Abstract
We have used a unique polymorphic 3′ transduction to show that a human L1, or LINE-1 (long interspersed nucleotide element–1), retrotransposition event most likely occurred in the maternal primary oocyte during meiosis I. We characterized a truncated L1 retrotransposon with a 3′ transduction that was inserted, in a Dutch male patient, into the X-linked gene CYBB, thereby causing chronic granulomatous disease. We used the unique flanking sequence to localize the precursor L1 locus, LRE3, to chromosome 2q24.1. In a cell culture assay, the retrotransposition frequency of LRE3 is greater than that for any other element that has been tested to date. The patient’s mother had two LRE3 alleles that differed slightly in the 3′-flanking genomic DNA. The patient had a single LRE3 allele that was identical to one of the maternal alleles; however, the patient’s insertion matched the maternal LRE3 allele that he did not inherit. Other data indicate that there is only a small chance that the father (unavailable for analysis) carries the precursor LRE3 allele. In addition, paternal origin of the insertion would have required that an LRE3 mRNA transcribed before meiosis II be carried separately from its precursor LRE3 allele in the fertilizing sperm. Since the mother carries a potential precursor allele and the insertion was on the patient’s maternal X chromosome, it is highly likely that the insertion originated during maternal meiosis I.
Introduction
Preliminary analysis of the human genome has shown that transposable elements compose ⩾45% of its mass (Lander et al. 2001). L1, or LINE-1 (long interspersed nucleotide element–1), retrotransposons are one of the most successful transposable elements, comprising 17% of the genome (Lander et al. 2001). Most of the ∼500,000 L1 copies present in the human genome are truncated at their 5′ end, rearranged by inversion, and/or mutated. Only a small percentage are full length (Lander et al. 2001), and only 40–80 potentially are retrotranspositionally active (Sassaman et al. 1997). In humans, the youngest and most active elements belong to the Ta-1d subset (Boissinot et al. 2000). The insertion reported here is the 14th-known recent L1 insertion in humans (Ostertag and Kazazian 2001). However, >70% of these insertions were ascertained by the presence of X-linked disease in male patients (Ostertag and Kazazian 2001). Estimates of the actual frequency of L1 insertions range from 1 in 10 to 1 in 120 individuals (Kazazian 1999; Li et al. 2001).
An intact, full-length human L1 element is ∼6.1 kb and contains (1) a 5′ UTR that is followed by two nonoverlapping ORFs, (2) a 3′ UTR with a polyadenylation signal, and (3) a polyA tail (Scott et al. 1987). The element encodes endonuclease and reverse-transcriptase activities in ORF2 (Mathias et al. 1991; Feng et al. 1996) and duplicates by the following mechanism (the details of which are largely unknown): First, the original retrotransposon is maintained, in situ, where it is transcribed (Whitcomb and Hughes 1992; Luan et al. 1993). Then, the transcript is reverse transcribed and integrated into a new genomic location by a process termed “target-primed reverse transcription” (Luan et al. 1993). Finally, on integration, the L1 element typically is flanked by target-site duplications (TSDs) of 6–20 bp; the integration site has a weak consensus sequence (5′-Py-Py/AAAA-3′) (Cost and Boeke 1998), and L1 elements appear to be inserted at random locations in the genome (Ovchinnikov et al. 2001).
There are exceptions to the canonical retrotransposition event. For example, L1 elements often transduce DNA flanking their 3′ ends. In ∼20% of cases, the L1 polyA signal is skipped in favor of a downstream polyA signal (Holmes et al. 1994; Goodier et al. 2000; Pickeral et al. 2000). In one previous instance, unique transduced sequence was used to find and characterize the precursor to a human L1 insertion (Holmes et al. 1994). Also, L1 elements can be inserted into the genome without a TSD (Narita et al. 1993; Jensen et al. 1994).
Little is known about either the timing of human retrotransposition events or the cell types in which such events occur. From an evolutionary standpoint, retrotransposition in a germline or a germline precursor is necessary. In tissue culture, it has been shown that retrotransposition from an episome is possible in several transformed cell lines—including feline G355.5 cells (Esnault et al. 2000); human HeLa (Moran et al. 1996), 143B TK− (thymidine kinase negative; American Type Culture Collection number CRL-8303), and H460 cells (E.O., unpublished data); and mouse LTK− cells (Moran et al. 1996; Naas et al. 1998). Our laboratory has failed to demonstrate retrotransposition in primary cell lines, including mouse embryonic fibroblasts and human fibroblasts (authors' unpublished data). In a transgenic mouse model, we have, by reverse-transcriptase PCR, found L1 mRNA in spermatocytes, and we have shown retrotransposition in male germ cells (Ostertag et al. 2000). It has also been shown that ORF1 is expressed in mouse testis (spermatocytes, myoid cells, and Leydig cells) and ovaries, as well as in steroidogenic cells (Branciforte and Martin 1994; Trelogan and Martin 1995). Finally, in one instance, an L1 insertion into the APC gene caused colon cancer in a human (Miki et al. 1992). However, to date, it has not been possible to define the timing and cell type of a heritable human retrotransposition event. Here, we present data that strongly suggest that an L1 insertion occurred during maternal meiosis I.
Subject and Methods
Patient History
The patient was born in 1980 and presented, in the 1st year of life, with multiple infectious complications—including recurrent pneumonia—as well as abscesses of the perianal region, liver, and lymph nodes. After diagnosis of chronic granulomatous disease (CGD [MIM 306400]) at 15 mo of age, prophylactic treatment with cotrimoxazole was initiated. Pneumonia caused by Aspergillus pneumoniae at the age of 22 mo led to treatment by the addition of itraconazole to the prophylactic protocol. Under this regimen, the patient remained infection free until 1999, when he presented again with pneumonia, leading to his hospitalization, that was caused by A. pneumoniae. Ensuing complications included several episodes of bacterial sepsis; fungal lesions in lungs, liver, and spleen; osteomyelitis; and proctitis. Currently, he is infection free and is followed as an outpatient.
Diagnosis of CGD
Diagnosis of CGD was achieved by analysis of granulocyte function, nitroblue tetrazolium (NBT) test, determination of cytochrome b558 content in whole and Triton X100–extracted cells, and monocyte hybridization, which were performed as described elsewhere (Weening et al. 1985).
PCR
Amplification of CYBB.—The 13 exons of CYBB, including their adjacent intronic sequences (namely, exon 1 and promoter sequences), were amplified as described elsewhere (Meischl et al. 2000). In brief, genomic DNA (50–500 ng) was amplified by use of the Rapid Cycler (Idaho Technology), with 50 cycles of 95°C for 5 s, 60°C for 30 s, and 72° C for 15 s and slope S9. The reaction volume of 15 μl contained 2 U of Taq DNA polymerase (Promega), 2 U of TaqStart antibody (Clontech Laboratories), 50 ng of each primer, 200 μM of each of the deoxyribonucleoside triphosphates (Promega), and reaction buffer (50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X100, 10 mM Tris, and 8% dimethyl sulfoxide, with pH 9.0 at 25°C) in 10-μl glass capillaries (Idaho Technology).
Amplification of insertion-containing exon 4 of CYBB.—Products of the insertion-containing exon 4 from the patient were obtained, with primers 5′-GTTAACAATTACTATTCCATTCTTTCCCCC-3′ and 5′-CTATGAATAGAGGGAACTCCCTGGTTCCAAG-3′, by use of the Expand Long Template PCR System (Roche), according to the manufacturer's recommendations. In brief, the region of interest was amplified by Expand Long Template PCR on a PTC-0220 Peltier Thermocycler (MJ Research), with 50–200 ng of genomic DNA as a template, in a total reaction volume of 20 μl. The PCR was run with the following settings: denaturing at 94°C for 30 s, annealing at 56°C for 40 s, and 30 cycles of extension at 68°C for 2 min.
Amplification of LRE3 from both the mother and the patient.—The unique primers LRE3F (5′-ATTTGCGGCCGCTACCCCCTACTGTTGTGCCTTG-3′) and LRE3R (5′-CGTCTACCGAAAGTCCATACTACCC-3′) were designed using the chromosome 2 sequence from AC067958 (GenBank). The first primer has a NotI site at the 5′ end for cloning. Three additional unique pairs were also designed to verify that LRE3 was present in the sequence on AC067958 (GenBank). The ∼7,750-bp product was amplified by use of the Expand Long Template PCR System (Roche), according to the manufacturer's recommendations. In brief, the 7.7-kb region was amplified on a PTC-0220 Peltier Thermocycler (MJ Research), with 50–200 ng of genomic DNA as a template, in a total reaction volume of 20 μl. The PCR was run with the following settings: denaturing at 94°C for 30 s, annealing at 67°C for 40 s, and 30 cycles of extension at 68°C for 6 min.
Polymorphism Studies
The presence or absence of LRE3 in 23 individuals from five different ethnic groups was determined using a two-reaction, three-primer technique (Boissinot et al. 2000). The Expand Long Template PCRs were performed in a PTC-0220 Peltier Thermocycler (MJ Research), with 50–200 ng of genomic DNA as a template in a total reaction volume of 20 μl. The PCR was run with the following settings: denaturing at 94°C for 30 s, annealing at 64.8°C for 40 s, and 30 cycles of extension at 68°C for 2 min. In brief, in the first reaction, two unique flanking primers, A (5′-TTCCACCTCTGTGGTATCTATGGTC-3′) and C (5′-CATGACAATGGTCTGGGCAGTG-3′), were used to yield a ∼1,350-bp product, in the absence of the element; in the second reaction, the unique A primer and a reverse primer B in the 5′ section of the L1 element (5′-TGTGGGATATAGTCTCGTGGTGC-3′) were used to yield a ∼1,330-bp product, in the presence of the element.
Southern Blotting
Southern blotting, with 8 μg of genomic DNA, was performed as described elsewhere (Sambrook et al. 1989).
Cloning of LRE3
The single band obtained with primers LRE3F and LRE3R was cut from a 0.7% agarose gel and was purified using a Gene Clean Spin kit (BIO 101). The band was digested with NotI (in LRE3F) and BstZ17I (in the 3′ end of the L1 element). The full-length LRE3 fragment, as a NotI/BstZ17I fragment, was swapped into pL1RP-EGFP (with BstZ17I) (full-length L1RP element tagged with the EGFP (enhanced green fluorescent protein) retrotransposition cassette; the pL1RP-EGFP construct was the same as that described by Ostertag et al. [2000] except that it was modified to contain an intact BstZ17I site), to produce pLRE3-EGFP. This clone was then tested for retrotransposition activity in a cell culture assay.
Sequencing and Sequence Analysis
Clones and purified PCR products were sequenced using L1-specific or 3′ transduction–specific primers. Each sequencing reaction contained ∼50 ng of PCR product or ∼500 ng of plasmid DNA and ∼3.2 pmol of oligonucleotide primer. All sequencing was performed on ABI 3100 automated sequencers.
Retrotransposition Assay
Constructs pL1RP-EGFP, pL1RP(JM111)-EGFP (Ostertag et al. 2000), and pLRE3-EGFP were assayed for retrotransposition activity in human 143B TK− osteosarcoma cells. Cell culture, transfection of cells, antibiotic selection, and flow cytometry were performed as described elsewhere (Ostertag et al. 2000).
Results
CGD is a disorder that is caused by a deficiency in the enzyme NADPH oxidase, which creates the reactive oxygen species that are used by phagocytes to kill bacteria and fungi. CGD is characterized by repeated infections. Age at onset and phenotype are quite variable. NADPH oxidase is composed of subunits that are encoded by several different genes. Defects in the X-linked gene CYBB, which encodes the gp91phox subunit of cytochrome b558, are the most common cause, but other autosomal forms exist (Roos et al. 1996). Analysis of the patient's granulocyte function revealed a normal chemotactic response but a reduced intracellular killing of Staphylococcus aureus (in the patient's granulocytes, 57% of the bacteria were killed after 30 min, whereas, in granulocytes from a control individual, 95% were killed after 30 min) and a greatly reduced oxygen consumption after addition of serum-treated zymosan (in the patient, 0.6 nmol/106 cells; in the control individual, 7.2 nmol/106 cells). A negative NBT test and the absence of cytochrome b558 in both whole cells and Triton X100–extracted cells confirmed the diagnosis of CGD. Failure to complement the patient's oxidase system in monocytes with that of a known patient with X-linked CGD indicated the presence of a defect in the X-linked gene CYBB.
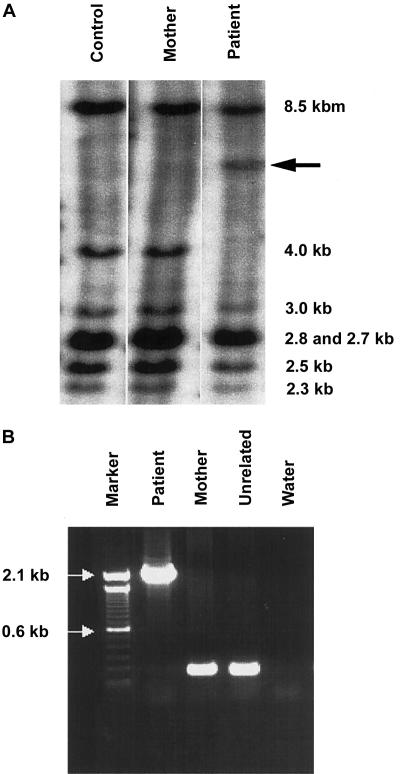
A Southern blot of EcoRI-digested genomic DNA probed with CYBB coding sequence (fig. 1A) revealed an increase of ∼2 kb in the fragment that comprises exon 4 of CYBB. The aberrant band was not seen in either the patient's mother or an unrelated control individual (fig. 1A); the patient’s father was unavailable for analysis.
Figure 1.
Insertion analyses. A, CYBB-specific Southern blot with EcoRI-digested DNA—from an unrelated control individual, the patient's mother, and the patient—demonstrating the shift, in the patient's sample, of the exon 4–containing fragment. The sizes of the wild-type bands are given; the arrow (←) indicates the aberrant band. The probe was constructed with coding cDNA of CYBB as template. B, Expand Long Template PCR of exon 4, in the patient, the patient’s mother, and an unrelated control. The arrows (→) indicate markers in the ladder.
To confirm the results of the Southern blot, we amplified exon 4 in the patient, his mother, and an unrelated control individual (fig. 1B). The patient lacked a normal exon 4 product but had an exon 4 that contained an extra 2,105 bp, which indicates that the patient had one X chromosome and that it contained an abnormal exon 4 of CYBB. Sequencing of the abnormal PCR product revealed an insertion, comprising the final 1,722–1,724 bp of an L1 element, followed by a 280-bp 3′ transduction. When compared with L1RP, the most active element yet tested in the cell culture assay, the L1 fragment was found to contain a single-nucleotide change in the 3′ UTR. The 3′ transduction was composed of a 21-bp polyA tail followed by 259 bp with no homology to L1 or CYBB sequences. This sequence was followed by a polyA tail of 101 bp that led directly to the remainder of exon 4 of CYBB. Since the 5′ two bases of the insertion were both A, it was impossible to determine whether they constitute a 2-bp TSD within a 2,103-bp insertion or whether they are part of a 2,105-bp insertion without a TSD (fig. 2). The 5′ portion of the transduced sequence contained an imperfect 29-repeat GAAA section. However, the final 153 bp contained unique sequence. A BLAST search of the human genome, for the entire 153-bp unique sequence, produced one exact match, AC067958 (GenBank), on chromosome 2q24.1.
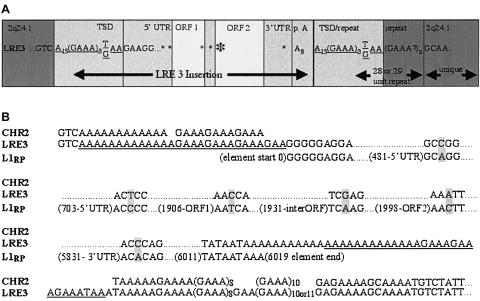
Figure 2.
Insertion sequence analyses. A, Schematic representation of the CYBB insertion. PT = important sequence of a PCR product from the patient’s exon 4. B, CYBB-insertion sequence alignments. CYBB = GenBank sequence at the point of insertion, into exon 4, of CYBB; PT = sequence of a PCR product from the patient’s CYBB exon 4 (which includes the disease-causing insertion); LRE3 = sequence of a PCR product from the patient’s chromosome 2q24.1; L1RP = GenBank sequence of the most active element known (the nucleotide number of the adjacent base is given in parentheses). A possible 2-bp TSD in the disease-causing insertion is underlined; differences between sequences are shaded; leader dots indicate sequence that is consistent with the L1RP sequence.
The AC067958 sequence (GenBank) contained a slightly different imperfect GAAA repeat section, upstream from the unique sequence, but no L1 sequence. Because active L1 elements are often polymorphic, we assumed that the precursor (i.e., LRE3) was absent from the Human Genome Project sequence. PCR using four unique primer pairs showed that, in this region, the patient’s mother was homozygous for a ∼6.1-kb LRE3 allele, whereas the patient was heterozygous (fig. 3). These results were confirmed using a third primer internal to the L1 element (data not shown).
Figure 3.
Expand Long Template PCR with flanking primers, demonstrating the presence or absence of LRE3 in the patient's mother, in the patient, and in an unrelated heterozygote. All bands were sequenced, to confirm findings.
On direct sequencing of the patient’s LRE3 PCR product, the final 1,720 bp of L1 sequence were an exact match of the insertion on the X chromosome (fig. 2). However, the imperfect GAAA repeat section 3′ from the patient’s LRE3 allele had 28 repeats, whereas his insertion had 29 repeats. We hypothesized that the mother had a 29-repeat LRE3 allele that was the actual precursor to the insertion and a 28-repeat allele that was inherited by the patient. After cloning the maternal LRE3 PCR product, we sequenced 12 clones and found that seven contained (GAAA)3TAAATAAAAAGAAAA(GAAA)8GAA(GAAA)11GAGAAAAGCAAAA (29 repeats) and that five contained (GAAA)3TAAATAAAAAGAAAA-(GAAA)8GAA(GAAA)10GAGAAAAGCAAAA (28 repeats). The sequence of the maternal 29-repeat LRE3 allele perfectly matched all 2,105 bp of the patient’s insertion.
A small possibility exists that a 29-repeat LRE3 transcript from the father was carried in the sperm and that the precursor to the insertion is paternal in origin. Since the father was unavailable for analysis, we observed the prevalence of the 29-repeat LRE3 allele in the white population, the most prevalent ethnic group in the Netherlands. Of 50 white haploid genomes, 9 contained an LRE3 element. We used PCR to obtain the 3′ flank. On sequencing, we discovered that all nine LRE3 elements were followed by different repeat sections and that only one of the nine matched the patient’s 29-repeat insertion. Thus, the allele frequency of the 29-repeat LRE3 allele in whites is ∼1 in 50 haploid genomes, or ∼2%. Since the patient is heterozygous for LRE3, his father has at least one chromosome 2 without an LRE3 allele. Therefore, if the father is white, then he has an ∼2% chance of having a 29-repeat LRE3 allele.
LRE3 is located on chromosome 2q24.1. Because of the polymorphic, repeating nature of this location, it is impossible to determine unambiguously the “empty site,” the exact point of insertion, or the length of the TSD. If we assume that an endonuclease-mediated insertion occurred, then the most likely point of insertion is 5′ from the (GAAA)n repeat at 5′-TGTC/AAAAA-3′. This point of insertion follows the consensus 5′-Py-Py/AAAA-3′ sequence of the L1-endonuclease site (Cost and Boeke 1998). The TSD would then be 28 bp—that is, 5′-AAAAAAAAAAAAAGAAAGAAAGAAA(G/T)AA-3′, with one G/T base change near the 3′ end (fig. 4).
Figure 4.
LRE3 sequence analyses. A, Schematic representation of the LRE3 insertion into chromosome 2q24.1. LRE3 = important sequence from LRE3. Potential TSDs are underlined; asterisks (*) are placed at locations where differences exist between LRE3 and L1RP. The larger asterisk indicates an amino acid change. B, LRE3 sequence alignments. CHR2 = GenBank sequence's “empty site” at the LRE3 locus; LRE3 = sequence of a PCR product from both the patient’s chromosome 2q24.1 and the patient's mother’s chromosome 2q24.1; L1RP = GenBank sequence of the most active element known (the nucleotide number of the adjacent base is given in parentheses). A possible 28-bp TSD of LRE3 is underlined; nucleotide differences are shaded; leader dots indicate sequence that is consistent with the L1RP sequence.
LRE3 has two ORFs, is a member of the Ta-1d subset of human retrotransposons, and is 99.9% identical to L1RP. Sequence comparison of LRE3 with L1RP showed the following six differences: two in the 5′ UTR; one in ORF1; one in the inter-ORF region; one, leading to an amino acid substitution, in ORF2; and one in the 3′ UTR (fig. 4). The mother’s LRE3 alleles are identical in sequence except for the 3′ flank.
Since LRE3 is 99.9% identical to L1RP, we predicted that the two elements would have similar retrotransposition activity in the cell culture assay. To test this prediction, we cloned both the mother’s and the patient’s PCR products directly into the pCEP4 expression vector, which contained an antisense EGFP sequence that was disrupted by an intron in the same orientation as the L1 element. On transfection into 143B TK− osteosarcoma cells, the EGFP retrotransposition marker is expressed only when the L1 is transcribed, the intron is spliced out, and the EGFP gene is reverse transcribed and integrated into the genome (Ostertag et al. 2000). When tested, under the direction of their endogenous L1 promoters, both the mother’s and the patient’s LRE3 alleles had retrotransposition activities ∼50% greater than that of L1RP (fig. 5). LRE3 is therefore the most active human L1 element yet characterized in a cell culture assay.
Figure 5.
Example of flow-cytometry results. A live/dead gate was set by use of forward and sideward scatter of cells, and ∼10,000 live cells were assayed for fluorescence. Cells within the M1 marker were counted as positive. The M1 marker was zeroed using a single JM111 clone (L1RP, with two disabling ORF1 point mutations) and a single L1RP clone (not shown) that lacked the 3,832-bp AflII fragment. Both negative controls were assayed in triplicate; one L1RP clone was assayed in triplicate, as a positive control; two LRE3 clones, one derived from maternal genomic DNA and another derived from patient genomic DNA, were assayed in triplicate. Numbers give the average number, over three assays, of positive cells for each clone ± 1 SD.
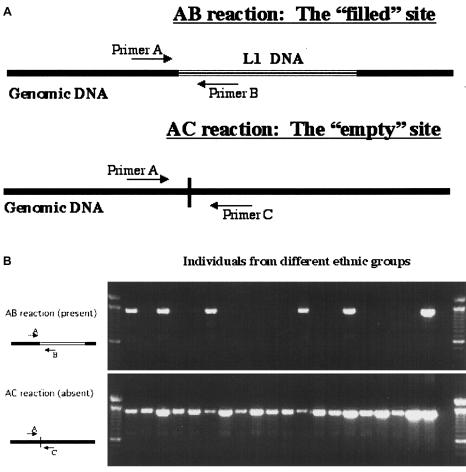
We assessed the frequency of LRE3 in the general population, using DNA from 5 Indo-Pakistanis, 9 sub-Saharan Africans, 5 Pacific Islanders, 4 Chinese, and 25 white individuals. One or two individuals of all ethnic groups except the sub-Saharan Africans, in whom LRE3 was not found, were heterozygous for LRE3. In sum, LRE3 was present in 16 of the 96 genomes, giving an allele frequency of 0.17 and further confirming the recent evolutionary nature of the Ta-1d subset (Boissinot et al. 2000) (fig. 6).
Figure 6.
Assay and gel analyses. A, Schematic representation of the polymorphism assay that was used to determine the absence or presence of an element. B, Sample gel, showing results from 20 individuals of diverse ethnic backgrounds.
Discussion
We used a unique polymorphic 3′ transduction to derive evidence that LRE3 retrotransposed, into CYBB, in the primary oocyte during maternal meiosis I. The patient’s 29-repeat CYBB insertion does not match his 28-repeat LRE3 allele but does match the mother’s 29-repeat LRE3 allele. Since the patient’s mother does not appear to have the CYBB insertion, the simplest explanation is that the 29-repeat LRE3 allele in the mother was transcribed and retrotransposed into an X chromosome before the conclusion of maternal meiosis I. On ovulation, the separation of homologous chromosomes occurred, and both the 28-repeat LRE3 allele and the X chromosome with the 29-repeat insertion became part of the secondary oocyte, whereas the actual precursor 29-repeat LRE3 allele and the X chromosome without the insertion were lost in the extruded polar body. At fertilization, the sperm carried a Y chromosome and a copy, lacking LRE3, of chromosome 2. The fertilized egg produced a male offspring with a single 28-repeat LRE3 allele on chromosome 2 and a 29-repeat insertion on the X chromosome.
Retrotransposition during maternal meiosis I provides the simplest explanation for our data. Since maternal meiosis I occurs in every ovulating female and lasts from birth to ovulation, a rare retrotransposition event could take place at any time during this period. However, other scenarios that require the occurrence of an additional rare event are formally possible:
-
1.
If the mother were mosaic, then the actual insertion into the X chromosome could have taken place as early as maternal embryogenesis. However, by PCR, we did not find any evidence for maternal mosaicism of lymphocyte DNA. In addition, at this time, there is no evidence that retrotransposition occurs during embryogenesis.
-
2.
The patient’s insertion could have come from his own precursor; instability during retrotransposition or replication at the polymorphic repeat locus could have resulted in an altered LRE3 allele that matched a maternal sequence. However, any instability would have to be very precise, since one specific maternal allele would have been changed to match the other. The wide variety of repeat formats found in the nine individuals tested suggests that this is unlikely. Also, present knowledge indicates that the process of retrotransposition appears to be accurate. Two precursors of human L1 insertions have been exact matches to their respective insertions over a total of ∼6 kb (Dombroski et al. 1991; Holmes et al. 1994).
-
3.
The father had a small chance of carrying a 29-repeat LRE3 allele and could have passed a lingering 29-repeat LRE3 message, in his sperm, that retrotransposed into the maternal X chromosome. This scenario would require the confluence of three rare events: (a) he would need to have the correct LRE3 message; (b) he would need to pass a stable full-length L1 mRNA, in his sperm, separate from its precursor chromosome; and (c) that message would need to be reverse transcribed and inserted into the mother’s X chromosome postfertilization.
Because of the relative rareness of these alternative scenarios, a retrotransposition event during maternal meiosis I is the most straightforward explanation for our data.
From an evolutionary perspective, only retrotransposition in germ cells or germ cell precursors can be passed on to progeny; somatic retrotransposition, on the other hand, is an evolutionary dead end. The new insertion cannot be passed on, and, if the insertion harms the host, then the precursor to the insertion will not be passed on. Although ORF1 expression has been detected in somatic cells in mouse testis (Branciforte and Martin 1994), the only known example of somatic retrotransposition is an L1 insertion, into an APC gene, resulting in colon cancer (Miki et al. 1992). Oncogenesis, however, is a multistep process, and the colon cell may have been partially transformed at the time of retrotransposition. Although there is no in vitro evidence of retrotransposition in primary cell lines, retrotransposition is readily demonstrated in cultured transformed cell lines (Moran et al. 1996).
LRE3, the precursor to the insertion, is the most active human L1 element yet tested in our cell culture assay. When active L1 elements are used to create an amino acid consensus sequence for ORF1 and ORF2 (data not shown), the proteins encoded by LRE3 ORF1 and ORF2 are exact matches. Therefore, LRE3 is the first element tested that has a perfect amino acid consensus through both ORF1 and ORF2. LRE3 and L1RP differ by one silent change in ORF1, four changes in noncoding regions, and one T→N amino acid change at position 5 of ORF2; however, LRE3 is 50% more active. Further analysis may provide insight into domains that are important in the mechanism of retrotransposition.
The insertion, into exon 4, of the patient’s CYBB gene appears to lack TSDs that have commonly been found in retrotransposition events. TSDs result when the L1 endonuclease cuts the two DNA strands at the insertion site in a staggered manner. At least two explanations for the apparent lack of TSDs are possible: First, L1-endonuclease–mediated insertion could produce very short TSDs of two bases (AA) or even no TSDs (Jensen et al. 1994); insertion, in this case, would have occurred at 5′-TC^AA-3′, a potential L1-endonuclease site (fig. 2) (Cost and Boeke 1998). Second, it has been proposed, as an explanation for the rare cases of TSD-less L1 insertions, that L1 fragments play a role in the repair of preexisting double-strand breaks (Mager et al. 1985; Van de Water et al. 1998; Kondo-Iida et al. 1999; Browning et al. 2001). Either hypothesis could explain the findings in the patient.
LRE3 is the second precursor L1 element that has been characterized using flanking sequence transposed in a de novo insertion (Holmes et al. 1994). Indeed, LRE3 is only the third active precursor L1 locus that has been characterized overall (hence, its designation as “L1 retrotransposable element–3”). With the inclusion of the full-length elements that were isolated from genome analysis and that have retrotransposition activity in the cell culture assay (Moran et al. 1996; Ostertag et al. 2000), there are now 10 active human L1 elements known (Sassaman et al. 1997; Schwahn et al. 1998; Kimberland et al. 1999).
Overall, 211 CYBB mutations are listed in the Human Gene Mutation Database. One deletion was a nonhomologous recombination that involves an L1 element that lies 5 kb upstream from CYBB (Kumatori et al. 1998). In addition, there has been a previous L1 insertion into an intron of CYBB (Meischl et al. 2000). More than half of known recent human L1 insertions have occurred in only three genes: CYBB, factor VIII, and dystrophin (Ostertag and Kazazian 2001). It would be interesting to determine whether these three genes are L1 hotspots or whether this seeming cluster of L1 activity is the result of an ascertainment bias.
The polymorphic 3′ transduction of this insertion has allowed us to predict, with great confidence, both the timing and the parental origin of an in vivo retrotransposition event. LRE3, presently the most active human L1 element in cell culture, is also the first element with an active amino acid consensus sequence for both ORFs. It will be interesting to see whether departures from consensus or changes in noncoding nucleotides will further increase the retrotransposition activity of L1 elements. As the most active L1 element currently known, LRE3 is now the best candidate for mutagenesis systems and other molecular biology applications.
Acknowledgments
We thank Dr. P. M. van Hagen (Academic Hospital Dijkzigt, Rotterdam) for supplying patient material and relevant information. C.M. was supported by grant Me 1528/1-1 from the Deutsche Forschungsgemeinschaft and by a grant from the Stichting Fondsenwervingsacties Volksgezondheid (Foundation for Fund Raising in Public Health). H.H.K. was supported by a grant from the National Institutes of Health.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for Homo sapiens BAC clone RP11-81F8 [accession number AC067958])
- Human Gene Mutation Database, The, http://archive.uwcm.ac.uk/uwcm/mg/search/120513.html (for CYBB mutations)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CGD [MIM 306400])
References
- Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 [DOI] [PubMed] [Google Scholar]
- Branciforte D, Martin SL (1994) Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol 14:2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning VL, Chaudhry SS, Planchart A, Dixon MJ, Schimenti JC (2001) Mutations of the mouse Twist and sy (fibrillin 2) genes induced by chemical mutagenesis of ES cells. Genomics 73:291–298 [DOI] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD (1998) Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37:18081–18093 [DOI] [PubMed] [Google Scholar]
- Dombroski B, Mathias S, Nanthakumar E, Scott A, Kazazian HH Jr (1991) Isolation of an active human transposable element. Science 254:1805–1808 [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T (2000) Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24:363–367 [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH Jr (2000) Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9:653–657 [DOI] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr (1994) A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7:143–148 [DOI] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Heidmann T (1994) Retrotransposition of the Drosophila LINE I element can induce deletion in the target DNA: a simple model also accounting for the variability of the normally observed target site duplications. Biochem Biophys Res Commun 202:111–119 [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr (1999) An estimated frequency of endogenous insertional mutations in humans. Nat Genet 22:130 [DOI] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH Jr (1999) Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet 8:1557–1560 [DOI] [PubMed] [Google Scholar]
- Kondo-Iida E, Kobayashi K, Watanabe M, Sasaki J, Kumagai T, Koide H, Saito K, Osawa M, Nakamura Y, Toda T (1999) Novel mutations and genotype-phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD). Hum Mol Genet 8:2303–2309 [DOI] [PubMed] [Google Scholar]
- Kumatori A, Faizunnessa NN, Suzuki S, Moriuchi T, Kurozumi H, Nakamura M (1998) Nonhomologous recombination between the cytochrome b558 heavy chain gene (CYBB) and LINE-1 causes an X-linked chronic granulomatous disease. Genomics 53:123–128 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 (errata: Nature 411:720; Nature 412:565) [DOI] [PubMed] [Google Scholar]
- Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, Careri D, Pinto MT, Kasper CK, Sommer SS (2001) Frequency of recent retrotransposition events in the human factor IX gene. Hum Mutat 17:511–519 [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605 [DOI] [PubMed] [Google Scholar]
- Mager DL, Henthorn PS, Smithies O (1985) A Chinese G gamma + (A gamma delta beta)zero thalassemia deletion: comparison to other deletions in the human beta-globin gene cluster and sequence analysis of the breakpoints. Nucleic Acids Res 13:6559–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254:1808–1810 [DOI] [PubMed] [Google Scholar]
- Meischl C, de Boer M, Ahlin A, Roos D (2000) A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet 8:697–703 [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y (1992) Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52:643–645 [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 [DOI] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH Jr (1998) An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17:590–597 (erratum: EMBO J 20:2608 [2001]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N, Nishio H, Kitoh Y, Ishikawa Y, Minami R, Nakamura H, Matsuo M (1993) Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest 91:1862–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr (2001) Biology of mammalian L1 retrotransposons. Annu Rev Genet 35:501–538 [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr (2000) Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res 28:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Troxel AB, Swergold GD (2001) Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res 11:2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD (2000) Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res 10:411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos D, de Boer M, Kuribayashi F, Meischl C, Weening RS, Segal AW, Ahlin A, Nemet K, Hossle JP, Bernatowska-Matuszkiewicz E, Middleton-Price H (1996) Mutations in the X-linked and autosomal recessive forms of chronic granulomatous disease. Blood 87:1663–1681 [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis A (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr (1997) Many human L1 elements are capable of retrotransposition. Nat Genet 16:37–43 [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19:327–332 [DOI] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L (1987) Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelogan SA, Martin SL (1995) Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA 92:1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Water N, Williams R, Ockelford P, Browett P (1998) A 20.7 kb deletion within the factor VIII gene associated with LINE-1 element insertion. Thromb Haemost 79:938–942 [PubMed] [Google Scholar]
- Weening RS, Corbeel L, de Boer M, Lutter R, van Zwieten R, Hamers MN, Roos D (1985) Cytochrome b deficiency in an autosomal form of chronic granulomatous disease: a third form of chronic granulomatous disease recognized by monocyte hybridization. J Clin Invest 75:915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb JM, Hughes SH (1992) Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol 8:275–306 [DOI] [PubMed] [Google Scholar]