engel et al. 2001, 2002). Gene conversion may contribute to as much as 10%–20% of the sequence variation between recently integrated Alu elements (Roy et al. 2000). It is likely that the same process may also alter the sequence diversity of L1 elements, since they are also part of a large, nearly identical multigene family and since they have previously been shown to have undergone limited gene conversion (Hardies et al. 1986; Burton et al. 1991). Unfortunately, the vast majority of primate L1 subfamily structure has only been deduced computationally and has not been verified at the wet bench, to precisely define the expansion of L1 elements in a phylogenetic context. Therefore, it is currently not possible to accurately estimate the level of gene conversion between L1 elements within the genome.
Sequence Diversity
One hallmark of L1 integration is the generation of target-site duplications flanking newly integrated elements. Two thousand base pairs of flanking sequence on each side of the element were searched for target-site duplications. Direct repeats >10 bp long are considered to be clear target-site duplications. Of the 399 elements (i.e., a total of 468 elements minus the 69 elements located at the end of sequencing contigs), we were able to identify clear target-site duplications for 272 elements. All elements with clear target-site duplications had endonuclease sites that matched those described elsewhere (Feng et al. 1996; Jurka 1997; Cost and Boeke 1998). A total of 13 elements (L1HS45, -70, -172, -178, -284, -372, -415, -416, -442, -443, -448, -513, and -558) apparently lacked target-site duplications or contained short target-site duplications. To further investigate these elements, PCRs specific for the pre-integration sites for those elements listed were performed on the common chimpanzee, pygmy chimpanzee, and, when possible, human samples. The resulting amplicons were cloned and sequenced, to unambiguously define the pre-integration site for each element. The resulting pre-integration sites were then compared with the original GenBank sequence for each locus.
All 13 of the L1Hs elements lacked obvious target-site duplications when compared with the common and pygmy chimpanzee pre-integration-site sequences. In addition, L1HS178 and L1HS284 had no observable target-site duplications and atypical endonuclease-cleavage sites. One possible explanation for this observation is that these elements have integrated independent of endonuclease cleavage of target sequence, which has elsewhere been proposed as a mechanism for the repair of double-stranded breaks in DNA (Moore and Haber 1996; Teng et al. 1996; Morrish et al. 2002). Alternatively, these elements may represent forward gene-conversion events of preexisting L1 elements that, by mutation, have rendered their target-site duplications unrecognizable. However, because little is known about the rates of these events in mammalian cells, further studies are required in order to resolve the mechanism underlying these integration events.
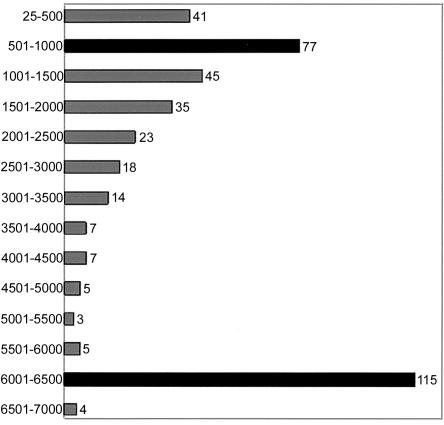
Another aspect of L1Hs Ta sequence diversity is created by variable 5′ truncation such that some of the elements in the human genome are only a few hundred base pairs long, whereas some full-length elements are >6,000 bp long. This phenomenon is classically attributed to the lack of processivity of the reverse-transcriptase enzyme in the creation of the L1 cDNA copy. The point of truncation is traditionally believed to occur as a function of length, where shorter inserts are more likely to occur in the human genome than are longer elements (Grimaldi et al. 1984). Our data show that there is an enrichment of full-length elements in the human genome and that many Ta elements have been faithfully replicated in their entirety and inserted into new genomic locations. Of the 399 elements examined, 119 were >6,000-bp long, representing an L1 Ta size class much larger than any other (fig. 4). By contrast, very few elements were found in the size class ranging between 3,500 and 5,500 bp, with only 22 of the 399 elements truncated to this particular size class. A bimodal distribution of the size of the elements is created, since there are a significant number of Ta L1 elements that are severely 5′ truncated and that are full length. One hundred ninety-eight elements were extremely small, having sizes <2,000 bp, and 118 of these elements were between 25 and 1,000 bp long. The distribution is noteworthy, although the mechanism by which these are enriched in the human genome remains to be determined. In addition, 20% (79/399) of the L1Hs elements examined are inverted at their 5′ end—which is an occurrence that is believed to be due to an event known as “twin priming” (Ostertag and Kazazian 2001), in which target-primed reverse transcription is interrupted by a second internal priming event, resulting in an inversion of the 5′ end of the newly integrated LINE. Although L1 truncation is most likely the result of the relatively low processivity of the L1 reverse transcriptase, processes, like twin priming, that form secondary structures in the RNA or DNA strands present at the integration site may also be associated with L1 truncation.
Figure 4.
Ta L1 element size classes (in bp), showing the size distribution of Ta L1Hs elements. Elements are grouped in 500-bp intervals ranging from <500 bp to 7,000 bp long. The two most common size intervals are shown in black.
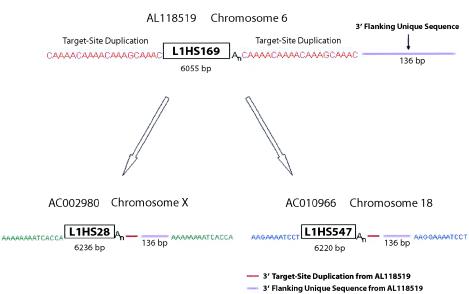
We also observed a significant amount of sequence diversity in the 3′ tails of members of the L1Hs Ta subfamily. The 3′ tails within this L1 subfamily range in size from 3 to >1,000 bp. Thirty-six percent contain AT-rich low-complexity sequence, 31% have homopolymeric A tails, 5% have simple sequence repeats with the most common repeat family TAAA, and 26% contain complex sequence that likely results from 3′ transduction events. The diversity in the tails of the L1 elements is not surprising, since previous studies have shown an association, as well as direct evidence that mobile-element–related simple-sequence-repeat motifs mutate to form nuclei for the generation of simple sequence repeats (Economou et al. 1990; Arcot et al. 1995; Ovchinnikov et al. 2001). Three-prime transduction by L1 elements is a unique duplication event that involves retrotransposons and that has elsewhere been described, in detail, in L1 elements (Boeke and Pickeral 1999; Moran et al. 1999; Goodier et al. 2000). We have identified a number of 3′ transduction events that are mediated by Ta L1Hs elements and believe that these elements have transduced a total of ∼8,500 bp of sequence. We have also taken advantage of the L1 element–mediated transduction to computationally identify a putative retrotransposition-competent L1 Ta source gene. L1HS169 has a 136-bp fragment that is located outside its direct repeats and that is adjacent to its 3′ tail; this fragment is also found adjacent to the 3′ tail of L1HS28 but inside its direct repeat (fig. 5). This suggests that L1HS28 is a daughter copy, or the progeny, of the full-length element L1HS169. In addition, AC010966 from chromosome 18 appears to be a transduction event that was also generated from an L1HS169 read-through transcript. Therefore, we conclude that L1HS169 is responsible for multiple transduction events in the human genome and has produced two independent L1 integrations located on chromosomes X and 18.
Figure 5.
L1HS169-mediated transduction, showing an L1Hs transduction event. L1HS169 marked by clear target-site duplications is the putative source gene for L1HS28. The L1HS28 insertion contains 3′ flanking sequences identical to that of L1HS169 and unique target-site duplications flanking this entire sequence—suggesting that L1HS28 was created from a read-through transcript of L1HS169 that, to give rise to L1HS28, integrated into a new location on chromosome X. In addition, a second transduction event—L1HS547, from chromosome 18—is also flanked by unique target-site duplications and was also derived from L1HS169.
Discussion
Here we report a comprehensive analysis of the dispersion and insertion polymorphism of the youngest known L1 subfamily (i.e., Ta) within the human genome. The computational approach described herein provides an efficient and high-throughput method for the recovery, from the human genome, of Ta L1Hs elements, many of which will be polymorphic for insertion presence/absence in individual human genomes. Individual L1 insertion polymorphisms that were identified are the products of unique insertion events within the human genome. Because each L1 element integrates into the human genome only once, individuals that share L1 insertions (and insertion polymorphisms) inherited them from a common ancestor, thereby making the L1 filled sites identical by descent. This distinguishes L1 insertion polymorphisms and other mobile-element insertion polymorphisms from other types of genetic variation—including microsatellites (Nakamura et al. 1987) and RFLPs (Botstein et al. 1980)—that are not necessarily homoplasy free. In addition, the ancestral state of an L1 insertion is known to be the absence of the L1 element. Knowledge about the ancestral state of L1 insertions facilitates the rooting of trees of population relationships by use of minimal assumptions. Therefore, the 115 new L1 insertion polymorphisms reported herein appear to have genetic properties that are similar to those of Alu insertion polymorphisms (Batzer et al. 1991, 1994; Perna et al. 1992; Hammer 1994; Stoneking et al. 1997; Jorde et al. 2000), and they will serve as an additional source of identical-by-descent genomic variability for the study of human population relationships.
It is noteworthy that the computational identification of L1 insertion polymorphisms introduces a selection for only those elements present in the draft-sequence database. As a result, elements that are not present in the database cannot be identified. This has important consequences with respect to the frequency spectrum of the elements identified. By use of this type of approach, a number of different types of L1 insertion polymorphisms are identified that vary in the frequency of the L1 insertion allele. By contrast, PCR-based display approaches provide an alternative method for the ascertainment of mobile-element insertion polymorphisms from the human genome (Roy et al. 1999; Sheen et al. 2000; Ovchinnikov et al. 2001). In these approaches, polymorphic mobile elements are directly identified; however, elements that are polymorphic but have higher allele frequencies (i.e., high-frequency insertion polymorphisms) are lost in the process, since most genomes will contain at least one filled allele that contains the mobile element and would not be scored as an insertion polymorphism. Therefore, more population-specific or private mobile-element insertion polymorphisms will be identified using PCR-based displays or other types of direct selection (Roy et al. 1999; Sheen et al. 2000; Ovchinnikov et al. 2001). Using our computational approach, we recovered only 14 of 49 Ta L1 elements that were elsewhere identified using PCR-based displays (Sheen et al. 2000; Ovchinnikov et al. 2001) and that had sufficient flanking unique DNA sequences for comparison to the data set that we studied. Thus, computational and experimental ascertainment of mobile-element insertion polymorphisms are quite complementary approaches for the identification of new mobile-element insertion polymorphisms.
The L1 Ta subfamily can be further subdivided—according to the nucleotides that are present, within ORF 2, at positions 5536 and 5539—into Ta-0 and Ta-1 (Boissinot et al. 2000). Ta-0 L1 elements are believed to be evolutionarily older, and they possess a G at position 5536 and a C at position 5539. Ta-1 L1 elements, however, have a T at position 5536 and a G at nucleotide 5539. Ta-1 L1 elements are considered to be younger, and it is believed that all actively transposing elements in humans belong to the Ta-1 subset of L1 elements (Boissinot et al. 2000). One hundred ninety-two of the 459 Ta elements identified from the draft human genomic sequence belong to the younger Ta-1 subset, and 137 belong to the Ta-0 subset. Another 105 of the elements either are 5′ truncated such that they terminated before these positions at 5536 and 5539 or are inverted or rearranged in the region in question. An additional 25 elements are sequence intermediates between Ta-1 and Ta-0.
Inspection of the insertion polymorphism data for each of these Ta subsets showed that only 35% of the Ta-0 L1 elements analyzed by PCR were polymorphic, with the remaining 65% being fixed present in the human populations screened. Consistent with the idea that Ta-0 L1 elements are older, 9 of the polymorphic elements were high-frequency insertion polymorphisms, 10 were intermediate-frequency insertion polymorphisms, and only 5 were low-frequency insertion polymorphisms. None of the Ta-0 L1 elements were fixed absent or very low frequency in the populations that were analyzed. By contrast, 56% of the Ta-1 L1 elements were polymorphic with respect to presence—with 18 high-frequency, 27 intermediate-frequency, and 11 low-frequency insertion polymorphisms. In addition, we can use the non-CpG mutation density in Ta-0 and Ta-1 L1 elements to calculate the estimated age of each of the Ta-derivative subfamilies. The non-CpG mutation density for the Ta-0 and Ta-1 L1 elements was 0.003103 and 0.002560, respectively. Using a neutral rate of evolution of 0.15% per million years (Miyamoto et al. 1987), we derive estimates of 2.07 (i.e., 0.003103/0.0015) million years and 1.71 (i.e., 0.002560/0.0015) million years from the Ta-0 and Ta-1 subsets, respectively. Although these estimates are not significantly different from each other, they do support the notion that the Ta-0 L1 elements are slightly older than the Ta-1 L1 elements, as do the differences in insertion polymorphism. In addition, they provide direct evidence that the Ta-0 and Ta-1 subsets have simultaneously amplified within the human genome.
Forty-four of the 124 full-length Ta L1Hs elements that were identified have both ORFs intact and are presumably retrotransposition-competent elements. This compares favorably with previous estimates of the number of potentially active L1 elements in the human genome (Sassaman et al. 1997). In addition, it is also important that those full-length elements that no longer have intact ORFs might have previously acted as active “source,” or driver, genes for the expansion of Ta L1 elements but might have accumulated mutations over time that inactivated them. These data, as well as data from the previous studies involving the isolation and amplification of some of these full-length Ta L1 elements within tissue-culture systems, demonstrate that multiple L1 elements have expanded within the human genome in an overlapping time frame. It is interesting to compare the amplification of the L1 elements to that of the Alu SINEs within the human genome. In the case of the L1 elements, one major family (Ta) with two subdivisions (Ta-0 and Ta-1) has expanded to a copy number of ∼500 elements in the past four to six million years since the divergence of humans and African apes. By contrast, the expansion of Alu elements is characterized by the amplification of at least three major lineages, or subfamilies of elements, that have collectively generated ∼5,000 copies (Batzer and Deininger 2002). On the basis of these copy numbers alone, it would appear that Alu elements have been 10 times more successful than L1 elements have been with respect to duplicating themselves, within primate genomes, over the past four to six million years. However, if we make the estimate relative to the total family size of 500,000 L1 elements or 1.1 million Alu elements (Lander et al. 2001), then the relative difference is merely fivefold. This difference in amplification is also apparent across the entire expansion of these repeated DNA sequence families, since the L1 elements have expanded to only 500,000 copies in 150 million years, whereas the Alu elements have expanded to 1.1 million copies in only 65 million years.
Since Alu and L1 elements are thought to utilize the same enzymatic machinery for their mobilization, the differential amplification of both young and old Alu and L1 elements within primate genomes is quite interesting (Boeke 1997). The two different classes of repeats putatively compete for access to the same reverse transcriptase and endonuclease; thus, it is possible that Alu elements are currently more effective than the L1 elements at attracting the replication machinery within the human genome. If this competition between interspersed elements is important, then we may expect to see differential rates of L1 and Alu expansion in different nonhuman primate genomes as the elements compete for the common components involved in mobilization. Differential mobilization of SINEs and LINEs has been elsewhere reported in rodent genomes (Kim and Deininger 1996; Ostertag et al. 2000). Therefore, it would not be surprising to see something similar in nonhuman primate genomes. Alternatively, the differential amplification may reflect differences in selection against new L1 and Alu insertions within the human genome (Lander et al. 2001). Since L1 elements are typically much larger than Alu repeats, it is easy to envision that the larger insertions would be much more disruptive to the genome than the shorter Alu insertions are. This type of selection has been suggested as one potential explanation for the differential distributions of L1 elements (Boissinot et al. 2001) and of Alu and L1 elements (Lander et al. 2001; Ovchinnikov et al. 2001) throughout the human genome. However, the argument that selection is responsible for the differential distribution of Alu sequences has recently been questioned (Brookfield 2001). Further studies of the expansion of interspersed elements within the genomes of nonhuman primates will be required in order to definitively address these questions.
Our analysis of mosaic Ta L1Hs elements suggests that gene conversion alters the sequence diversity within these elements. This is not surprising, since previous studies have indicated that gene conversion plays a role in the generation of sequence diversity in Alu repeats (Maeda et al. 1988; Batzer et al. 1995; Kass et al. 1995; Roy et al. 2000; Carroll et al. 2001; Roy-Engel et al. 2002), as well as the generation of sequence diversity in L1 elements, within the genome (Hardies et al. 1986; Burton et al. 1991; Tremblay et al. 2000). Unfortunately, an accurate estimate of L1-based gene conversion is not yet possible, because primate L1 subfamily structure is not yet clearly defined. However, gene conversion appears to play a significant role in the sculpting of human genomic diversity (Ardlie et al. 2001; Frisse et al. 2001). Because of the hierarchical subfamily structure of Alu and LINEs and because of the defined pattern of ancestral mutations, these elements provide a unique opportunity for the estimation of gene conversion throughout the genome. It is also important to consider that the gene conversion between large multigene families, such as SINEs and LINEs, may occur by a mechanism that is completely different from that which occurs at other unique and low-repetition sequences within the human genome. Nevertheless, large-scale studies of orthologous sequences from the same L1 element in different human genomes will begin to quantitatively address this issue and also will provide insight into the molecular mechanism that drives the process. In addition, detailed pedigree analyses or studies of germ cell–derived L1 diversity will provide insight into the germ line rate of gene conversion between L1 elements. Clearly, L1 elements continue to have a significant impact on human genetic diversity—through recombination, insertional mutagenesis, gene conversion, sequence transduction, and the generation of other simple-sequence-repeat motifs (Kazazian and Moran 1998; Goodier et al. 2000; Ovchinnikov et al. 2001).
Acknowledgments
This research was supported by National Institutes of Health grants R01 GM59290 (to L.B.J. and M.A.B.), R21 CA87356-02 (to G.D.S.), and R01 GM60518 (to J.V.M.); by support from the W. M. Keck Foundation (to J.V.M.); by Louisiana Board of Regents Millennium Trust Health Excellence Fund grants (2000-05)-05, (2000-05)-01, and (2001-06)-02 (to M.A.B.); and, through award 2001-IJ-CX-K004 (to M.A.B.), by the Office of Justice Programs, National Institute of Justice, U.S. Department of Justice. Points of view expressed in this article are those of the authors and do not necessarily represent the official position of the U.S. Department of Justice.
Appendix A: Supplementary Data
Table A1.
L1Hs Ta PCR Primers, Chromosomal Locations, and PCR Product Sizes[Note]
|
Primer Sequence(5′→3′) |
PCR Product Sizesd(bp) |
||||||||
| Element | GenBankAccessionNumber | ChromosomalLocationa | 5′ | 3′ | AnnealingTemperatureb | HumanDiversityc | Filled | Empty | SubfamilySpecific |
| L1HS1 | AC010739 | 2 | AGGGAATGCTTATATTGTTGATGAG | ACTTCCTTCAGGGTTAATAGCAAAG | 60 | FP | 3,877 | 159 | 224e |
| L1HS2 | AC010305 | 16 | ACCAAATATCTGGACACTTTCTGG | GAAGTCAGCAGTGGTTAATTTTACA | 60 | IF | 6,131 | 74 | 171 |
| L1HS3 | AC008572 | 5 | GCTTCTAGAATTGGAAGTAATATGG | AGTAGCCTTGAATCATCTTTTG | 56 | FP | 656 | 95 | 422e |
| L1HS4 | AC009494 | Y | Inserted in repeats | Inserted in repeats | … | R | 467 | … | … |
| L1HS5 | AC020647 | 12 | TCAACTACAAAGTTGAAGAATAGG | GTTTCCATCAACAAGATCATGTCAAG | 58 | LF | 546 | 376 | 455e |
| L1HS6 | AC016138 | 3 | TTTATTTCCCTGCATCTGATTA | CCTGTTATTAGATAATGAGTTCTAGTC | 54 | HF | 402 | 122 | 219e |
| L1HS7 | AC004773 | 7q11 | CCTTAGACATATTCTTGGAAATAG | CCAGAATATTTGGGTATTTCATCTG | 58 | HF | 326 | 169 | 256e |
| L1HS8f | AC004491 | 7q | Inserted in repeats | Inserted in repeats | … | R | 1,689 | … | … |
| L1HS9f | AC004694 | 7p | TCTTTCAATGGAAACAAGAGGTATC | AGGGAGAGGGACACTGAGTTTAT | 59 | FP | 6,126 | 74 | 178 |
| L1HS10g | AF149774 | 7p | Inserted in repeats | Inserted in repeats | … | R | 6,076 | … | … |
| L1HS11 | AL049842 | 6q | Inserted in repeats | Inserted in repeats | … | R | 667 | … | … |
| L1HS12g | AC007538 | Xq28 | GTTAAAGCAATCAAGCAATCTACTG | TAACAAGGCCACTGTAGAAAAGATT | 59 | FP | 6,188 | 104 | 209 |
| L1HS13f | AC007938 | 7q31 | ATGGGAAGGAACCCCATCTAT | AATTACTCCTCTCTTTGGCCTGTT | 59 | HF | 745 | 128 | 220e |
| L1HS14 | L05367 | 17q | AAGTGGATTAACAGTAACATACAGA | CCAAGCTGATAACTGATTATCTCA | 55 | IF | 601 | 251 | 158 |
| L1HS15f | AC007556 | 2 | AATGCATACCCATGAGGACAA | ATGGTGTTGCACAACAAAAGAA | 60 | HF | 6,167 | 126 | 197 |
| L1HS16 | AP000220 | 21q | CCCTCACAGAGTGCTTGGTAA | GGGAAGGTAGGAAAACAGATT | 56 | IF | 368 | 101 | 207e |
| L1HS17f | AC007486 | X | GCATCCCTAAAGCAATAATCCA | GGAATTTTCCACTTGTGGTGTC | 60 | Paralog | 4,286 | 90 | 170e |
| L1HS18 | AC005798 | 4 | TTGAACAGCTTAGACTCGTCAGATA | GCAGTTAGACAGGAAAACAGAAAGA | 60 | HF | 6,174 | 87 | 212 |
| L1HS19 | AC007876 | Y | Inserted in repeats | Inserted in repeats | … | R | 6,115 | … | … |
| L1HS20 | AC009241 | 2 | AATGGAAGAGCTCTCAAATTCCTTA | GCAACCATTCAAAAATTTACAACAG | 61 | IF | 2,302 | 62 | 181 |
| L1HS21 | AC008277 | 2 | GTGTTGGCATATTTCTATTCG | TAAAGGCTGAACTTTGCATTG | 57 | LF | 2,606 | 84 | 178e |
| L1HS22 | AC010682 | Y | GCTCTCGGGTTCTTCTACCTCT | TCTACTGTTCCATGCAATAGATGTG | 60 | NR | 3,216 | 266 | 249e |
| L1HS24f,g | AC004554 | Xp22 | GTGTATTTTGCCTTTTGAACCAA | CAAAAACTTGTTTCACTTGATTTTTAG | 59 | IF | 6,148 | 101 | 181e |
| L1HS25f,g | AC002385 | 7q31 | GAGGACCTTATTCATTTATTGC | CCATCTGAGCTTTAGTTTTGTCATA | 60 | FP | 6,140 | 94 | 191e |
| L1HS26f | AC003689 | 11q12 | GCTTCAAGCTTAAAAGATGTAGACT | CCTACCCAAGTATCCACTGTCC | 60 | IF | 2,652 | 589 | 420e |
| L1HS27 | AC007736 | 2 | AGAACGTTGCCACATTATTTTGA | GTAGGAAGGTCTGGACTGGAGTATT | 58 | FP | 3,667 | 68 | 214e |
| L1HS28g,h | AC002980 | Xp22 | CTTTTGTGACACTGGATTTCTAGC | CACTGTATATTGGAGCTGTTTTTCC | 58 | IF | 6,531 | 282 | 373 |
| L1HS29f | AC005090 | 7p | Inserted in repeats | Inserted in repeats | … | R | 1,476 | … | … |
| L1HS30f | AL022166 | Xp11 | CCCTAAACAGAAAGGAAAATGAGAC | TCCTCATTGTGGTTCAAGGTTATAC | 60 | IF | 4,795 | 97 | 175e |
| L1HS31h | AC019212 | X | GACAACACAAAGAAAACCCAAGAT | CTTATGTCCCAAAGCTAGTGAGTGA | 56 | FP | 2,317 | 86 | 176 |
| L1HS32f | AC004911 | 7q | TCTCTAATCCAGCCTTTCAATTC | TGTTTCTTTTCCTGTGTGTTTCC | 57 | IF | 463 | 280 | 384 |
| L1HS34h | AC002122 | 5p15 | ATGTCTGTCTTGACATTCCTAAGC | AATATGTAGAATGGCACAGGCTTC | 58 | IF | 2,177 | 284 | 328 |
| L1HS35g | AC010081 | Y | CTACCACATAACTGAGTGACAGTTT | CAATGTGCATCCATATAGCTGTGTT | 61 | FP | 6,308 | 233 | 239 |
| L1HS36f | AC004000 | Xq23 | Inserted in repeats | Inserted in repeats | … | R | 6,038 | … | … |
| L1HS37 | AC003080 | 7q31 | Inserted in repeats | Inserted in repeats | … | R | 6,017 | … | … |
| L1HS38f | AC004142 | 7q31 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS39 | AC005690 | 4 | AGAACCAATCTTGCCCACAC | TGAGGAGTTTCTGAGTAACCTGGTA | 60 | HF | 6,337 | 155 | 189 |
| L1HS41 | AF222686 | Xp11 | Inserted in repeats | Inserted in repeats | … | R | 1,959 | … | … |
| L1HS42 | AC020925 | 5 | Inserted in repeats | Inserted in repeats | … | R | 580 | … | … |
| L1HS43 | AF172277 | 7q21 | TTTATTGCACCTCCTGGTAAAGTAG | AGAGCACCATTAAACAACACAAGAT | 58 | IF | 6,157 | 89 | 191 |
| L1HS44f | AC004883 | 7q | TAGCTGTGCTTGTTATGTCCAGTT | GAATGAGTTTTGTGTGGTTCTGTG | 57 | VLF | 2,288 | 478 | 615e |
| L1HS45f | AC004865 | 1 | AATAGGCCCAGCTATTAGATTTAGC | CCTTTAAACCTTTGAACACGATTT | 53 | FP | 329 | 81 | 150e |
| L1HS46f,g | AC006027 | 7p | CCTGTGTTCCTTTTGTAATCC | CAAATGTCTCTTCAAGGACTG | 55 | HF | 6,382 | 326 | 183e |
| L1HS47 | AC006986 | Y | AGTCAAATGATTTTTAACTGCTG | GAGGGCAAGATCATGAAACA | 58 | Paralog | 6,177 | 86 | 230e |
| L1HS48f | AC005105 | 7p | CGAAAAGCTTAGGAAACTGTTTGT | TAAGCAATCTTCAGTTTAGGAAA | 58 | FP | 1,242 | 810 | 420 |
| L1HS49 | AC010202 | 12q | Inserted in repeats | Inserted in repeats | … | R | 612 | … | … |
| L1HS50 | AF198097 | Xp11 | Inserted in repeats | Inserted in repeats | … | R | 6,308 | … | … |
| L1HS51 | AC008055 | 12q22 | GCCCCTTACGTTAGAATAGAAAC | TGGATTGGTCCATACTACTGT | 55 | FP | 1,094 | 272 | 239e |
| L1HS55f,g | AC004704 | 4q25 | Inserted in repeats | Inserted in repeats | … | R | 6,063 | … | … |
| L1HS56f | AC005908 | 12p13 | CCATTCATCAGCCATTTGCTA | GTGGCTTTAAAACAACGAGATG | 59 | FP | 6,545 | 459 | 494e |
| L1HS57f | AC006222 | 4 | CAGCAAGACTCTGTCTCTAAAATGAT | GGACTTGAATTTGGTCTTGTTTCTA | 59 | LF | 589 | 195 | 284e |
| L1HS58f | AC005939 | 17 | Inserted in repeats | Inserted in repeats | … | R | 6,101 | … | … |
| L1HS59 | AC003678 | 11q12 | Inserted in repeats | Inserted in repeats | … | R | 2,081 | … | … |
| L1HS60f | AC006465 | 7p | GAAGTATGGAAATTGAGTCACA | CCCTAAGCTGTATCACTTTAAAACA | 56 | FP | 445 | 104 | 246e |
| L1HS61f | AC002288 | 16p12 | ACGTTTGTGCTTCACTCTAAGTTCT | CAAAATACCGGGATTATAGTTGTGA | 57 | FP | 353 | 68 | 175e |
| L1HS62 | AC006840 | 4 | ATTAAAAGGAATGGACATGCAACAC | AATCTCAAAAGCTTCCTTGCACT | 60 | FP | 6,282 | 182 | 256e |
| L1HS63 | AC023423 | Y | AAGAAAGTGTTGTCAGAGAGTGTGA | AGGCCATTGGTCAGTCATAATTT | 60 | Paralog | 6,160 | 115 | 200 |
| L1HS65f | AC004053 | 4q25 | Inserted in repeats | Inserted in repeats | … | R | 1,781 | … | … |
| L1HS68f,g | AC004200 | 6p21 | Inserted in repeats | Inserted in repeats | … | R | 6,242 | … | … |
| L1HS69f,h | AC004220 | 5 | GGATGTTGATGATGGAGTCAGTC | TAACCATTTGAAACCATTAGAGGTC | 60 | FP | 1,410 | 76 | 180 |
| L1HS70f,h | AL049588 | Xq | GTTCATTTGAGTGAGGGTACTGTCT | TAAGTCCCAAAAATTGCATCC | 59 | IF | 3,174 | 175 | 256e |
| L1HS72 | AL133413 | 9q | CTGAGATGAGACAGCAGGTCTTC | TCTGCTGAGATTCTTCCATTTACC | 60 | FP | 825 | 147 | 221 |
| L1HS73 | AC018822 | 3p | ATAAGGAGCCTAGGGAAGAACTTTT | CAAGCATGCCTGAAACATCTAT | 55 | HF | 1,126 | 462 | 162e |
| L1HS74g | AC011990 | 17 | CTGGACGTATTTCTTACAGAGTTGA | CCCTAAGTTATTTTCCTTGAGGCTA | 60 | LF | 6,163 | 125 | 186e |
| L1HS76 | U08211 | X | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS77f,h | AB020867 | 8p | TTCCTAAATGGCCTTACTATCCTTT | TCAGAAGTGCTAACAACTCTAGTAGGA | 58 | HF | 990 | 78 | 233 |
| L1HS78f | AP000084 | 21q22 | TAGTACCTCCCTTAAAGAGCTG | GAGGAAAAGAAAAGTGCCTGATA | 59 | IF | 374 | 107 | 175e |
| L1HS80f | AC017051.4 | UL | Inserted in repeats | Inserted in repeats | … | R | 1,823 | … | … |
| L1HS81 | AP000962 | 21q21 | AAGTGTTATATATTGGAGCAATTC | ACAAGACAATGCCAATTTTAAGAGA | 60 | FP | 848 | 148 | 401 |
| L1HS83f | AJ001189 | Xq12 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS85 | AC008132 | 22q11 | TTTGTATGCCTTGTGTTTTGTATTG | AGGAGAGTCTCATCTCCAGAGTTAC | 58 | LF | 593 | 79 | 183e |
| L1HS86g | AL121825 | 22 | GCAGTATCAGGAAATGCAATACAC | GGGATTCAGTCACCTTTATTAGACA | 60 | HF | 6,154 | 410 | 180e |
| L1HS87g | AL078622 | 22 | Inserted in repeats | Inserted in repeats | … | R | 6,065 | … | … |
| L1HS91f | Z84572 | 13q12 | ATACGTGCAAAACAGGAGATTTGA | TGTTTATGGTGAAGGATAAGTCTCA | 59 | FP | 1,619 | 78 | 167 |
| L1HS92 | AL022153 | Xq | ACAATCCCTACTTCAGAAAGTT | CAACACTTTGATCATGAATAATAGCTC | 57 | FP | 859 | 121 | 206 |
| L1HS93 | Z95325 | Xq21 | Inserted in repeats | Inserted in repeats | … | R | 4,882 | … | … |
| L1HS94f,g | AL031586 | Xq | TCGTATGAATAACCTTGTGTTCTTG | TTTAGATCCTCGTCACTCAAAGTGT | 57 | FP | 6,250 | 151 | 264 |
| L1HS95f | AL023284 | 6q | GGAAATTCTCAAGCTCAAGTTAAAA | CTTTTAAAGTGTGTTCTCACAGTGG | 60 | FP | 717 | 119 | 320e |
| L1HS97f | AL030998 | Xq | AACCAAACCCACAATCAGTAGAA | CTAGCTAAAGGTTTGCTATTTTT | 58 | FP | 1,640 | 182 | 407e |
| L1HS98f | AL022099 | 6p | ATCTGCATTGGGCCAAGTTTT | TCTCCTGTAAGACAGCACCATA | 60 | FP | 1,561 | 129 | 242e |
| L1HS99f | AL022726 | 6p | Inserted in repeats | Inserted in repeats | … | R | 6,290 | … | … |
| L1HS100 | Z98754 | Xq | Inserted in repeats | Inserted in repeats | … | R | 6,161 | … | … |
| L1HS101f | Z72519 | X | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS102f | AL096677 | 20p | CCATTTGCCATAAATAAAGGCATC | ACTGTTACAAGTTTCCCCAAATGT | 59 | FP | 6,741 | 611 | 542 |
| L1HS103g | AL121591 | 20 | Inserted in repeats | Inserted in repeats | … | R | 6,019 | … | … |
| L1HS104f | AL096799 | 20 | GAGATGTGGTTTTGTTTGAACTG | GCAGCTCACATAGTTTAGAGAAGAT | 59 | IF | 6,196 | 131 | 219e |
| L1HS106 | AL117339 | 10 | CTGACTGTTGAAACTTCTCCATTG | CAATAGACATGAAGGCATGGAAG | 57 | FP | 3,103 | 378 | 345 |
| L1HS108g | AL031768 | 6p | Inserted in repeats | Inserted in repeats | … | R | 6,091 | … | … |
| L1HS109g | AL137191 | 14 | GCCTTTCTATCTTTTGCTCTTGGT | GACACATACCAATTACAGGCAAAG | 59 | FP | 6,549 | 501 | 381e |
| L1HS110f,g | AL078623 | 20 | GGATTCTGACCTTATTCTAACAGCA | AGTTGACTGTTGGTGTTGATTGTGT | 56 | HF | 6,263 | 212 | 253 |
| L1HS111f | AC002069 | 7q21 | Inserted in repeats | Inserted in repeats | … | R | 535 | … | … |
| L1HS112 | AC018755 | 19 | AGGTTCCATCTCTAATACTGGATAA | TGATCACTTTGTTGTTAAGATGGAG | 60 | LF | 1,686 | 102 | 170 |
| L1HS113h | AL133386 | 6p | AGTTTTGGCCTGAGAGAGAAGTAGA | GGTAGGCTAGAGATCCCTTCAATTA | 55 | FP | 405 | 184 | 328e |
| L1HS115 | AL132639 | 14 | Inserted in repeats | Inserted in repeats | … | R | 182 | … | … |
| L1HS116 | AC024610 | 18 | CTGTGCACTTTTCCATATGTTTGAC | TCTAATCTATGGTGGATGCTCTTTC | 56 | FP | 252 | 76 | 189 |
| L1HS117f,g | AC005885 | 12q | TGCAGTGTTCTATTTATGTCGTAGGT | CGAGAGAGGGAGGAAAGTGAG | 57 | IF | 6,629 | 535 | 176e |
| L1HS118 | AC020599 | 4 | ATGCCAGAAATACCTCTTTTACCTT | CTAAGTGCAATTCTCTCAGATTTTG | 60 | IF | 6,321 | 286 | 277 |
| L1HS119f | AC005739 | 5 | GGCTTATTTAGAGCACCTGGATTTA | GAGATCCAAAGCTTATGCTGTAAGT | 60 | FP | 904 | 243 | 257e |
| L1HS123f | AC005350 | 5q | Inserted in repeats | Inserted in repeats | … | R | 397 | … | … |
| L1HS124f | AC004499 | 20q | TGACATAATTAATGGAGAAAACCAG | GAGATCCCTGTCCTTGTGTGAT | 60 | FP | 749 | 515 | 373e |
| L1HS125 | AF001905 | Xq25 | CCTCACGTTTCTCCACATTGTA | TTCTGGCCTTCATAGTGTTTTA | 60 | HF | 332 | 96 | 169 |
| L1HS126f | AC004784 | 19q13 | Inserted in repeats | Inserted in repeats | … | R | 1,552 | … | … |
| L1HS127 | AC004384 | X | Inserted in repeats | Inserted in repeats | … | R | 225 | … | … |
| L1HS129f | AC003100 | 4q25 | Inserted in repeats | Inserted in repeats | … | R | 1,132 | … | … |
| L1HS130 | AL133320 | 1p | Inserted in repeats | Inserted in repeats | … | R | 6,066 | … | … |
| L1HS131 | AL163152 | 14 | TTGACTGTGTACTGCCAGTCTCT | GTAACCTACCAGTTTACAGTTACC | 58 | IF | 381 | 179 | 212 |
| L1HS132 | AP001693 | 21 | CCCTGATACACCAGTATATCTTA | GAAAAGAAAAGTGCCTGATA | 56 | IF | 753 | 486 | 173e |
| L1HS133 | AC008716 | 5 | CATGGTGTCCCAGTGTTAAAAA | TATCTCTTACCTCTTCTTGCCCATA | 59 | FP | 3,351 | 821 | 738e |
| L1HS134 | AF265340 | 16 | CACAGTCAACTCAACCACTGAATAA | AAGGAGATGGAAGTAAGTGCAAAC | 60 | FP | 751 | 433 | 603e |
| L1HS135 | AL137804 | 11p | TTTTTGAAGGGAGTACAGTAATAGGT | GCCTTCCATAGTTCCTATTTGC | 58 | FP | 6,475 | 429 | 500e |
| L1HS136 | AL157791 | 14 | Inserted in repeats | Inserted in repeats | … | R | 175 | … | … |
| L1HS137 | AL157879 | 5 | Inserted in repeats | Inserted in repeats | … | R | 6,057 | … | … |
| L1HS150 | AP000966 | 21q21 | CAAGAACAACTGAAAAATGCAGAT | CCCCTCAGTCTCTGGTTACCTA | 58 | FP | 642 | 89 | 141e |
| L1HS151 | AC019205 | 6 | CTTTGATCAGTTCTTGGAACTAGGA | CCTCTATGCCTTATTCATGCTTATC | 60 | FP | 573 | 405 | 476e |
| L1HS153 | Z84814 | 6p | CCAATTCACTTTGTCTCCTAGAAAT | AGTTCACGAAGTTGAAAGCTTATGT | 60 | IF | 931 | 169 | 219 |
| L1HS155 | AC019050 | 2 | TGGCATGTCAATATATACCTGAAGA | GGAAAACAGAAATAAAAGACGGACA | 60 | FP | 7,004 | 596 | 720 |
| L1HS157f | ALO49842 | 6q | ATTCAAGTTCCAGTAAGCTGTGTTT | GAACTTTGGAAAATTCACAACTACC | 60 | HF | 892 | 143 | 245 |
| L1HS158f | AC008467 | 5 | CAGCCCAGAGTAGTTCATGTTTT | GAAGGAAAAGGAGCTGCTTAGATA | 59 | IF | 6,194 | 147 | 207e |
| L1HS159 | AC009976 | Y | Inserted in repeats | Inserted in repeats | … | R | 1,439 | … | … |
| L1HS160 | AL121938 | 6q | CTAAATAGGCAGAGGAAAGGAAAAC | TAAACTTCCAAGAGATCAGCACTTC | 60 | HF | 1,071 | 99 | 225e |
| L1HS162 | AC009404 | 2 | Inserted in repeats | Inserted in repeats | … | R | 463 | … | … |
| L1HS163 | AL139114 | 9p | GGGACAGGGGTTAAGATTTTATTTT | AGTTCTCAACTGTAAAGGCAGTGTC | 60 | IF | 2,898 | 85 | 251 |
| L1HS164 | AB045357 | 1q | GGAAGGAAGTGGGGATAATAAGTAA | CCCAATTCAGTTTCTTCATTCTATG | 60 | FP | 1,507 | 193 | 267e |
| L1HS165 | AC011666 | 1q21 | CACAGTGATGGAGTTACAATCTTTG | GCTTTAAAGTCAGACAGGCTTGAGT | 62 | FP | 1,509 | 200 | 276e |
| L1HS166g | AC021017 | 8 | TGCCTGAAATGCTATTGGTAGTATC | GTGCCCAGCCCATAATATAAA | 60 | IF | 6,204 | 102 | 251 |
| L1HS167 | AC018637 | 7 | Inserted in repeats | Inserted in repeats | … | R | 2,975 | … | … |
| L1HS168 | AC009492 | 2 | CTTTTTCAAGGCCATCTGTGAG | AATCCTTACAATGAAAAGGGTGT | 61 | FP | 666 | 97 | 180 |
| L1HS169 | AL118519 | 6q | TATTGAGGTGTAACCAGCATACAAT | CCACACGAAAGATATATGAATTGC | 60 | IF | 6,289 | 214 | 288 |
| L1HS171 | AL137145 | 10 | GAAAGTTCATGAAAGTTGTGATGC | ACAAGAGAATCTATCTCCTGAAGAA | 60 | IF | 6,157 | 91 | 198 |
| L1HS172 | AL133479 | 9p | CTAAGATCAGTCACAGGCTTAATGA | CAGGTGCAAGTGGTTTAATTTTC | 60 | IF | 1,326 | 111 | 193e |
| L1HS173 | AL359218 | 14 | CACCATCTAGTGATTTTATGTTCTGC | AATAATCCCCATTGACTGTGTACTG | 55 | HF | 319 | 123 | 217e |
| L1HS174 | AJ271735 | Xq | Inserted in repeats | Inserted in repeats | … | R | 3,252 | … | … |
| L1HS175 | AL136382 | 1p | Inserted in repeats | Inserted in repeats | … | R | 717 | … | … |
| L1HS176 | AC025819 | Y | Inserted in repeats | Inserted in repeats | … | R | 1,522 | … | … |
| L1HS177 | AC017015 | 18 | CAAGTTCCTCACCAAATGAAACTAC | TCCATTTTACTGATGTTGAATAGGC | 58 | HF | 693 | 165 | 273e |
| L1HS178 | AC023480 | 3p | GAATATTGAGCTTTCTTCACCTTT | CAAGCATGCCTGAAACATCTAT | 60 | HF | 508 | 54 | 162e |
| L1HS179 | AC017089 | 4 | Inserted in repeats | Inserted in repeats | … | R | 3,573 | … | … |
| L1HS180 | AC009276 | 7 | GGAGTGTAGAATACTGGGGAAAATC | CTTATTTCCCAATGAGCCCTGTA | 56 | IF | 507 | 84 | 225e |
| L1HS181 | AC025759 | 5 | Inserted in repeats | Inserted in repeats | … | R | 1,179 | … | … |
| L1HS183f | AC000100 | 19 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS184 | AL450108 | X | Inserted in repeats | Inserted in repeats | … | R | 6,094 | … | … |
| L1HS185 | AL157837 | 1q | CTGGCAGTTCCCTCAATGTAA | GAGTAGCTAGCAAAACAGGTAATGAA | 60 | FP | 604 | 108 | 214e |
| L1HS186 | AL359332 | 14 | GGTCTAACAATATTCATGATGC | CCTCTTTTACCCTGTGAAGAAAAT | 60 | FP | 6,313 | 249 | 205e |
| L1HS187 | AL357153 | 14 | Inserted in repeats | Inserted in repeats | … | R | 6,059 | … | … |
| L1HS189 | AL512407 | 6 | Inserted in repeats | Inserted in repeats | … | R | 907 | … | … |
| L1HS190 | AC073893 | Y | TCTACTGTTCCATGCAATAGATGTG | GGGTTCTTCTACCTCTGCATAACT | 57 | NR | 3,243 | 190 | 331 |
| L1HS191 | AC007972 | Y | TCCTCCAAGACCCTCTAAAATAAAT | TTTTGTCTTCCCTGAGTAAATTCTG | 60 | FP | 2,645 | 122 | 251 |
| L1HS192g | AC018680 | 4 | TTTCACTTTTTCTATGGTGATGAGG | CTTAGAATGTTACACTTTTCCGACA | 60 | FP | 6,218 | 155 | 196 |
| L1HS193 | AC018503 | 3 | CTACAGTGGCATTTCTTTAGGACAA | TATACAACAGAACTGAATCACTGAC | 60 | FP | 6,296 | 239 | 288 |
| L1HS195 | AC044791 | 15 | GCTTACATCTCAAATTCTGGTACCTT | TGTAAGAGCCAAAGCCTTTTAAACT | 60 | FP | 1,521 | 150 | 209 |
| L1HS196 | AC025263 | 12 | Inserted in repeats | Inserted in repeats | … | R | 6,071 | … | … |
| L1HS197 | AC027332 | 5 | TGGAGTAGAATTCAAGCAAACTGAA | AGAGTTTATGATAGGTCCCCATTCT | 60 | HF | 6,226 | 97 | 260e |
| L1HS200 | AC009892 | 19 | Inserted in repeats | Inserted in repeats | … | R | 1,686 | … | … |
| L1HS202 | AL391097 | 20 | TTGTACCTATGATTTGTGTGATAGGC | GCTCTACATAAAAAGATGTTCACCA | 60 | FP | 990 | 754 | 435 |
| L1HS203 | AL354750 | 10 | Inserted in repeats | Inserted in repeats | … | R | 152 | … | … |
| L1HS204 | AL157815 | 13q | ACTAGTTGATGACAAACTGGATGTG | GAGTGGCATAATCAATTGCTAGAGA | 60 | FP | 647 | 126 | 182e |
| L1HS206 | AL355382 | 6 | GTTTGTCAAGTGACAGGAATCTCTT | GCTAAGTCATCAATAAGCCCCTAAT | 60 | FP | 2,704 | 154 | 186 |
| L1HS207g | AL354861 | 9 | CTTTGCATATCTCTGTCATCCTACA | GATGAGATCATTCACACACTTTCTG | 60 | FP | 6,208 | 164 | 170 |
| L1HS208 | AL354793 | X | AACATTGGGAGAAGTTTGCAGTAT | CCAAGTTGTTAAGCACTCCATAGTT | 60 | FP | 6,639 | 570 | 689e |
| L1HS209 | AL158159 | 9 | GATGAGTTATCTTTGACGCTTTGAC | TGATAGATGAATGAGCTTTATGGTC | 57 | FP | 508 | 118 | 213e |
| L1HS210 | AL135908 | 6 | ATGTGGGGAAGATGAAGAAATC | GAAAACCCCACTATAGGAGTAAATTG | 59 | NR | 5,322 | 132 | 564 |
| L1HS211 | AC079598 | 12 | TCTATCGTCTCTGTCTTCTTAATGC | AATGACACTCTGCCTTCAGACTTAG | 57 | NR | 3,001 | 275 | 407e |
| L1HS212 | AL157700 | Xq | TTCTAGCCCTCTACTAATGTCCTTG | TTCTAAGGTAGCTGCAGATAAGTGG | 60 | FP | 1,045 | 184 | 234e |
| L1HS213 | AC087432 | 3p | AATGCCTGATAAAAGTAGACACACC | GTGGGAATATATCTTCTTGGGTTT | 60 | HF | 1,710 | 89 | 188e |
| L1HS214 | AC007483 | 3 | TAGCTGAGAAACCATAAGCCTAGAA | ACCTGAATGTCCACTCATTCACT | 60 | HF | 4,159 | 328 | 330e |
| L1HS215 | AC037423 | 9 | Inserted in repeats | Inserted in repeats | … | R | 1,162 | … | … |
| L1HS216 | AC023880 | 7 | CTATACCAAATGCAGTCAGGATGTT | TCCCATAACTCTGTCACACTAGAAA | 59 | FP | 714 | 197 | 228 |
| L1HS217 | AC073148 | 7 | Inserted in repeats | Inserted in repeats | … | R | 6,063 | … | … |
| L1HS218 | AC016910 | 2 | TCTTACAGCACTATTCAGTGTTTGC | TTCCTCTCAAGGAACTCAAACC | 60 | FP | 6,136 | 82 | 174 |
| L1HS219 | AC021020 | 3 | Inserted in repeats | Inserted in repeats | … | R | 6,096 | … | … |
| L1HS220g | AC016635 | 5 | ATTGGCCTTCAGAAGTGATTAAGAC | TAGATAGCCAGACAAACAAACCTTG | 60 | LF | 6,244 | 135 | 260e |
| L1HS222 | AL445932 | 6 | TCTTTCTCCTCTTGTAATGTCTCAG | AAGATACTGTGCTTCACTCTTCTGG | 60 | LF | 6,195 | 118 | 238 |
| L1HS223 | AL450488 | X | Inserted in repeats | Inserted in repeats | … | R | 4,210 | … | … |
| L1HS224 | AL358934 | 9 | GATCTGAATCTTTGCTCTCCAGATA | ACGTGGTACAAAAGAAAACACTGTC | 60 | FP | 1,121 | 126 | 215 |
| L1HS225 | AL445523 | X | Inserted in repeats | Inserted in repeats | … | R | 3,537 | … | … |
| L1HS226h | AL353153 | 6 | CCCTAAGCCTGTCAGAAGTTAGTATC | GCCATGAAAGATAAGGAGATAAGAG | 60 | LF | 2,114 | 120 | 359 |
| L1HS227 | AL157701 | X | Inserted in repeats | Inserted in repeats | … | R | 518 | … | … |
| L1HS228 | AL353657 | 13q | AATATCCACTACCCAATTCCATAGG | GCTGCAATTTAGCAGGATTTCT | 60 | HF | 1,383 | 184 | 205 |
| L1HS230 | AL359174 | 6 | Inserted in repeats | Inserted in repeats | … | R | 1,291 | … | … |
| L1HS231 | AL354896 | 13 | GAGTATGAGAGCTCTGCTTTCTGTC | CTTGAAGGACTGGGATACTTGAAA | 60 | HF | 2,289 | 379 | 481 |
| L1HS232 | AL365367 | 1p32 | TGTCACTCCAGTGATAGAAGCTAGA | ACAGTTAACTTCAAGGCAGGTTGAC | 60 | FP | 1,181 | 69 | 214e |
| L1HS233 | AL357507 | 6 | TAGTTGTCTACAACCAAGTGCTGAG | TCTGCATAGATCAGGAATTCTAAGG | 59 | IF | 1,232 | 81 | 174 |
| L1HS234g | AL356438 | 6 | Inserted in repeats | Inserted in repeats | … | R | 6,092 | … | … |
| L1HS235g | AL158193 | 13 | ACAGGATCTTAAGGTTGAAGGTTTG | GGTTCTACCCAAAGTAGTCAAGAAA | 59 | IF | 6,441 | 420 | 179 |
| L1HS236 | AL365400 | X | Inserted in repeats | Inserted in repeats | … | R | 1,711 | … | … |
| L1HS238 | AL357519 | 6 | GCAGGTAGGATACATGTAAGCATTT | ATCACAGCAATGGCATATCATC | 60 | FP | 2,155 | 374 | 360e |
| L1HS240g | AL137845 | X | Inserted in repeats | Inserted in repeats | … | R | 6,103 | … | … |
| L1HS241 | AP003112 | 8q23 | GATAATCAGGTGATTGTGAACTGTG | CTACCACCCTTTTTACTCCCTTTAC | 60 | FP | 366 | 148 | 206e |
| L1HS242f | Z80899 | 6p21 | AGTTCACGGTCTCTATCTCTCCTTT | AACCTGTCTTTGACTGTTGAGC | 58 | IF | 576 | 150 | 277e |
| L1HS243 | AC019041 | 2 | CACTAACATTCTGCATCTCACAATC | GTGGGAGGACATGAATAACACAT | 58 | FP | 6,148 | 96 | 202 |
| L1HS244 | AC009269 | 15 | Inserted in repeats | Inserted in repeats | … | R | 5,512 | … | … |
| L1HS245 | AC017040 | 2 | AAGGCTCTTTATCACAGGAAGTACC | ACGTTAATCACCGATCATTGC | 60 | FP | 2,141 | 294 | 263e |
| L1HS246g | AC068723 | 15q21 | Inserted in repeats | Inserted in repeats | … | R | 6,224 | … | … |
| L1HS247 | AC009274 | 7 | GTGTGAAGTATTACCTCGGTGTTG | CTGTGTGGAGCAATAGTAACCAGAT | 60 | FP | 2,238 | 286 | 275 |
| L1HS248g | AL360236 | 6 | AGAACAAGTGAGTGGCTAAAACCTC | AGCCAACAATTTTCCCATCTC | 60 | FP | 6,705 | 658 | 710 |
| L1HS249 | AL355852 | X | Inserted in repeats | Inserted in repeats | … | R | 1,297 | … | … |
| L1HS250 | AL162373 | 13 | AGTACCTGGTGAGTTCTCCTCAAC | GGTCTTTTGTGAGATGTCATACCTG | 57 | FP | 2,055 | 110 | 194e |
| L1HS251 | AL445429 | 6 | Inserted in repeats | Inserted in repeats | … | R | 757 | … | … |
| L1HS252g | AP002768 | 11q | Inserted in repeats | Inserted in repeats | … | R | 6,026 | … | … |
| L1HS253 | AP001955 | 4q | Inserted in repeats | Inserted in repeats | … | R | 1,780 | … | … |
| L1HS254 | AC013546 | 8 | Inserted in repeats | Inserted in repeats | … | R | 5,961 | … | … |
| L1HS255 | AC022731 | 8 | Inserted in repeats | Inserted in repeats | … | R | 1,104 | … | … |
| L1HS256 | AC019218 | 8 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS257 | AC016756 | 8 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS258 | AC024905 | 3 | GATTGGACTCCATTTCCTCTTGTAT | ATAAATTCTGGGACCTCTGCTTAAT | 57 | FP | 1,717 | 1,011 | 643 |
| L1HS259 | AC020707 | 9 | Inserted in repeats | Inserted in repeats | … | R | 1,893 | … | … |
| L1HS260 | AL354982 | 9 | GGCAACGGAATAATAGCTTCA | GTCAGCACTCCCATCTTAAATGTCT | 57 | HF | 6,461 | 358 | 510e |
| L1HS261 | AL161631 | 9 | Inserted in repeats | Inserted in repeats | … | R | 1,904 | … | … |
| L1HS262 | AC013579 | 1 | GATCCCTGTGTCTGGAGCACT | GGAATTCATGGAGAAGGTGAGTT | 60 | FP | 1,148 | 97 | 186 |
| L1HS263 | AL356139 | 9q | Inserted in repeats | Inserted in repeats | … | R | 889 | … | … |
| L1HS264 | AL391643 | 9 | GAGGAGGAAGAAGGCTGATAATATG | GACAGCCACTAAGTTAATGAGATCC | 60 | FP | 284 | 133 | 174e |
| L1HS265g | AC018938 | 9 | GCATTATTTCTGGAGCACTCACT | GTCTTGTGCTATTAAGCCTGGTCT | 60 | FP | 6,087 | 105 | 207 |
| L1HS266 | AL137021 | 9q31 | Inserted in repeats | Inserted in repeats | … | R | 207 | … | … |
| L1HS268 | AC025428 | 10 | CTTTGCTCTCTTGCTCCATGTAT | TATCTGTTTACCAACCCATCTCACC | 60 | FP | 6,235 | 90 | 283e |
| L1HS269 | AC020642 | 10 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS270 | AC026989 | 14 | Inserted in repeats | Inserted in repeats | … | R | 313 | … | … |
| L1HS271 | AC020644 | 10 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS272 | AL157787 | 10 | CTATGTCCTAGCCTTCCCAGATG | AGAAAAGACAAGACAGGATAGGG | 58 | FP | 1,125 | 201 | 223e |
| L1HS273 | AL354951 | 10 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS274 | AC027118 | 10 | GCACATGGCTTCTTAGCTAACTT | CTTTCTTGCATAAATGACTCTGTCC | 57 | FP | 2,081 | 611 | 317 |
| L1HS275 | AL590378 | 10 | Inserted in repeats | Inserted in repeats | … | R | 1,414 | … | … |
| L1HS277 | AC026393 | 10 | Inserted in repeats | Inserted in repeats | … | R | 312 | … | … |
| L1HS278g | AC027591 | 11 | Inserted in repeats | Inserted in repeats | … | R | 6,020 | … | … |
| L1HS280 | AC078971 | 11 | Inserted in repeats | Inserted in repeats | … | R | 6,063 | … | … |
| L1HS281 | AC037434 | 11 | Inserted in repeats | Inserted in repeats | … | R | 343 | … | … |
| L1HS282 | AP001002 | 11q | CTTACCTCCAGAGCATGCACATTAT | CCCCTCCTTCTCAATTTAAGGTTAC | 61 | FP | 6,448 | 156 | 249e |
| L1HS283 | AP000409 | 11 | Inserted in repeats | Inserted in repeats | … | R | 2,294 | … | … |
| L1HS284 | AC018619 | 11 | AGATAGGAGAATCCTCTGGTCTTCT | CTATTGTTGGGTACTTGGGTCACT | 58 | FP | 1,877 | 174 | 268e |
| L1HS285 | AC015772 | 11 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS286 | AC011829 | 11 | Inserted in repeats | Inserted in repeats | … | R | 1,189 | … | … |
| L1HS287 | AC021304 | 11 | CCTTTTATCTGAAATAAGTGGTTGG | CTTCCTTTAGCTGGGCTGTTCTAAG | 61 | VLF | 1,693 | 95 | 216e |
| L1HS288g | AC016775 | 11 | Inserted in repeats | Inserted in repeats | … | R | 6,081 | … | … |
| L1HS289 | AC021245 | 11 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS290 | AP001179 | 11q | CCTGTCAGTCTTATCTTTGCTCTACA | GGCATAGAGACAAATCCAAATTAAG | 60 | NR | 6,537 | 285 | 235 |
| L1HS291g | AC025410 | 6 | CTCCCACTACTTTATGGGAAGGT | AGGACTTCCAATTCCTAGTATGCAG | 58 | HF | 5,658 | 216 | 271e |
| L1HS292 | AC073915 | 12q | GACTCCACACTAGCTTCTTTGACTT | GAGACTCAGTTGACAAGGAGTTACC | 60 | FP | 1,117 | 117 | 213 |
| L1HS293 | AC026831 | 12 | TTACAATGGATACGTTAGACAGCTC | CCATAATTGGTTAGGATGATGAGAC | 60 | LF | 2,517 | 417 | 317e |
| L1HS294 | AC027442 | 12 | CTTTACCTGTTCCACTAATCAC | GGCACAAGATGGATATAAAGGA | 57 | FP | 6,154 | 103 | 168 |
| L1HS295 | AC012144 | 13 | GAGGAATGGTTGAACAGCTTG | ATGTGGCTGGAGAAATACCTCTAAG | 61 | FP | 713 | 100 | 208e |
| L1HS297h | AC064857 | 12 | GTCCAGAGTGATGCATTTTATTTGG | GCATAGTCATTTAATGCATGTCAGC | 58 | FP | 771 | 461 | 549e |
| L1HS298 | AC025880 | 12 | ATATACCATACTCCTTTCCCCTTCC | TGAGCCCTGTATTTTAATCACTTGT | 60 | LF | 1,037 | 80 | 235e |
| L1HS299 | AC027287 | 12 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS300 | AC026577 | 1 | Inserted in repeats | Inserted in repeats | … | R | 3,364 | … | … |
| L1HS301 | AC027382 | 1 | CTATCCCATAGATGGTGGGTAGAAT | GAGGAAATAGCACAGGTATGGTAAA | 61 | IF | 1,770 | 411 | 431 |
| L1HS302 | AL365220 | 1p21 | Inserted in repeats | Inserted in repeats | … | R | 2,391 | … | … |
| L1HS303 | AL451063 | 1 | CTATGTTCTGGGAGAAGAGCTGAT | CTAGGGTCAGAAAGAACTTTGATGT | 62 | FP | 780 | 87 | 170 |
| L1HS304 | AL354885 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS305 | AC016371 | 1 | CAAAAAGCAGCCCTATATTAGC | GCCTGCCTCATTATCTTTCATT | 58 | FP | 3,998 | 415 | 409e |
| L1HS306 | AL136459 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS307 | AL390860 | 1 | Inserted in repeats | Inserted in repeats | … | R | 6,066 | … | … |
| L1HS308 | AL390200 | 1 | CCTACTAGGCCCTCTTCTTTTGTAT | GTCTTGTTGTGCCAGACACTTTA | 62 | IF | 3,441 | 455 | 652e |
| L1HS309 | AL391904 | 1 | Inserted in repeats | Inserted in repeats | … | R | 2,161 | … | … |
| L1HS310 | AL157946 | 1p31 | Inserted in repeats | Inserted in repeats | … | R | 286 | … | … |
| L1HS311 | AL162402 | 1p13 | Inserted in repeats | Inserted in repeats | … | R | 693 | … | … |
| L1HS312 | AL139225 | 1p13 | Inserted in repeats | Inserted in repeats | … | R | 783 | … | … |
| L1HS313 | AC034157 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS314 | AL357975 | 1 | TGGCTAGCAAAAAGGTGGAC | AGGGCAGAGAAAAATGGTCA | 58 | IF | 6,215 | 109 | 255e |
| L1HS315 | AL139137 | 1 | AAGTCCCAATTCCCTAGTCTGTCT | GACACAGAATCATGTCACAATACCC | 61 | FP | 6,286 | 77 | 332 |
| L1HS316 | AC026905 | 1 | CTTTAGCAGTTTTCATGCCTCCT | AGGTTGATGGTAACCTGTAGGAAC | 59 | FP | 6,240 | 173 | 245 |
| L1HS317 | AL356323 | 1 | CTCTGCCTCAAGTGTGTCTTGACTA | GAGAACACACCCTTGCTCAGTAAAT | 59 | FP | 901 | 711 | 626e |
| L1HS318 | AL365225 | 1 | Inserted in repeats | Inserted in repeats | … | R | 5,243 | … | … |
| L1HS320 | AL357973 | 1 | GGGATTCAAATGGGAAACAAG | CTCCTTTCCAGTATCTGCTCTTATG | 60 | IF | 1,748 | 140 | 305 |
| L1HS321 | AL356455 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS323 | AC068071 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS324 | AL139284 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS325 | AL360154 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS326g | AC025702 | 1 | CTCACCGTTATCAAAGGGTAGAAAC | CTAGCCCCAAATTTGAGAAACAG | 60 | FP | 6,250 | 156 | 289e |
| L1HS327 | AC018874 | 1 | GGTACAATGTAATCATGGGTTGG | GAGTTAACCGTTAGTCCACAAGATG | 58 | FP | 4,695 | 172 | 413 |
| L1HS328 | AL135842 | 1q21 | Inserted in repeats | Inserted in repeats | … | R | 2,188 | … | … |
| L1HS329 | AC058795 | 1 | CTTCACCTCTGAATGACACACAT | GGCTTCATAATGCATCGCTAA | 60 | FP | 1,188 | 454 | 365e |
| L1HS330 | AL139285 | 1p31 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS331 | AL138777 | 1q31 | Inserted in repeats | Inserted in repeats | … | R | 1,064 | … | … |
| L1HS332 | AC008110 | 1 | CATGTTAGAACTGGCTCAAGTATCC | CCTGCAGAAATTTGCCTTTAG | 58 | IF | 2,850 | 87 | 227e |
| L1HS333 | AC023026 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS334 | AC026253 | 2 | ACACTTCTGAGAATTTCCCTGTG | TTACTCCCTCTTTACTGTCTTGGTG | 60 | FP | 1,095 | 199 | 341 |
| L1HS335 | AC023434 | 1 | CATGCATCTCTGAACTACTGACTTG | ATAAAAACCTGTTTAGGCCAAGG | 60 | IF | 1,276 | 395 | 284e |
| L1HS336 | AC013264 | 1 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS337 | AC010890 | 2 | GGTACAATATGAGGCATCACGTA | GTAGCATCCTTTATAGCTTTGCTGA | 60 | HF | 3,174 | 210 | 329e |
| L1HS338 | AC068953 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS339 | AC017035 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS341 | AC069384 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS342 | AC018591 | 2 | GAGACTCAGTTGACAAGGAGTTACC | AAACAGGACCTGCTGTCCATAA | 60 | FP | 1,087 | 78 | 183e |
| L1HS343 | AC068572 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS344 | AC048375 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS345 | AC073509 | 2 | CACAGCATTTACCAAAGCACTC | CTCAGTTCATTGCACAGTTTGG | 60 | LF | 2,587 | 192 | 229e |
| L1HS346 | AC016674 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS348 | AC018378.3 | 2 | GAAATGGGAAGAGGAGTTGACA | CCTATTTTTATCTCAGCTGATGTCG | 60 | HF | 748 | 283 | 526e |
| L1HS349 | AC009963 | 2 | GGAGCTGGGAGAATTATTGAAAC | CCACTCTCAACTACTGTCCAACAAG | 60 | HF | 229 | 114 | 182 |
| L1HS350 | AC022605 | 2 | TGGTATATAGTTCTAAGGACCCACAG | GCTACTTTTGCTTCTGGGTGTT | 58 | FP | 725 | 243 | 331e |
| L1HS351 | AC013262 | 2 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS352 | AC073874 | 2 | Inserted in repeats | Inserted in repeats | … | R | 970 | … | … |
| L1HS353 | AC019324 | 2 | TCCATGATAGAACACACTCTTCC | AATCCCTGTCAAAACCAATCC | 59 | HF | 1,822 | 426 | 167 |
| L1HS354 | AC012442 | 2 | Inserted in repeats | Inserted in repeats | … | R | 6,217 | … | … |
| L1HS355 | AC011901 | 2 | Inserted in repeats | Inserted in repeats | … | R | 6,067 | … | … |
| L1HS356 | AC009290 | 2 | CATCCTGTTGAAGAACAGAGAGATG | ATAGAGTGACCAGAAACTCCAGAGA | 60 | FP | 6,290 | 156 | 250e |
| L1HS358 | AC019130 | 2 | GAGACTCTTTGGACTCAGAGTATAACC | AGTCCTGTCATACCAGTTATTGGAC | 59 | FP | 6,621 | 128 | 673 |
| L1HS359 | AC024062 | 2 | Inserted in repeats | Inserted in repeats | … | R | 4,808 | … | … |
| L1HS360 | AC023416 | 2 | GAGGTCTTTGTGCAGAGGTATAAGA | CTCACCAACATCAGTTTCCTTTG | 60 | IF | 3,222 | 153 | 218e |
| L1HS361 | AC073642 | 2 | AGCCCATTAGATATATGTGGCTGT | CTTTTTATATTGGTCACCCCCAAC | 61 | FP | 6,319 | 281 | 372e |
| L1HS363h | AC010913 | 2 | GTTAGACAGCGACATGCACAG | ACCTCTGTGCCTTACCAAAAAC | 60 | FP | 577 | 106 | 198e |
| L1HS364 | AC026860 | 3 | CTTAGCCTCTGTCTTTAGGGAAAAC | CATGACCAACGGTGCATAATA | 60 | HF | 6,139 | 97 | 170e |
| L1HS365 | AC068355 | 3 | Inserted in repeats | Inserted in repeats | … | R | 888 | … | … |
| L1HS366 | AC083853 | 3 | AGAAAACTTCCAGACACCTATCC | CTATGTCCTAGCCTTCCCAGATG | 60 | FP | 1,088 | 163 | 183 |
| L1HS367 | AC078805 | 3 | GACTCATATTACCCTGGACAACAAC | AGTCTCTCCTTGCTCAGTTTGGTAG | 60 | FP | 6,784 | 83 | 401e |
| L1HS368 | AC023144 | 3 | Inserted in repeats | Inserted in repeats | … | R | 168 | … | … |
| L1HS369 | AC076971 | 3q | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS370 | AC068365 | 3 | GCAATCAGTTTCACACTCAACTG | CATGTGATCTATTGTGTACCATCAGG | 58 | FP | 3,436 | 146 | 323e |
| L1HS371 | AC026611 | 3 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS372 | AC022077.13 | 3 | GAAGAGAAAGAGGAAATAGCACAGG | CTATCCCATAGATGGTGGGTAGAAT | 60 | IF | 1,779 | 599 | 431e |
| L1HS373g | AC022838 | 3 | GAAAGAGAGTTCTCTGTACCACACC | GTCATGTCCCAACAGGACATTT | 60 | VLF | 6,294 | 215 | 231 |
| L1HS374 | AC063919 | 3 | Inserted in repeats | Inserted in repeats | … | R | 6,265 | … | … |
| L1HS375 | AC023139 | 3 | TGTGGTACAGTCACACTACAAAG | GATAGCATACACCATCATGCACT | 60 | IF | 3,862 | 430 | 469e |
| L1HS376 | AC069203 | 3 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS377 | AC078856 | 3q | GGGAGATGTAGAGTTTTATGTGACC | CTAATGTGCTGGGCAAACATAAGAT | 57 | FP | 577 | 139 | 201 |
| L1HS378 | AC069225 | 3 | CTCCCCTTTTTGCCTTACTTCT | CTTACTTGCAATAGCCCATTCAC | 60 | IF | 5,569 | 646 | 369e |
| L1HS380 | AC024470 | 3 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS382 | AC055732 | 3 | GCAGACACTAGAAGCTTTTGCAT | GCCACAAAATCTGGCACTTATAG | 58 | FP | 3,357 | 426 | 185 |
| L1HS383g | AC017085 | 3 | ATTAGTCAGTAATAGAGCCCCCTGT | AAAGACTTCTTTCCAGCTCTACCC | 60 | FP | 6,493 | 267 | 515 |
| L1HS385 | AC078808 | 3 | Inserted in repeats | Inserted in repeats | … | R | 6,068 | … | … |
| L1HS386 | AC023438 | UL | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS387 | AC069417 | 3 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS388 | AC025818 | 3 | Inserted in repeats | Inserted in repeats | … | R | 713 | … | … |
| L1HS389 | AC024216 | 3 | CATGTAGAGATGATCTTCAAAGCTG | GCCTGATAAAAGTAGACACACCTG | 60 | FP | 1,782 | 162 | 263 |
| L1HS390 | AC036128 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS391 | AC022040 | 4 | GTGGACATCAGAGTATCCCTTTCT | AGAAGGGTACATGACAACTGGTTAG | 60 | HF | 889 | 113 | 203 |
| L1HS393 | AC013336 | 4 | TACACAGAATCTGATGCTAGGAGAG | CGGGAACATAAAGTCATAGCGTAAC | 61 | LF | 751 | 277 | 412e |
| L1HS395 | AC067804 | 4 | GTTGCATTTTGGAAAGGAAGG | TAGTGGAAAGACAGACAGTTTAGGG | 61 | IF | 1,218 | 119 | 214 |
| L1HS396 | AC007512 | 4 | AGACTCAAACTCAAAACTCCTGTGT | TCACAAGCAGACATTTCTTACTGAA | 60 | FP | 6,643 | 562 | 373e |
| L1HS397 | AL161439 | 6 | ACTCATCCTAGAGCTTTACCCAGTT | CACAAAGTCAACAGGTTTGATCC | 58 | FP | 1,085 | 259 | 231e |
| L1HS398 | AC069349 | 8 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS399 | AC027502 | 4 | Inserted in repeats | Inserted in repeats | … | R | 614 | … | … |
| L1HS401 | AC068037 | 4 | Inserted in repeats | Inserted in repeats | … | R | 1,342 | … | … |
| L1HS402 | AC020593 | 4 | Inserted in repeats | Inserted in repeats | … | R | 361 | … | … |
| L1HS403h | AL158816 | 6 | Inserted in repeats | Inserted in repeats | … | R | 360 | … | … |
| L1HS404 | AC021700 | 4 | CCACCTTACGTTCAGCTGTTAAT | CGGTGATTAGGTGACAGCTTTT | 60 | LF | 3,262 | 163 | 231e |
| L1HS405 | AC032017 | 4 | ATCAAAAGTCCTGTGTGTTTGTCTT | GAAATTTTGCTAGACATAGCTGTCC | 60 | FP | 1,206 | 396 | 202e |
| L1HS406 | AC067842 | 4 | GCAAGTTTTACCCATAGTACACAGG | GTATGTAGAAGGCAGGGGTACACT | 60 | HF | 3,589 | 209 | 302 |
| L1HS407 | AC041010 | 4 | CTCACCAGTACGAGAAGCAAGTT | TCTGACCTAGGGATGATTCTTCA | 60 | FP | 413 | 227 | 217 |
| L1HS408 | AC019133 | 4 | TTTTAGCCAAGCTCTTTGTTCC | CATTATGGCAGCGTAGACATTG | 56 | FP | 2,059 | 106 | 209 |
| L1HS409 | AC027782 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS410g | AC011633 | 4 | GCTAAGCAATGGAGGAAAATATCG | TGTACATGGTGTGAGGTATGAA | 57 | IF | 6,211 | 100 | 244e |
| L1HS411 | AC073338 | 4 | ACACACACACGATGGAAAGTATCT | AGCACATCCTAAATCTTCCTCTCT | 60 | FP | 2,670 | 136 | 246 |
| L1HS412 | AC067901 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS413g | AC023332 | 4 | TCATGAGCATCACTCTTACCATGT | ACTCAGCTGACTTGCCATAAATGT | 60 | IF | 6,199 | 127 | 191 |
| L1HS414 | AC025955 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS415 | AC009816 | 4 | TCAGACCCATATATGAGCATAACC | GCTTAGAAGAATTTTTAGCCAGGTG | 56 | HF | 1,360 | 590 | 476e |
| L1HS416 | AC068256 | 4 | TTAGTCACTATGACTTGAGCCACTT | TAGTGATAGTGTAGAGAGGGGGTTG | 61 | FP | 822 | 238 | 284 |
| L1HS417 | AP001860 | 4 | Inserted in repeats | Inserted in repeats | … | R | 865 | … | … |
| L1HS418g | AC011981 | 2 | CGATTTCTGTCTTTGTGAACGTAGT | CCTTACAGAGTAGAAATCTCACGAT | 60 | IF | 6,380 | 328 | 358 |
| L1HS419 | AC061978 | 4 | Inserted in repeats | Inserted in repeats | … | R | 6,034 | … | … |
| L1HS420 | AC041038 | 4 | Inserted in repeats | Inserted in repeats | … | R | 6,066 | … | … |
| L1HS421 | AC024974 | UL | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS422 | AC009577 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS423 | AC022672 | 11 | CTCCCTGTCTTCTGGGTTAAAATA | GGAAGTCCCACTTTTTCAGTAGAG | 60 | HF | 5,680 | 201 | 248e |
| L1HS424 | AC080124 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS425 | AC013724 | 4 | Inserted in repeats | Inserted in repeats | … | R | 6,120 | … | … |
| L1HS426 | AC023921 | 5 | AGATTCCCTTTGGTATCCAAATCAC | GTTGCCATACTCCGCATAAAGTC | 60 | IF | 3,394 | 204 | 252 |
| L1HS427 | AC015990 | 4 | TACGGGCAAAGACTGAGAGTACTAA | TTCAGCCTTCTGACATCAAACT | 57 | IF | 2,230 | 139 | 220e |
| L1HS429 | AC060816 | 4 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS430 | AC024963 | 4 | CAGAGAACCAACATGTAGGAACAA | GTTACAGGTCAAAGGAGGTCTGAG | 60 | LF | 4,034 | 127 | 223e |
| L1HS432 | AC011399 | 5 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS433 | AC027339 | 5 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS434 | AC010437 | 5 | ACCTGGGCCACATTTATTTTTC | TGTAGAAGAAGACACCGTCGTTAG | 60 | FP | 2,637 | 250 | 246e |
| L1HS435 | AC026403 | 5 | GACTCAGTTGACAAGGAGTTACCA | ACACTAGCTTCTTTGACTTCACCA | 55 | FP | 1,115 | 111 | 211e |
| L1HS437 | AC023526 | 5 | ATCTATCATTTATCTGCCCCGTCT | ACAAGGATTAGCAGGAAGTCTGTT | 60 | IF | 2,954 | 256 | 201e |
| L1HS438 | AC011433 | 5 | TCCTCTCACCAACCACATAAAGTA | ATCCCTTGGATACAAAGATGTGC | 60 | FP | 1,909 | 570 | 345 |
| L1HS439 | AC016573 | 5 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS440 | AC010409 | 5 | Inserted in repeats | Inserted in repeats | … | R | 6,133 | … | … |
| L1HS441 | AC026444 | 5 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS442 | AC027325 | 5 | GACGGTTACTCAGAAAAACACAAG | GTAGATGCCACTGTTACCCTGACT | 60 | IF | 907 | 224 | 185e |
| L1HS443 | AC021600 | 5 | GCTAGACTCTCTACCTTTGGCTTT | TGATACCTGACTCTATGCACCACT | 56 | FP | 891 | 261 | 382 |
| L1HS444 | AC027315 | 5 | TTATTGGAATAGCTTCTCCTGTCAC | GCTGTTCCTAACTCTAGTCCTCCA | 60 | FP | 464 | 303 | 296e |
| L1HS445 | AC008374 | 5 | Inserted in repeats | Inserted in repeats | … | R | 551 | … | … |
| L1HS446 | AC010314 | 5 | CTCGTGACATTTCCATCATATAGC | TTAAGTCACCTAAGGGTTGTAAGTG | 56 | LF | 6,142 | 109 | 182e |
| L1HS447 | AC018759 | 5 | GTACATCTCTTTGGACACTTCCACT | GTTTAAGTCCAACATCCTGTTCTG | 59 | IF | 691 | 560 | 386 |
| L1HS448 | AC016545 | 5 | GTCAATTAGAGCATGAAGAAACCAC | GTACATCTCTTTGGACACTTCCACT | 60 | IF | 652 | 525 | 382e |
| L1HS449 | AC011378 | 5 | CTAGGGAGGTGAAAATTCAGATGT | GCATGTTGCACAACAGTATGTA | 60 | FP | 1,797 | 281 | 315e |
| L1HS450 | AC011413 | 5 | GTGAAGACTGTTGGTCAGTTACTTGT | GTCATTGAGATTGGCAGGTAAAAG | 60 | HF | 6,179 | 128 | 189e |
| L1HS451 | AC010490 | 5 | Inserted in repeats | Inserted in repeats | … | R | 994 | … | … |
| L1HS453 | AL360232 | 6 | Inserted in repeats | Inserted in repeats | … | R | 6,064 | … | … |
| L1HS455 | AC027643 | 6 | CATACACAAGGGCGAAGAGTTAAA | GCCTCTTTTACATCAGTTACCACTC | 60 | FP | 259 | 110 | 213e |
| L1HS456 | AC026966 | 6 | TAACACTTAGTGATTGCTGGGAGAG | GGACAAGGTGAAGTGGAAAACTAGA | 60 | FP | 1,641 | 121 | 215 |
| L1HS457 | AC025887 | 18 | Inserted in repeats | Inserted in repeats | … | R | 286 | … | … |
| L1HS460 | AL355489 | 6 | Inserted in repeats | Inserted in repeats | … | R | 6,044 | … | … |
| L1HS461 | AL358992 | 6 | ATCCAGCAAAAGTATCCCTTAAGTA | TCCTGTCCCAATTCTTTGTATTAT | 60 | LF | 4,143 | 324 | 417 |
| L1HS462 | AC069403 | 11 | Inserted in repeats | Inserted in repeats | … | R | 4,163 | … | … |
| L1HS463 | AL391336 | 6 | ATTAAATCTGTGTGGGAGTGG | AGGGTGACTTCAGTGATATCTTCA | 60 | FP | 6,304 | 247 | 346 |
| L1HS465 | AL356601 | 6 | Inserted in repeats | Inserted in repeats | … | R | 1,936 | … | … |
| L1HS469h | AC020586 | UL | GGTACTGGCTGTTCAGTATTTTT | GTCTCAAAGCCCATTTCATAGTTC | 60 | FP | 6,458 | 101 | 212e |
| L1HS472 | AC018400 | UL | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS476 | AC079756 | 7 | Inserted in repeats | Inserted in repeats | … | R | 897 | … | … |
| L1HS477 | AC024730 | 7 | Inserted in repeats | Inserted in repeats | … | R | 1,271 | … | … |
| L1HS478 | AC069008 | 7 | Inserted in repeats | Inserted in repeats | … | R | 991 | … | … |
| L1HS479 | AC079855 | 7 | CACTCGAAGGGTAAGTGAGATTTT | CCACTAGCGCACCATTTTTCTAAT | 58 | FP | 6,223 | 146 | 276 |
| L1HS480 | AC021836 | 4 | AGAGGTAACCACTACCTTGCAACT | GCCTCATGACAGGAGAAGAGATAAA | 60 | IF | 2,701 | 272 | 265 |
| L1HS483 | AC026011 | 8 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS484g | AC073647 | 7 | Inserted in repeats | Inserted in repeats | … | R | 6,692 | … | … |
| L1HS485 | AC027189 | 8 | CTCAGTTCCACATAAACCTTGACA | GAAGCAATTAACCTAGCAGTAGGAC | 60 | FP | 548 | 74 | 183e |
| L1HS486 | AL356516 | 9 | CCCTCATCACCAAATATCTGAGAA | AGCTGACAGTCTAGTGAATGAGGTC | 60 | IF | 905 | 139 | 196 |
| L1HS487 | AL162731 | 9 | Inserted in repeats | Inserted in repeats | … | R | 6,079 | … | … |
| L1HS488g | AL353649 | 9 | CAAATTGTCAATGCTAACCACTCC | GGAAAAAGGCACTTTGGCTTATC | 62 | FP | 6,787 | 724 | 472e |
| L1HS489 | AC009284.2 | 9 | TCTCCAGAAACCATCACAGTAAGA | AGGAGTTGAAAGTAGGATGGGTTT | 60 | FP | 322 | 104 | 202e |
| L1HS490h | AL358937 | 9 | CAGCTGTCTTGCTAAGAATCCAT | AGACCACAGACTCTTTGAGGGTAAG | 60 | FP | 2,289 | 397 | 206 |
| L1HS491 | AL355303 | 10 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS492 | AL450466 | 10 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS493 | AL138764 | 10 | GACTACCTTTCTGCGTATTCCTTTC | GTCTAACAGGTACACGAGACTCCAT | 61 | IF | 1,603 | 111 | 241e |
| L1HS494 | AC068972 | 8 | Inserted in repeats | Inserted in repeats | … | R | 2,974 | … | … |
| L1HS495 | AC083848 | 8 | Inserted in repeats | Inserted in repeats | … | R | 1,341 | … | … |
| L1HS496 | AC024929 | 8 | CCTTTGGAAGAGAAAGAGGATATG | CTCCCAATGGAAAGGAACTTGTAT | 60 | FP | 617 | 70 | 177 |
| L1HS497 | AC060775 | 8 | GCCTAGTGGGAAGACAAAAAGTATT | GCTGTAATGTTAACCTCGAAGTCGT | 60 | FP | 950 | 346 | 439e |
| L1HS498g | AC067844.3 | 8 | AGGTTTCCCCAAAATTTACCC | CTGATGTGTGGATTCACTGTTCTT | 58 | FP | 6,281 | 184 | 295 |
| L1HS499 | AC024649 | 8 | Inserted in repeats | Inserted in repeats | … | R | 1,045 | … | … |
| L1HS500 | AC009630.5 | 8 | GTGTTGCCTTCACCACAATAGTA | TTTCTCCGAGTACAGGTTACGAG | 60 | FP | 1,145 | 206 | 227e |
| L1HS501 | AC022207 | 12 | GTTGGCAACTTACTCTCAAATGG | AAATACACTCGACTGGCCACTAA | 60 | FP | 6,254 | 199 | 306e |
| L1HS502 | AC011881 | UL | Inserted in repeats | Inserted in repeats | … | R | 537 | … | … |
| L1HS503 | AC055118 | 13 | GTGAGGAATGGTTGAACAGCTT | TGTGGCTGGAGAAATACCTCTAA | 60 | FP | 713 | 101 | 206e |
| L1HS504 | AL158045 | 13 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS505 | AL162716 | 13 | Inserted in repeats | Inserted in repeats | … | R | 384 | … | … |
| L1HS506 | AL138684 | 13 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS507 | AC064832 | 15 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS508 | AC048381 | 15 | ACAGAACCTTTTAGAGGGAATCG | CTCCGTGTGGTAAAATTAGCTGT | 58 | HF | 6,144 | 103 | 184 |
| L1HS509 | AL356017 | 14 | CACTCATGACTGCCTGACTTCT | CAGGGATTACTCTTCTGTTGTGG | 61 | FP | 443 | 131 | 220e |
| L1HS510 | AL390800 | 14 | Inserted in repeats | Inserted in repeats | … | R | 1,837 | … | … |
| L1HS511 | AL162632 | 14 | Inserted in repeats | Inserted in repeats | … | R | 6,088 | … | … |
| L1HS512 | AC021839 | 14 | AAAGAGACAATCCACAGCATAGTTG | GATTTATTCCTTCATGGAGATGTGC | 61 | HF | 2,071 | 722 | 266e |
| L1HS513 | AL160156 | 13 | CCAAACTTGAGCCTCCTGTAATC | CCTTGAAATAAGCAGGAAGAAGC | 61 | IF | 809 | 142 | 235e |
| L1HS514 | AL138961 | 13 | CCTCAGCTTTGGATCCTGTAGTT | AGAAGAATTGGGTCCTGTTGAA | 60 | FP | 6,670 | 334 | 361 |
| L1HS515 | AL163537 | 13 | GGATGGTAAAGGAGTGGCATAAT | TGTGGAGCCCAGATCTTTTAAT | 60 | FP | 637 | 106 | 193 |
| L1HS516 | AC044907 | 15 | CCACAGTTTACACAGAAGCTGAA | GAAGGAGTGGATGTGTTTCAGTAA | 60 | IF | 6,151 | 101 | 212 |
| L1HS518 | AC074236 | 15 | Inserted in repeats | Inserted in repeats | … | R | 2,636 | … | … |
| L1HS519 | AC074100 | 15 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS520g | AC015558 | 15 | Inserted in repeats | Inserted in repeats | … | R | 6,087 | … | … |
| L1HS521h | AC067951 | 15 | GCTTTGTTTACCTTTCTGCTCACT | CACCAAAAGGAGAAGCCAATAAAG | 60 | FP | 1,248 | 344 | 441e |
| L1HS522 | AC009555 | 15 | Inserted in repeats | Inserted in repeats | … | R | 190 | … | … |
| L1HS523 | AC009658.6 | 15 | CGTGGAAGATGTTACGAGGATTA | AGAGAATGCGATGTCGATTAGAG | 60 | FP | 570 | 105 | 204 |
| L1HS524 | AC020892 | 15 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS525 | AC009057 | 16 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS526g | AC025289 | 16 | ACCCTCCAAGGTAACTGAATCTTA | ATGCCCATGCTTGTTAGCTACTAC | 60 | IF | 6,076 | 223 | 324e |
| L1HS527 | AC026472 | 16 | Inserted in repeats | Inserted in repeats | … | R | 1,224 | … | … |
| L1HS528 | AC009021.4 | 16 | CGGATGGGAGCACAAAATTACTA | TGCCTACTAAGATACCTTGGAAATG | 61 | FP | 991 | 172 | 278 |
| L1HS529 | AC022164 | 16 | TGAGTAATGTGGCGGTTTAGTTC | AACCAGTCAAGAAGCCAAAGAG | 61 | FP | 6,143 | 116 | 193e |
| L1HS530 | AC009063 | 16 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS531 | AC055852 | 17 | Inserted in repeats | Inserted in repeats | … | R | 2,839 | … | … |
| L1HS532 | AL356138 | 20 | CCTCTAATCTATGGTGGATGCTCT | TGGTAGGGAGCTGGTAAAAGTCTA | 61 | FP | 308 | 175 | 242e |
| L1HS534 | AC007448 | 17 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS535 | AC034266 | 17 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS539 | AC034266 | 17 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS541 | AC068204 | 18 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS542 | AC023983 | 18 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS543 | AC009267 | 18 | TACATTAGTCTGCCTCTGATTCCA | GGCCATTCTTTTCATCTGTTGTAG | 61 | FP | 547 | 99 | 183 |
| L1HS545 | AC007768 | 18 | TGGGAACTCATGTTACAGTTTCAC | ATTTGTCATGATCACAGCCACCT | 59 | FP | 2,514 | 95 | 216 |
| L1HS546 | AP001460 | 18 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS547 | AC010966 | 18 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS548 | AP001113 | 18 | Inserted in repeats | Inserted in repeats | … | R | 6,237 | … | … |
| L1HS551 | AC021325 | 18 | Inserted in repeats | Inserted in repeats | … | R | 184 | … | … |
| L1HS552 | AP001564 | 18 | CAGTGAACTGCTTTCTCACAATTC | CAAGAAGTTTTCCTGGAGTCTCTC | 60 | IF | 4,144 | 123 | 235 |
| L1HS554 | AC027230 | 18 | Inserted in repeats | Inserted in repeats | … | R | 561 | … | … |
| L1HS556 | AC026898 | 18 | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS557 | AP001019 | 18 | ACAAAAGCACCTAGAAGCAGTCAT | CTTTTTCTCCTATGCTCGTGGTAT | 60 | FP | 2,277 | 85 | 229e |
| L1HS558 | AC015819 | 18 | TGCTTTCTTTCTTTCACATAGATCA | GCAGACACGAATCACAGTTTGTAT | 61 | HF | 983 | 128 | 203e |
| L1HS559 | AC023394 | 18 | Inserted in repeats | Inserted in repeats | … | R | 1,620 | … | … |
| L1HS561 | AC013620 | 14 | TACCCATTTAAAGGGCAAAGTG | CTACCCATTTAAACCACTAATGCTG | 61 | LF | 430 | 114 | 239e |
| L1HS562g | AC019175 | X | TGTCTGTTCAGTCCTTTCTCACAT | AGCAAAATGTATGCCGAAGACT | 59 | FP | 6,170 | 115 | 181 |
| L1HS564 | AC034155.5 | X | TGCAATTGACATAGATACTGCAGAG | CCCTTCCCTTTCTGTACATGTCTT | 61 | LF | 2,085 | 471 | 425e |
| L1HS565 | AL442646 | X | Inserted in repeats | Inserted in repeats | … | R | 6,029 | … | … |
| L1HS567 | AL158143 | X | End of sequencing contig | End of sequencing contig | … | EC | … | … | … |
| L1HS568 | AL356003 | X | Inserted in repeats | Inserted in repeats | … | R | 1,297 | … | … |
| L1HS569 | AC021992 | X | Inserted in repeats | Inserted in repeats | … | R | 596 | … | … |
Note.— Indeterminable data are denoted by ellipses.
Determined from accession information (GenBank) or by PCR analysis of monochromosomal hybrid cell-line DNA samples (National Institute of General Medical Sciences).
Amplification of each locus required 2 min 30 s at 94°C initial denaturing and 32 cycles for 1 min at 94°C, 1 min annealing temperature, and 1 min elongation at 72°C. A final extension time of 10 min at 72°C was also used.
EC = element at the end of sequencing contigs; R = element residing in other repeats; Paralog = element with a paralog; NR = element with inconclusive PCR results. Elements represented here are classified according to allele frequency as high-frequency (HF) (present in more than 2/3 [67%] but not in all alleles tested), intermediate-frequency (IF) (present in more than 1/3 [33%] of alleles tested but in no more than 2/3 [67%] of the alleles), low-frequency (LF) (present in no more than 1/3 [33%] alleles tested), or very-low-frequency (VLF) (or “private”) insertion polymorphisms or as fixed-present (FP) insertions (every individual tested had the L1 element in both chromosomes).
Empty product size is calculated computationally by removal of the Ta L1Hs elements and one direct repeat from the identified filled site. Subfamily-specific product size is calculated with an internal subfamily-specific primer located in the 3′ UTR to the proximal 3′ primer. For cases in which target-site duplication sequence was not found flanking the element, PCR product sizes may vary from those reported. Except as marked, all elements were assayed using the internal subfamily-specific primer and the flanking forward primer.
Found in 5′→3′ orientation in GenBank and assayed using the internal subfamily-specific primer and the flanking reverse primer.
Elements previously identified by Boissinot et al. (2000).
Full-length elements with intact ORFs.
Table A2.
Autosomal L1Hs Ta–Associated Human Genomic Diversity
|
African American |
Asian/Alaskan Nativea |
European German |
Egyptian |
||||||||||||||||||
| No. with Genotype |
No. with Genotype |
No. with Genotype |
No. with Genotype |
||||||||||||||||||
| Element | +/+ | +/− | −/− | fc | Hetd | +/+ | +/− | −/− | fc | Hetd | +/+ | +/− | −/− | fc | Hetd | +/+ | +/− | −/− | fc | Hetd | AvgHetb |
| L1HS2 | 1 | 7 | 11 | .24 | .37 | 11 | 6 | 0 | .82 | .30 | 8 | 9 | 3 | .63 | .48 | 7 | 7 | 2 | .66 | .47 | .40 |
| L1HS5 | 0 | 2 | 18 | .05 | .10 | 0 | 2 | 18 | .05 | .10 | 1 | 7 | 12 | .23 | .36 | 0 | 6 | 12 | .17 | .29 | .21 |
| L1HS6 | 17 | 1 | 0 | .97 | .06 | 18 | 0 | 0 | 1.00 | .00 | 18 | 0 | 1 | .95 | .10 | 14 | 0 | 0 | 1.00 | .00 | .04 |
| L1HS7 | 17 | 3 | 0 | .93 | .14 | 19 | 0 | 0 | 1.00 | .00 | 19 | 1 | 0 | .98 | .05 | 19 | 0 | 0 | 1.00 | .00 | .05 |
| L1HS13 | 15 | 0 | 0 | 1.00 | .00 | 15 | 0 | 0 | 1.00 | .00 | 18 | 0 | 0 | 1.00 | .00 | 18 | 1 | 0 | .97 | .05 | .01 |
| L1HS14 | 9 | 11 | 0 | .72 | .41 | 7 | 9 | 3 | .61 | .49 | 1 | 11 | 8 | .33 | .45 | 2 | 9 | 9 | .33 | .45 | .45 |
| L1HS15 | 13 | 4 | 2 | .79 | .34 | 20 | 0 | 0 | 1.00 | .00 | 18 | 2 | 0 | .95 | .10 | 15 | 5 | 0 | .88 | .22 | .17 |
| L1HS16 | 1 | 6 | 13 | .20 | .33 | 7 | 9 | 3 | .61 | .49 | 3 | 6 | 11 | .30 | .43 | 1 | 3 | 11 | .17 | .29 | .38 |
| L1HS18 | 19 | 1 | 0 | .98 | .05 | 19 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | 18 | 0 | 0 | 1.00 | .00 | .01 |
| L1HS20 | 3 | 15 | 2 | .53 | .51 | 9 | 7 | 3 | .66 | .46 | 14 | 6 | 0 | .85 | .26 | 15 | 5 | 0 | .88 | .22 | .36 |
| L1HS21 | 0 | 3 | 17 | .08 | .14 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 17 | .00 | .00 | .04 |
| L1HS26 | 5 | 4 | 9 | .39 | .49 | 8 | 1 | 3 | .71 | .43 | 11 | 2 | 2 | .80 | .33 | 11 | 4 | 3 | .72 | .41 | .42 |
| L1HS32 | 9 | 8 | 2 | .68 | .44 | 13 | 5 | 1 | .82 | .31 | 15 | 5 | 0 | .88 | .22 | 13 | 4 | 1 | .83 | .29 | .32 |
| L1HS34 | 0 | 10 | 10 | .25 | .38 | 3 | 14 | 3 | .50 | .51 | 1 | 10 | 6 | .35 | .47 | 1 | 5 | 12 | .19 | .32 | .42 |
| L1HS39 | 11 | 3 | 1 | .83 | .29 | 15 | 1 | 0 | .97 | .06 | 12 | 0 | 0 | 1.00 | .00 | 11 | 1 | 3 | .77 | .37 | .18 |
| L1HS43 | 4 | 10 | 6 | .45 | .51 | 8 | 11 | 1 | .68 | .45 | 12 | 7 | 1 | .78 | .36 | 7 | 9 | 1 | .68 | .45 | .44 |
| L1HS44 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 19 | .00 | .00 | .00 |
| L1HS46 | 16 | 3 | 0 | .92 | .15 | 16 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | 13 | 0 | 0 | 1.00 | .00 | .04 |
| L1HS57 | 0 | 3 | 17 | .08 | .14 | 0 | 2 | 18 | .05 | .10 | 0 | 3 | 17 | .08 | .14 | 6 | 4 | 9 | .42 | .50 | .22 |
| L1HS73 | 19 | 1 | 0 | .98 | .05 | 20 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | 18 | 0 | 0 | 1.00 | .00 | .01 |
| L1HS74 | 0 | 1 | 19 | .03 | .05 | 2 | 5 | 13 | .23 | .36 | 2 | 7 | 11 | .28 | .41 | 1 | 5 | 12 | .19 | .32 | .28 |
| L1HS77 | 6 | 12 | 2 | .60 | .49 | 19 | 1 | 0 | .98 | .05 | 18 | 2 | 0 | .95 | .10 | 17 | 2 | 1 | .90 | .18 | .21 |
| L1HS78 | 1 | 6 | 13 | .20 | .33 | 5 | 3 | 11 | .34 | .46 | 3 | 4 | 13 | .25 | .38 | 0 | 5 | 12 | .15 | .26 | .36 |
| L1HS85 | 0 | 0 | 9 | .00 | .00 | 0 | 3 | 17 | .08 | .14 | 0 | 2 | 18 | .05 | .10 | 0 | 2 | 14 | .06 | .12 | .09 |
| L1HS86 | 14 | 0 | 0 | 1.00 | .00 | 14 | 1 | 0 | .97 | .07 | 12 | 1 | 2 | .83 | .29 | 17 | 1 | 0 | .97 | .06 | .10 |
| L1HS104 | 7 | 8 | 5 | .55 | .51 | 9 | 5 | 4 | .64 | .47 | 5 | 12 | 3 | .55 | .51 | 10 | 5 | 3 | .69 | .44 | .48 |
| L1HS110 | 20 | 0 | 0 | 1.00 | .00 | 19 | 1 | 0 | .98 | .05 | 20 | 0 | 0 | 1.00 | .00 | 18 | 2 | 0 | .95 | .10 | .04 |
| L1HS112 | 0 | 2 | 17 | .05 | .10 | 0 | 5 | 14 | .13 | .23 | 1 | 4 | 15 | .15 | .26 | 1 | 1 | 7 | .17 | .29 | .22 |
| L1HS117 | 8 | 1 | 1 | .85 | .27 | 9 | 3 | 1 | .81 | .46 | 9 | 8 | 1 | .72 | .41 | 7 | 4 | 3 | .64 | .48 | .40 |
| L1HS118 | 0 | 6 | 13 | .16 | .27 | 3 | 8 | 8 | .37 | .48 | 0 | 7 | 13 | .18 | .30 | 0 | 3 | 15 | .08 | .16 | .30 |
| L1HS131 | 10 | 0 | 2 | .83 | .29 | 8 | 3 | 3 | .68 | .45 | 5 | 3 | 4 | .54 | .52 | 14 | 2 | 0 | .71 | .44 | .42 |
| L1HS132 | 2 | 12 | 6 | .40 | .49 | 4 | 13 | 2 | .55 | .51 | 3 | 8 | 9 | .35 | .47 | 0 | 9 | 11 | .23 | .36 | .46 |
| L1HS153 | 6 | 6 | 8 | .45 | .51 | 2 | 9 | 8 | .34 | .41 | 4 | 7 | 8 | .39 | .49 | 3 | 6 | 8 | .35 | .47 | .47 |
| L1HS157 | 17 | 0 | 0 | 1.00 | .00 | 17 | 1 | 0 | .97 | .06 | 18 | 1 | 0 | .97 | .05 | 18 | 0 | 0 | 1.00 | .00 | .03 |
| L1HS158 | 4 | 12 | 4 | .50 | .51 | 9 | 7 | 1 | .74 | .40 | 6 | 13 | 1 | .63 | .48 | 2 | 14 | 4 | .45 | .51 | .48 |
| L1HS160 | 18 | 0 | 0 | 1.00 | .00 | 18 | 0 | 0 | 1.00 | .00 | 19 | 1 | 0 | .98 | .05 | 16 | 0 | 0 | 1.00 | .00 | .01 |
| L1HS163 | 4 | 11 | 4 | .50 | .51 | 1 | 13 | 6 | .38 | .48 | 12 | 6 | 0 | .83 | .29 | 5 | 9 | 5 | .50 | .51 | .45 |
| L1HS166 | 0 | 3 | 17 | .08 | .14 | 4 | 7 | 9 | .38 | .48 | 3 | 10 | 7 | .40 | .49 | 1 | 5 | 12 | .19 | .32 | .36 |
| L1HS169 | 13 | 1 | 1 | .90 | .19 | 8 | 8 | 2 | .67 | .46 | 12 | 4 | 1 | .82 | .30 | 12 | 0 | 0 | 1.00 | .00 | .24 |
| L1HS171 | 3 | 9 | 8 | .38 | .48 | 0 | 6 | 13 | .16 | .27 | 1 | 15 | 3 | .45 | .51 | 1 | 2 | 10 | .15 | .27 | .38 |
| L1HS172 | 14 | 4 | 2 | .80 | .33 | 5 | 12 | 3 | .55 | .51 | 12 | 5 | 3 | .73 | .41 | 10 | 9 | 1 | .73 | .41 | .41 |
| L1HS173 | 15 | 1 | 0 | .97 | .06 | 17 | 0 | 0 | 1.00 | .00 | 12 | 0 | 0 | 1.00 | .00 | 4 | 1 | 3 | .56 | .53 | .15 |
| L1HS177 | 20 | 0 | 0 | 1.00 | .00 | 18 | 0 | 0 | 1.00 | .00 | 19 | 1 | 0 | .98 | .05 | 12 | 0 | 0 | 1.00 | .00 | .01 |
| L1HS178 | 17 | 3 | 0 | .93 | .14 | 19 | 0 | 0 | 1.00 | .00 | 19 | 1 | 0 | .98 | .05 | 12 | 1 | 0 | .96 | .08 | .07 |
| L1HS180 | 1 | 6 | 13 | .20 | .33 | 1 | 9 | 10 | .28 | .41 | 4 | 10 | 6 | .45 | .51 | 4 | 8 | 7 | .42 | .50 | .44 |
| L1HS197 | 11 | 1 | 1 | .88 | .21 | 8 | 1 | 0 | .94 | .11 | 12 | 0 | 1 | .92 | .15 | 14 | 0 | 0 | 1.00 | .00 | .12 |
| L1HS213 | 20 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | 18 | 2 | 0 | .95 | .10 | .02 |
| L1HS214 | 20 | 0 | 0 | 1.00 | .00 | 17 | 0 | 0 | 1.00 | .00 | 19 | 0 | 0 | 1.00 | .00 | 17 | 3 | 0 | .93 | .14 | .04 |
| L1HS220 | 0 | 0 | 20 | .00 | .00 | 1 | 1 | 18 | .08 | .14 | 0 | 2 | 18 | .05 | .10 | 0 | 4 | 16 | .10 | .18 | .10 |
| L1HS222 | 1 | 6 | 8 | .27 | .40 | 0 | 3 | 16 | .08 | .15 | 0 | 1 | 18 | .03 | .05 | 0 | 2 | 18 | .05 | .10 | .18 |
| L1HS226 | 0 | 3 | 17 | .08 | .14 | 0 | 1 | 18 | .03 | .05 | 2 | 6 | 12 | .25 | .38 | 1 | 4 | 15 | .15 | .26 | .21 |
| L1HS228 | 17 | 0 | 0 | 1.00 | .00 | 14 | 0 | 0 | 1.00 | .00 | 18 | 0 | 0 | 1.00 | .00 | 12 | 1 | 1 | .89 | .20 | .05 |
| L1HS231 | 20 | 0 | 0 | 1.00 | .00 | 17 | 2 | 0 | .95 | .10 | 20 | 0 | 0 | 1.00 | .00 | 18 | 1 | 1 | .93 | .14 | .06 |
| L1HS233 | 1 | 4 | 14 | .16 | .27 | 1 | 6 | 11 | .22 | .36 | 1 | 7 | 11 | .24 | .37 | 0 | 7 | 13 | .18 | .30 | .32 |
| L1HS235 | 1 | 15 | 3 | .45 | .51 | 1 | 9 | 7 | .32 | .45 | 1 | 11 | 8 | .33 | .45 | 3 | 12 | 5 | .45 | .51 | .48 |
| L1HS242 | 4 | 11 | 5 | .53 | .39 | 0 | 11 | 8 | .29 | .42 | 2 | 11 | 7 | .38 | .48 | 4 | 5 | 10 | .34 | .46 | .44 |
| L1HS260 | 20 | 0 | 0 | 1.00 | .00 | 19 | 0 | 0 | 1.00 | .00 | 18 | 2 | 0 | .95 | .10 | 19 | 0 | 0 | 1.00 | .00 | .02 |
| L1HS287 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | .00 |
| L1HS291 | 20 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | 18 | 2 | 0 | .95 | .10 | 20 | 0 | 0 | 1.00 | .00 | .02 |
| L1HS293 | 1 | 4 | 15 | .15 | .26 | 4 | 8 | 7 | .42 | .50 | 1 | 4 | 15 | .15 | .26 | 0 | 2 | 18 | .05 | .10 | .28 |
| L1HS298 | 2 | 1 | 15 | .14 | .25 | 0 | 1 | 16 | .03 | .06 | 0 | 4 | 16 | .10 | .18 | 0 | 0 | 8 | .00 | .00 | .12 |
| L1HS301 | 4 | 14 | 1 | .58 | .50 | 11 | 8 | 0 | .79 | .34 | 7 | 11 | 1 | .66 | .46 | 4 | 12 | 1 | .59 | .50 | .45 |
| L1HS308 | 1 | 5 | 13 | .18 | .31 | 2 | 5 | 11 | .25 | .39 | 1 | 7 | 10 | .25 | .39 | 4 | 9 | 5 | .47 | .51 | .40 |
| L1HS314 | 4 | 5 | 6 | .43 | .51 | 1 | 4 | 11 | .19 | .31 | 1 | 8 | 9 | .28 | .41 | 2 | 9 | 9 | .33 | .45 | .42 |
| L1HS320 | 5 | 12 | 2 | .58 | .50 | 0 | 4 | 14 | .11 | .20 | 0 | 4 | 16 | .10 | .18 | 2 | 7 | 8 | .32 | .45 | .33 |
| L1HS332 | 3 | 5 | 7 | .37 | .48 | 1 | 3 | 13 | .15 | .26 | 1 | 3 | 6 | .25 | .39 | 1 | 1 | 4 | .25 | .41 | .39 |
| L1HS335 | 8 | 9 | 2 | .66 | .46 | 13 | 5 | 1 | .82 | .31 | 10 | 10 | 0 | .75 | .38 | 14 | 4 | 1 | .84 | .27 | .36 |
| L1HS337 | 17 | 3 | 0 | .93 | .14 | 17 | 3 | 0 | .93 | .14 | 19 | 1 | 0 | .98 | .05 | 14 | 6 | 0 | .85 | .26 | .15 |
| L1HS345 | 0 | 1 | 19 | .03 | .05 | 0 | 1 | 18 | .03 | .05 | 0 | 2 | 18 | .05 | .10 | 0 | 1 | 18 | .03 | .05 | .06 |
| L1HS348 | 18 | 2 | 0 | .95 | .10 | 15 | 4 | 1 | .85 | .26 | 17 | 3 | 0 | .93 | .14 | 16 | 4 | 0 | .90 | .18 | .17 |
| L1HS349 | 19 | 1 | 0 | .98 | .05 | 20 | 0 | 0 | 1.00 | .00 | 14 | 3 | 3 | .78 | .36 | 15 | 2 | 0 | .94 | .11 | .13 |
| L1HS353 | 16 | 2 | 0 | .94 | .11 | 20 | 0 | 0 | 1.00 | .00 | 18 | 2 | 0 | .95 | .10 | 17 | 2 | 0 | .95 | .10 | .08 |
| L1HS360 | 0 | 10 | 10 | .25 | .38 | 3 | 10 | 6 | .42 | .50 | 2 | 11 | 7 | .38 | .48 | 3 | 6 | 7 | .38 | .48 | .46 |
| L1HS364 | 4 | 12 | 4 | .50 | .51 | 20 | 0 | 0 | 1.00 | .00 | 18 | 1 | 0 | .97 | .05 | 17 | 3 | 0 | .93 | .14 | .18 |
| L1HS372 | 8 | 10 | 2 | .65 | .47 | 11 | 8 | 1 | .75 | .38 | 4 | 13 | 3 | .53 | .51 | 8 | 11 | 1 | .68 | .45 | .45 |
| L1HS373 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | .00 |
| L1HS375 | 6 | 12 | 1 | .63 | .48 | 11 | 8 | 0 | .79 | .34 | 4 | 16 | 0 | .60 | .49 | 11 | 9 | 0 | .78 | .36 | .42 |
| L1HS378 | 18 | 2 | 0 | .95 | .10 | 8 | 10 | 2 | .65 | .47 | 14 | 3 | 3 | .78 | .36 | 13 | 5 | 1 | .82 | .31 | .31 |
| L1HS391 | 18 | 0 | 0 | 1.00 | .00 | 19 | 1 | 0 | .98 | .05 | 20 | 0 | 0 | 1.00 | .00 | 19 | 0 | 0 | 1.00 | .00 | .01 |
| L1HS393 | 1 | 2 | 14 | .12 | .21 | 0 | 0 | 19 | .00 | .00 | 0 | 0 | 19 | .00 | .00 | 0 | 0 | 14 | .00 | .00 | .05 |
| L1HS395 | 7 | 9 | 1 | .68 | .45 | 8 | 9 | 3 | .63 | .48 | 3 | 12 | 5 | .45 | .51 | 9 | 7 | 3 | .66 | .46 | .48 |
| L1HS404 | 1 | 9 | 10 | .28 | .41 | 0 | 0 | 18 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 2 | 16 | .06 | .11 | .13 |
| L1HS406 | 17 | 3 | 0 | .93 | .14 | 16 | 4 | 0 | .90 | .18 | 18 | 2 | 0 | .95 | .10 | 16 | 4 | 0 | .90 | .18 | .15 |
| L1HS410 | 0 | 10 | 10 | .25 | .38 | 5 | 10 | 5 | .50 | .51 | 3 | 10 | 6 | .42 | .50 | 7 | 11 | 1 | .66 | .46 | .47 |
| L1HS413 | 0 | 11 | 9 | .28 | .41 | 1 | 9 | 9 | .29 | .42 | 0 | 7 | 13 | .18 | .30 | 3 | 6 | 10 | .32 | .44 | .39 |
| L1HS415 | 17 | 1 | 0 | .97 | .06 | 18 | 2 | 0 | .95 | .10 | 18 | 0 | 0 | 1.00 | .00 | 20 | 0 | 0 | 1.00 | .00 | .04 |
| L1HS418 | 4 | 10 | 6 | .45 | .51 | 13 | 4 | 1 | .83 | .29 | 5 | 12 | 3 | .55 | .51 | 2 | 8 | 8 | .33 | .46 | .44 |
| L1HS423 | 18 | 2 | 0 | .95 | .10 | 17 | 0 | 0 | 1.00 | .00 | 17 | 1 | 1 | .92 | .15 | 15 | 1 | 1 | .91 | .17 | .10 |
| L1HS426 | 1 | 14 | 5 | .40 | .49 | 7 | 5 | 5 | .56 | .51 | 2 | 5 | 9 | .28 | .42 | 3 | 6 | 10 | .32 | .44 | .47 |
| L1HS427 | 5 | 13 | 2 | .58 | .50 | 15 | 5 | 0 | .88 | .22 | 8 | 9 | 3 | .63 | .48 | 11 | 8 | 0 | .79 | .34 | .39 |
| L1HS430 | 0 | 2 | 18 | .05 | .10 | 0 | 4 | 14 | .11 | .20 | 0 | 0 | 20 | .00 | .00 | 1 | 0 | 19 | .05 | .10 | .10 |
| L1HS437 | 1 | 14 | 5 | .40 | .49 | 0 | 3 | 17 | .08 | .14 | 1 | 4 | 15 | .15 | .26 | 2 | 10 | 7 | .37 | .48 | .34 |
| L1HS442 | 10 | 10 | 0 | .75 | .38 | 17 | 1 | 0 | .97 | .06 | 14 | 6 | 0 | .85 | .26 | 8 | 7 | 2 | .68 | .45 | .29 |
| L1HS446 | 0 | 2 | 18 | .05 | .10 | 0 | 2 | 17 | .05 | .10 | 1 | 6 | 12 | .21 | .34 | 0 | 0 | 17 | .00 | .00 | .14 |
| L1HS447 | 12 | 7 | 1 | .78 | .36 | 11 | 3 | 3 | .74 | .40 | 14 | 5 | 1 | .83 | .30 | 13 | 4 | 2 | .79 | .34 | .35 |
| L1HS448 | 9 | 2 | 7 | .56 | .51 | 3 | 13 | 2 | .53 | .51 | 14 | 5 | 1 | .83 | .30 | 7 | 8 | 2 | .65 | .47 | .45 |
| L1HS450 | 12 | 4 | 4 | .70 | .43 | 20 | 0 | 0 | 1.00 | .00 | 19 | 0 | 1 | .95 | .10 | 18 | 1 | 1 | .93 | .14 | .17 |
| L1HS461 | 0 | 3 | 14 | .09 | .17 | 0 | 1 | 19 | .03 | .05 | 0 | 1 | 18 | .03 | .05 | 0 | 0 | 17 | .00 | .00 | .07 |
| L1HS480 | 3 | 8 | 9 | .35 | .47 | 4 | 8 | 6 | .44 | .51 | 5 | 10 | 5 | .50 | .51 | 4 | 10 | 6 | .45 | .51 | .50 |
| L1HS486 | 3 | 7 | 10 | .33 | .45 | 7 | 9 | 4 | .58 | .50 | 1 | 2 | 17 | .10 | .18 | 0 | 1 | 18 | .03 | .05 | .30 |
| L1HS493 | 5 | 8 | 6 | .47 | .51 | 5 | 8 | 7 | .45 | .51 | 9 | 7 | 3 | .66 | .46 | 9 | 2 | 4 | .67 | .46 | .49 |
| L1HS508 | 16 | 4 | 0 | .90 | .18 | 17 | 3 | 0 | .93 | .14 | 11 | 8 | 1 | .75 | .38 | 17 | 2 | 0 | .95 | .10 | .20 |
| L1HS512 | 19 | 1 | 0 | .98 | .05 | 18 | 0 | 0 | 1.00 | .00 | 19 | 0 | 0 | 1.00 | .00 | 17 | 0 | 0 | 1.00 | .00 | .01 |
| L1HS513 | 0 | 4 | 16 | .10 | .18 | 6 | 10 | 3 | .58 | .50 | 4 | 7 | 9 | .38 | .48 | 2 | 6 | 10 | .28 | .41 | .39 |
| L1HS516 | 2 | 8 | 9 | .32 | .44 | 1 | 2 | 16 | .11 | .19 | 6 | 9 | 5 | .53 | .51 | 3 | 7 | 6 | .41 | .50 | .41 |
| L1HS526 | 5 | 13 | 2 | .58 | .50 | 13 | 6 | 0 | .84 | .27 | 3 | 12 | 4 | .47 | .51 | 3 | 7 | 9 | .34 | .46 | .44 |
| L1HS552 | 0 | 6 | 11 | .18 | .30 | 5 | 7 | 8 | .43 | .50 | 2 | 14 | 3 | .47 | .51 | 1 | 5 | 12 | .19 | .32 | .41 |
| L1HS558 | 16 | 4 | 0 | .90 | .18 | 16 | 3 | 1 | .88 | .22 | 17 | 3 | 0 | .93 | .14 | 18 | 2 | 0 | .95 | .10 | .16 |
| L1HS561 | 0 | 1 | 19 | .03 | .05 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | 0 | 0 | 20 | .00 | .00 | .01 |
Asian and Alaskan native samples were used interchangeably as a geographically unique human population.
Average heterozygosity for all populations.
Frequency of the element.
Unbiased heterozygosity.
Table A3.
X-Linked L1Hs Ta–Associated Human Genomic Diversity
|
African American |
Asian/Alaskan Nativea |
European German |
Egyptian |
||||||||||||||||||||||||||
| No. with Genotype |
No. with Genotypes |
No. with Genotypes |
No. with Genotypes |
||||||||||||||||||||||||||
| Female |
Male |
Female |
Male |
Female |
Male |
Female |
Male |
||||||||||||||||||||||
| Element | +/+ | +/− | −/− | + | − | fc | Hetd | +/+ | +/− | −/− | + | − | fc | Hetd | +/+ | +/− | −/− | + | − | fc | Hetd | +/+ | +/− | −/− | + | − | fc | Hetd | AvgHetb |
| L1HS24 | 1 | 5 | 3 | 1 | 8 | .30 | .40 | 3 | 2 | 1 | 8 | 2 | .73 | .43 | 5 | 3 | 1 | 7 | 3 | .71 | .44 | 5 | 8 | 4 | 1 | 2 | .51 | .50 | .44 |
| L1HS28 | 5 | 4 | 0 | 6 | 3 | .74 | .42 | 0 | 3 | 3 | 3 | 7 | .27 | .44 | 1 | 5 | 3 | 9 | 1 | .57 | .49 | 9 | 6 | 1 | 2 | 1 | .74 | .43 | .44 |
| L1HS30 | 0 | 5 | 5 | 4 | 5 | .31 | .48 | 1 | 4 | 2 | 7 | 3 | .54 | .53 | 2 | 4 | 3 | 6 | 4 | .50 | .53 | 3 | 10 | 3 | 3 | 0 | .54 | .39 | .48 |
| L1HS125 | 7 | 1 | 1 | 7 | 1 | .85 | .26 | 6 | 0 | 0 | 10 | 0 | 1.00 | .00 | 9 | 0 | 0 | 9 | 0 | 1.00 | .00 | 16 | 0 | 0 | 3 | 0 | 1.00 | .00 | .07 |
| L1HS562 | 1 | 5 | 3 | 1 | 8 | .30 | .40 | 3 | 2 | 1 | 8 | 2 | .73 | .43 | 5 | 3 | 1 | 7 | 3 | .71 | .44 | 5 | 8 | 4 | 1 | 2 | .51 | .50 | .44 |
| L1HS564 | 0 | 3 | 7 | 2 | 7 | .17 | .32 | 0 | 0 | 6 | 1 | 9 | .05 | .10 | 0 | 2 | 7 | 1 | 9 | .11 | .20 | 0 | 3 | 13 | 0 | 3 | .09 | .09 | .18 |
Asian and Alaskan native samples were used interchangeably as a geographically unique human population.
Average heterozygosity for all populations.
Frequency of the element.
Unbiased heterozygosity.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Batzer Lab, http://batzerlab.lsu.edu/
- BLAST, http://www.ncbi.nlm.nih.gov/blast/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the DNA sequences from the common and pygmy chimpanzee orthologs of L1HS72 [accession numbers AF489459 and AF489460]; diverse DNA sequences from L1HS72 [accession numbers AF489450–AF489458]; and Ta L1 element pre-integration site sequences, namely, L1HS45 [accession numbers AF461364 and AF461365], L1HS172 [accession numbers AF461368 and AF461369], L1HS178 [accession numbers AF461370 and AF461371], L1HS284 [accession numbers AF461372 and AF461373], L1HS372 [accession numbers AF461374 and AF461375], L1HS416 [accession numbers AF461376 and AF461377], L1HS442 [accession numbers AF461378 and AF461379], L1HS443 [accession numbers AF461386 and AF461387], L1HS513 [accession numbers AF461380–AF461382], and L1HS558 [accession number AF461383])
- Genetic Information Research Institute Censor Server, http://www.girinst.org/Censor_Server-Data_Entry_Forms.html
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
- RepeatMasker Web Server, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA (1995) Alu repeats: a source for the genesis of primate microsatellites. Genomics 29:136–144 [DOI] [PubMed] [Google Scholar]
- Ardlie K, Liu-Cordero SN, Eberle MA, Daly M, Barrett J, Winchester E, Lander ES, Kruglyak L (2001) Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet 69:582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausabel FM, Brent R, Kingston ME, Moore DD, Seidman JG (1987) Current protocols in molecular biology. John Wiley & Sons, New York [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Gudi VA, Mena JC, Foltz DW, Herrera RJ, Deininger PL (1991) Amplification dynamics of human-specific (HS) Alu family members. Nucleic Acids Res 19:3619–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Rubin CM, Hellmann-Blumberg U, Alegria-Hartman M, Leeflang EP, Stern JD, Bazan HA, Shaikh TH, Deininger PL, Schmid CW (1995) Dispersion and insertion polymorphism in two small subfamilies of recently amplified human Alu repeats. J Mol Biol 247:418–427 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Stoneking M, Alegria-Hartman M, Bazan H, Kass DH, Shaikh TH, Novick GE, Ioannou PA, Scheer WD, Herrera RJ, Deininger PL (1994) African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci USA 91:12288–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8:1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD (1997) LINEs and Alus—the polyA connection. Nat Genet 16:6–7 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Pickeral OK (1999) Retroshuffling the genomic deck. Nature 398:108–109 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV (2001) Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 18:926–935 [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331 [PMC free article] [PubMed] [Google Scholar]
- Brookfield JF (2001) Selection on Alu sequences? Curr Biol 11:R900–R901 [DOI] [PubMed] [Google Scholar]
- Burton FH, Loeb DD, Edgell MH, Hutchison CA 3d (1991) L1 gene conversion or same-site transposition. Mol Biol Evol 8:609–619 [DOI] [PubMed] [Google Scholar]
- Carroll ML, Roy-Engel AM, Nguyen SV, Salem AH, Vogel E, Vincent B, Myers J, Ahmad Z, Nguyen L, Sammarco M, Watkins WS, Henke J, Makalowski W, Jorde LB, Deininger PL, Batzer MA (2001) Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J Mol Biol 311:17–40 [DOI] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD (1998) Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37:18081–18093 [DOI] [PubMed] [Google Scholar]
- Cost GJ, Golding A, Schlissel MS, Boeke JD (2001) Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res 29:573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA 3d, Edgell MH (1992) Master genes in mammalian repetitive DNA amplification. Trends Genet 8:307–311 [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr (1991) Isolation of an active human transposable element. Science 254:1805–1808 [DOI] [PubMed] [Google Scholar]
- Economou EP, Bergen AW, Warren AC, Antonarakis SE (1990) The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome. Proc Natl Acad Sci USA 87:2951–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng B, Ainsworth P, Waye JS (1994) Anomalous migration of PCR products using nondenaturing polyacrylamide gel electrophoresis: the amelogenin sex-typing system. J Forensic Sci 39:1356–1359 [PubMed] [Google Scholar]
- Fanning TG, Singer MF (1987) LINE-1: a mammalian transposable element. Biochim Biophys Acta 910:203–212 [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916 [DOI] [PubMed] [Google Scholar]
- Fitch DH, Bailey WJ, Tagle DA, Goodman M, Sieu L, Slightom JL (1991) Duplication of the γ-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci USA 88:7396–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A (2001) Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet 69:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH Jr (2000) Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9:653–657 [DOI] [PubMed] [Google Scholar]
- Grimaldi G, Skowronski J, Singer MF (1984) Defining the beginning and end of KpnI family segments. EMBO J 3:1753–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF (1994) A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 11:749–761 [DOI] [PubMed] [Google Scholar]
- Hardies SC, Martin SL, Voliva CF, Hutchison CA 3d, Edgell MH (1986) An analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol Biol Evol 3:109–125 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA (2000) The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am J Hum Genet 66:979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J (1997) Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA 94:1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P (1996) CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem 20:119–121 [DOI] [PubMed] [Google Scholar]
- Kass DH, Batzer MA, Deininger PL (1995) Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol Cell Biol 15:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr (1998) Mobile elements and disease. Curr Opin Genet Dev 8:343–350 [DOI] [PubMed] [Google Scholar]
- ——— (2000) L1 retrotransposons shape the mammalian genome. Science 289:1152–1153 [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr, Moran JV (1998) The impact of L1 retrotransposons on the human genome. Nat Genet 19:19–24 [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164–166 [DOI] [PubMed] [Google Scholar]
- Kim J, Deininger PL (1996) Recent amplification of rat ID sequences. J Mol Biol 261:322–327 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605 [DOI] [PubMed] [Google Scholar]
- Maeda N, Wu CI, Bliska J, Reneke J (1988) Molecular evolution of intergenic DNA in higher primates: pattern of DNA changes, molecular clock, and evolution of repetitive sequences. Mol Biol Evol 5:1–20 [DOI] [PubMed] [Google Scholar]
- Miyamoto MM, Slightom JL, Goodman M (1987) Phylogenetic relations of humans and African apes from DNA sequences in the psi eta-globin region. Science 238:369–373 [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE (1996) Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature 383:644–646 [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH Jr (1999) Exon shuffling by L1 retrotransposition. Science 283:1530–1534 [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato T, Taccioli G, Batzer MA, Moran JV (2002) DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31:159–165 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Leppert M, O'Connell P, Wolff R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E, White R (1987) Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235:1616–1622 [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr (2001) Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res 11:2059–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr (2000) Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res 28:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Troxel AB, Swergold GD (2001) Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res 11:2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Batzer MA, Deininger PL, Stoneking M (1992) Alu insertion polymorphism: a new type of marker for human population studies. Hum Biol 64:641–648 [PubMed] [Google Scholar]
- Prak ET, Kazazian HH Jr (2000) Mobile elements and the human genome. Nat Rev Genet 1:134–144 [DOI] [PubMed] [Google Scholar]
- Rothbarth K, Hunziker A, Stammer H, Werner D (2001) Promoter of the gene encoding the 16 kDa DNA-binding and apoptosis-inducing C1D protein. Biochim Biophys Acta 1518:271–275 [DOI] [PubMed] [Google Scholar]
- Roy AM, Carroll ML, Kass DH, Nguyen SV, Salem AH, Batzer MA, Deininger PL (1999) Recently integrated human Alu repeats: finding needles in the haystack. Genetica 107:149–161 [PubMed] [Google Scholar]
- Roy AM, Carroll ML, Nguyen SV, Salem AH, Oldridge M, Wilkie AO, Batzer MA, Deininger PL (2000) Potential gene conversion and source genes for recently integrated Alu elements. Genome Res 10:1485–1495 [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, Carroll ML, El-Sawy M, Salem AE, Garber RK, Nguyen SV, Deininger PL, Batzer MA (2002) Non-traditional Alu evolution and primate genomic diversity. J Mol Biol 316:1033–1040 [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL (2001) Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159:279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FR, Pandya A, Kayser M, Mitchell RJ, Liu A, Singh L, Destro-Bisol G, Novelletto A, Qamar R, Mehdi SQ, Adhikari R, de Knijff P, Tyler-Smith C (2000) A polymorphic L1 retroposon insertion in the centromere of the human Y chromosome. Hum Mol Genet 9:421–430 [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr (1997) Many human L1 elements are capable of retrotransposition. Nat Genet 16:37–43 [DOI] [PubMed] [Google Scholar]
- Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD (2000) Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res 10:1496–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF (1988) Unit-length LINE-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 8:1385–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9:657–663 [DOI] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J (1995) Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol 246:401–417 [DOI] [PubMed] [Google Scholar]
- Stoneking M, Fontius JJ, Clifford SL, Soodyall H, Arcot SS, Saha N, Jenkins T, Tahir MA, Deininger PL, Batzer MA (1997) Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res 7:1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Kim B, Gabriel A (1996) Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383:641–644 [DOI] [PubMed] [Google Scholar]
- Tremblay A, Jasin M, Chartrand P (2000) A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol Cell Biol 20:54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R (1998) Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem 273:891–897 [DOI] [PubMed] [Google Scholar]