Abstract
Prior evidence has supported the existence of multiple susceptibility genes for schizophrenia. Multipoint linkage analysis of the 270 Irish high-density pedigrees that we have studied, as well as results from several other samples, suggest that at least one such gene is located in region 6p24-21. In the present study, family-based association analysis of 36 simple sequence-length–polymorphism markers and of 17 SNP markers implicated two regions, separated by ∼7 Mb. The first region, and the focus of this report, is 6p22.3. In this region, single-nucleotide polymorphisms within the 140-kb gene DTNBP1 (dystrobrevin-binding protein 1, or dysbindin) are strongly associated with schizophrenia. Uncorrected, empirical P values produced by the program TRANSMIT were significant (P<.01) for a number of individual SNP markers, and most remained significant when the data were restricted to include only one affected offspring per nuclear family per extended pedigree; multiple three-marker haplotypes were highly significant (P=.008–.0001) under the restricted conditions. The pattern of linkage disequilibrium is consistent with the presence of more than one susceptibility allele, but this important issue is unresolved. The number of markers tested in the adjacent genes, all of which are negative, is not sufficient to rule out the possibility that the dysbindin gene is not the actual susceptibility gene, but this possibility appears to be very unlikely. We conclude that further investigation of dysbindin is warranted.
Introduction
Although genetic factors are known to be important in the etiology of schizophrenia (Gottesman and Shields 1982; Kendler 2000), unambiguous identification of the important susceptibility genes for this common and debilitating condition has not yet been accomplished. Various causes for this barrier to progress, both established and speculated, have been amply discussed, and, in recognition of the magnitude of the problem, improvements on various fronts have been initiated; these include (a) dramatic expansion of sample sizes by inclusion of triad and case-control studies and (b) a renewed emphasis on additional biologically based intermediate phenotypes.
As has been detailed in dozens of extensive reviews (Karayiorgou and Gogos 1997; Tsuang et al. 1999; Riley and McGuffin 2000; Baron 2001; Bray and Owen 2001), after several well-publicized false leads, many plausible candidate regions for schizophrenia are now under intense scrutiny; these include regions 1q21-22, 1q32-42, 5q21-34, 6p24-21, 6q13-26, 8p22-21, 10p15-11, 13q14-32, 15q13-15, and 22q11-13. After the initial reports (Straub et al. 1995; Wang et al. 1995), chromosome 6p (locus SCZD3 [MIM 600511]) has had support from a number of follow-up studies, with markers in a variety of locations showing at least some degree of positive evidence. It has been difficult, though, to distinguish true positives from false positives, because of the large number of markers tested under many genetic and phenotypic models using many samples, most of which were only of moderate size, that were ascertained under different conditions. In addition, it is still not clear to what extent (if any) a positive marker or multipoint peak >20 cM from the original should be interpreted as a “replication.” Statistical significance alone is clearly neither a sufficient nor a particularly well-chosen metric (Morton 1998), since (1) very significant results (e.g., a heterogeneity LOD score [H-LOD] >4) are not necessarily true and (2) supposedly weaker results (e.g., H-LOD 1.5–3) have led to gene identification and so must not be prematurely discounted. Independent replication attempts, although theoretically a powerful tool, are actually only rarely performed, because of numerous inherent and fundamental differences between studies (Straub et al. 1996; Vieland 2001).
On 6p, the markers positive for schizophrenia are distributed widely over the 25 Mb between D6S296 and D6S291. They do appear, however, to be concentrated within four subregions: 6p25-24, near D6S296/D6S309 (Antonarakis et al. 1995; Maziade et al. 1997, 2001; Hovatta et al. 1998; Lindholm et al. 1999; Bailer et al. 2000; Hwu et al. 2000); 6p24, near D6S940/D6S470 (Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6 and 8 1996); 6p23-22, near D6S260 (Turecki et al. 1997; Hovatta et al. 1998; Hwu et al. 2000); and 6p21, both proximal to (Arolt et al. 1996) and within (Wright et al. 2001) the locus for human leukocyte antigen. The sample studied by Schwab et al. (1995) shows positive markers and multiple multipoint peaks from 6p24 through 6p21 (Schwab et al. 2000), as does the Irish high-density sample that we have studied. In addition, there have been association studies of SCA1 (on 6p22) (Wang et al. 1996; Pujana et al. 1997; Joo et al. 1999) and of NOTCH4 (Wei and Hemmings 2000; Imai et al. 2001; McGinnis et al. 2001; Sklar et al. 2001; Ujike et al. 2001).
In two previous reports from the Irish Study of High Density Schizophrenia Families (ISHDSF), we have presented evidence in support of the linkage of region 6p24-21 to schizophrenia and schizophrenia-related disorders (Straub et al. 1995, 2002). In the present study, we describe the results of family-based association analysis of simple sequence-length polymorphism (SSLP) markers, additional genotyping of SNPs in 6p22, and analysis of SNP haplotypes. We find that genetic variation in the gene for DTNBP1, which is the human ortholog of mouse dysbindin, is associated with schizophrenia and related phenotypes.
Subjects and Methods
Pedigree Ascertainment and Diagnostic Assessment
A detailed description of the ISHDSF has been published elsewhere (Kendler et al. 1996). The sample used here contains 270 families and 1,425 genotyped individuals (Straub et al. 2002). The main diagnostic instruments utilized in the fieldwork were the Structured Interview for DSM-III-R Diagnosis (Spitzer et al. 1992) and the Structured Interview for Schizotypy (Kendler et al. 1989). Diagnosis was based on DSM-III-R criteria and all available information (personal history, hospital record, and family-history report), by individuals blind to knowledge of genotypes and to psychopathology of relatives. This research was approved by the Ethical Review Boards of Virginia Commonwealth University, the Health Research Board (Dublin), and Queen’s University (Belfast). The diagnostic definitions are as follows.
Narrow: categories D1–D2 (625 affected individuals, 565 of whom were genotyped)
These categories comprise the “core schizophrenic phenotypes,” which include schizophrenia, poor-outcome schizoaffective disorder, and simple schizophrenia (Kendler et al. 1994).
Intermediate: categories D1–D5 (804 affected individuals, 691 of whom were genotyped)
These categories comprise a restricted definition of the schizophrenia spectrum, which includes only disorders that repeatedly have been shown to coaggregate in families with narrowly defined schizophrenia (Kendler and Diehl 1993); to the “narrow” definition, the three additional categories (i.e., D3–D5) add schizotypal personality disorder and all other nonaffective psychotic disorders (i.e., schizophreniform disorder, delusional disorder, atypical psychosis, and good-outcome schizoaffective disorder).
Broad: categories D1–D8 (888 affected individuals, 744 of whom were genotyped)
These categories comprise a broad definition of the schizophrenia spectrum, which includes all disorders that aggregate significantly in relatives of schizophrenic probands in the Roscommon Family Study (Kendler et al. 1993a, 1993b, 1993c), an epidemiologic, case-controlled family study conducted, in parallel, in the west of Ireland; to the “intermediate” definition, the three additional categories (i.e., D6–D8) add mood-incongruent and mood-congruent psychotic affective illness, and paranoid, avoidant, and schizoid personality disorder.
Very broad: categories D1–D9 (1,172 affected individuals, 952 of whom were genotyped)
These categories comprise all psychiatric disorders; to the “broad” definition, the additional category (i.e., D9) adds all other psychiatric disorders (e.g., nonpsychotic affective disorders, anxiety disorders, alcoholism, and other non–schizophrenia-spectrum personality disorders).
SNP Identification
SNPs were identified by sequencing of amplicons from pools of 6–12 unrelated affected individuals from families showing a linkage signal in 6p22 but not in 6p24. We sequenced in both directions, using ABI 377 sequencers (Perkin Elmer Biosystems) and dye-terminator chemistry. Many of the novel SNPs that we discovered were assigned NCBI reference identification numbers (see the NCBI Single Nucleotide Polymorphism Web site) after being submitted to NCBI by other laboratories, and these “rs” numbers are shown in table 2. Information on SNP P1635 can be accessed via its NCBI “Assay ID” (i.e., “ss” number), which is 4473786.
Table 2.
Markers and Map of 6p22
| Gene (Exon Size) | Marker (NCBI Numbera) | Polymorphism | Frequencyb | Chromosomal Locationc(Nucleotide Position) | Distancefrom D6S260(bp) | Size of Interval(bp) |
| Jumonji | D6S260 | SSLP | NA | 17854849–17855203 | … | … |
| P1730 (rs1474588) | G/C | .330 | 17912083 | 56,880 | 56,880 | |
| P1283 (rs760659) | G/A | .196 | 17954963 | 99,760 | 42,880 | |
| Dysbindin gene:d | ||||||
| Exon 10 (413 bp) | 17973412–17973824 | |||||
| Exon 9 (144 bp) | 17975131–17975274 | |||||
| P1328 (rs742106) | C/T | .376 | 17974854 | 119,651 | 19,891 | |
| Exon 8 (156 bp) | 17983845–17984000 | |||||
| P1333 (rs742105) | C/T | .480 | 18023448 | 168,245 | 48,594 | |
| P1287 (rs760666) | C/T | .246 | 18039495 | 184,292 | 16,047 | |
| Exon 7 (23 bp) | 18043664–18043686 | |||||
| Exon 6 (133 bp) | 18065872–18066004 | |||||
| P1655 (rs2619539) | G/C | .537 | 18071229 | 216,026 | 31,734 | |
| Exon 5 (133 bp) | 18077948–18078080 | |||||
| P1635 | G/A | .102 | 18078476 | 223,273 | 7,247 | |
| P1325 (rs1011313) | T/C | .088 | 18083806 | 228,603 | 5,330 | |
| Exon 4 (61 bp) | 18088349–18088409 | |||||
| P1765 (rs2619528) | A/G | .167 | 18100203 | 245,000 | 16,397 | |
| P1757 (rs2005976) | A/G | .167 | 18101176 | 245,973 | 973 | |
| P1320 (rs760761) | C/T | .178 | 18101506 | 246,303 | 330 | |
| Exon 3 (51 bp) | 18101918–18101968 | |||||
| Exon 2 (54 bp) | 18102692–18102745 | |||||
| P1763 (rs2619522) | C/A | .155 | 18104023 | 248,820 | 2,517 | |
| P1578 (rs1018381) | C/T | .076 | 18107444 | 252,241 | 3,421 | |
| P1583 (rs909706) | C/T | .377 | 18111245 | 256,042 | 3,801 | |
| Exon 1 (160 bp) | 18113419–18113578 | |||||
| Unknown | P1586 (rs885773) | A/G | .093 | 18119860 | 264,657 | 8,615 |
| P1294 (rs441539) | A/G | .380 | 18215012 | 359,809 | 95,152 | |
| P1140 (rs1000117) | C/A | .301 | 18324363 | 469,160 | 109,351 | |
| D6S1676 | SSLP | NA | 18525332–18525609 | 670,406 | 201,246 | |
| AFM189YE3 | SSLP | NA | 18534500–18534767 | 679,564 | 9,158 |
Reference identification number for each SNP, from the dbSNP database (see the NCBI Single Nucleotide Polymorphism Web site).
Of the second allele shown. NA = not applicable.
From the UCSC August 2001 draft assembly of the genome (see the UCSC Human Genome Project Working Draft Web site).
Exons and their genomic locations are based on transcript BC011912.
SNP Genotyping
The fluorescence-polarization template–directed incorporation method (Chen and Kwok 1999; Chen et al. 1999), which is a single-base extension with allele-specific ddNTP termination, was used, with minor modifications (Sullivan et al. 2001).
Statistical Analysis
The GENEHUNTER multipoint methods and a graph of the results are contained in our genome-scan report (Straub et al. 2002). The Penetrance-Additive model is an intermediate, heterozygotic genetic model that is additive on the penetrance scale. For example, for the phenotypic model using categories D1–D2, we set AA to 0.55, Aa to 0.275, and aa to 0.0006, with a phenocopy proportion of 0.10 and a schizophrenia-allele frequency of 0.0098. The statistical methods used in the present study are as follows. SIMWALK2 (Weeks et al. 1995; Sobel and Lange 1996) was used for detection of errors in the data. Before data cleaning, the recombinant profiles produced by SIMWALK haplotyping showed 72 single recombinants, 104 double recombinants, and 4 quadruple recombinants. There were 26,337 genotypes that were checked, and SIMWALK flagged 324 recombinants that had a >.25 posterior probability of being due to genotyping error; for this group of 324 recombinants, the probability was >.5 for 196 and >.75 for 119. As a rule, only those recombinants for which the probability is >.90 are highly likely to be true errors, so, to be conservative, we eliminated the 119 recombinants (0.45% of all genotypes) having probabilities >.75. Version 2.5 (April 1999) of the program TRANSMIT (Clayton 1999; Clayton and Jones 1999; Dudbridge et al. 2000) was used for family-based transmission/disequilibrium test (TDT) analysis. In all TRANSMIT runs, the program performed ⩾100,000 replicates, and, from these, derived the P values empirically. For both the pairwise analysis of SSLPs and the haplotype analysis, to protect against misleading results due to rare alleles or haplotypes, the command-line switches (i.e., flags) “-agg3” and “-c3” were used to aggregate all alleles or haplotypes with frequencies <.03 before haplotype construction. For some tests, the switches “-1” and “-nomf” were used together, to simultaneously restrict the analysis to one affected offspring per nuclear family per extended pedigree, each chosen at random by the program. To assess the effect of the particular (random) choices that were made, we ran each test 20 times, and we present here the median P value. For the most significant results, we also performed an additional 50 runs, and the median was consistently the same as that for the initial 20 runs. We note that, given the linkage evidence, there is an increased a priori probability that small P values represent true positive results. Since the markers are tightly linked, the P values may be highly correlated, and thus a simple Bonferroni correction is unduly conservative, and we report only these empirical P values as determined by TRANSMIT. In our Irish data set, only 60 families have both parents of the largest sibship genotyped, another 90 families have one parent genotyped, and the remaining 120 families have neither parent genotyped. Thus, a program such as TRANSMIT, which is “vertical” in that it attempts to reconstruct the missing parental information, is more powerful (see Cervino and Hill 2000) than either (a) programs such as SIBASSOC and STDT, which utilize only information within sibships, or (b) the traditional TDT, which relies heavily on the usually scarce heterozygous parents. The standardized (i.e., Lewontin [1988]) measure of linkage disequilibrium (LD), denoted as “D′,” and the measure d2 were both calculated from the TRANSMIT haplotype frequencies for alleles at all possible pairs of SNP loci. MEGA2 (Mukhopadhyay et al. 1999), version 2.2, was used to calculate allele and genotype frequencies and to test for departure from Hardy-Weinberg equilibrium, in affected individuals, in unaffected individuals, and in the two groups combined.
Results
The results from GENEHUNTER multipoint analysis (see Straub et al. 2002) of data from 36 SSLP markers are shown in table 1. In both the H-LOD (shown) and nonparametric linkage (NPL) (not shown) results, there are two predominant, sharp peaks, one at ∼8 cM (in 6p24.3, near D6S940) and another at ∼17 cM (in 6p22, near D6S260). A third peak, smaller than these two, is present near D6S422 (in 6p22, distal to HLA). For the two larger peaks, the H-LODs are greatest when the broad-definition diagnostic categories (i.e., D1–D8) are used. Whether these peaks indicate the presence of multiple susceptibility loci or are statistical artifacts could not be determined on the basis of the linkage data alone. In an attempt to narrow the target region(s), family-based TDT analysis of the 36 SSLPs was performed, by TRANSMIT (Clayton 1999; Clayton and Jones 1999; Dudbridge et al. 2000). In the first analysis (denoted “All”), data from all individuals in all families were used. The second analysis, designed to mimic aspects of the traditional TDT test, used data from only one affected offspring per nuclear family per extended pedigree. We did not run simple TDT or haplotype relative-risk analysis, since, for family structures such as those in the ISHDSF, each has the considerable disadvantage of discarding valuable information (Cervino and Hill 2000). For all tests, the program generates empirical P values by simulation. Under the first analysis (i.e., All), of the 36 SSLPs, only the 6p24.3 marker D6S940 showed a global P value <.01. The P values were .033, .023, .016, and .008, for diagnostic categories D1–D2, D1–D5, D1–D8, and D1–D9, respectively. The adjacent SSLPs, GATA23E10 (892 kb distal) and D6S470 (182 kb proximal), were both negative. For D6S940, when the analysis was performed on the restricted data set without aggregation of rare alleles, the P values were .024, .114, .08, and .08, respectively. With both the restricted data set and aggregation of alleles having frequency ⩽3%, all four tests were negative. This LD finding in region 6p24.3 was not pursued further in the present study.
Table 1.
GENEHUNTER Multipoint Linkage Results[Note]
|
Location |
Multipoint H-LOD |
|||||||
| Region | Marker Name | Locus | cM | Mba | D1–D2 | D1–D5 | D1–D8 | D1–D9 |
| GATA3H05 | D6S477 | 0 | 7.19 | .10 | .55 | 1.12 | .05 | |
| F13A1 | F13A1 | 1.8 | 7.20 | .15 | .79 | 1.40 | .14 | |
| D6S309 | D6S309 | 4.9 | 9.28 | .26 | 1.02 | 1.40 | .28 | |
| D6S277 | D6S277 | 6 | 9.56 | .22 | .99 | 1.44 | .31 | |
| D6S263 | D6S263 | 6.01 | 9.60 | .33 | 1.20 | 1.50 | .34 | |
| MS236 | D6S226 | 6.02 | 9.64 | .34 | 1.05 | 1.33 | .29 | |
| D6S296F | D6S296 | 6.03 | 9.91 | .33 | 1.05 | 1.33 | .28 | |
| D6S410 |
D6S410 |
6.33 |
10.05 |
.27 |
1.08 |
1.39 |
.34 |
|
| 6p24.3 | GATA23E10 | 7.53 | 10.54 | .46 | 1.50 | 1.90 | .45 | |
| 6p24.3 | UT6077F | D6S940 | 7.73 | 11.44 | .43 | 1.29 | 1.69 | .42 |
| 6p24.3 | D6S470L | D6S470 | 8.03 | 11.62 | .82 | 1.75 | 2.15 | .46 |
| 6p24.3 | AFMc031yd9 |
8.04 |
11.68 |
.46 |
1.36 |
1.75 |
.44 |
|
| ATA33A11 | D6S1263 | 12.34 | 14.56 | .23 | .64 | 1.08 | .31 | |
| ATA50C05 | D6S2434 | 14.04 | 15.83 | .33 | .54 | 1.02 | .27 | |
| D6S443 | D6S443 | 14.34 | 15.87 | .33 | .59 | 1.00 | .29 | |
| B337WG9L | D6S1653 | 14.54 | 16.33 | .33 | .56 | .95 | .31 | |
| D6S259 | D6S259 | 15.74 | 16.62 | .49 | 1.16 | 1.60 | .79 | |
| a244wf5 |
D6S1578 |
16.04 |
16.90 |
.56 |
1.33 |
1.78 |
.79 |
|
| 6p22.3 | D6S260F | D6S260 | 16.64 | 17.85 | .67 | 1.60 | 2.22 | .90 |
| 6p22.3 | AFM321tc5 | D6S1676 | 17.34 | 18.53 | .64 | 1.55 | 2.08 | .91 |
| 6p22.3 | AFM189ye3 | 17.35 | 18.53 | .64 | 1.56 | 2.08 | .91 | |
| 6p22.3 | AFMb008wb5 |
D6S1605 |
18.05 |
18.76 |
.59 |
1.81 |
2.20 |
1.07 |
| SCA1 (CAG) | SCA1 | 18.25 | 18.77 | .32 | 1.45 | 1.68 | .98 | |
| SCA78a | 18.35 | 18.78 | .26 | 1.33 | 1.56 | .90 | ||
| D6S288 | D6S288 | 18.36 | 18.84 | .30 | 1.42 | 1.66 | .96 | |
| a288zg9 | D6S1584 | 19.16 | 19.16 | .05 | 1.05 | 1.02 | .43 | |
| TW9 | 19.17 | 19.18 | .05 | 1.04 | 1.01 | .38 | ||
| D6S274L | D6S274 | 19.18 | 19.20 | .04 | 1.00 | .99 | .36 | |
| a219zd9 | D6S1567 | 20.28 | 19.94 | .00 | .41 | .56 | .07 | |
| D6S285 | D6S285 | 20.78 | 21.43 | −.01 | 1.01 | 1.15 | .20 | |
| D6S422L | D6S422 | 21.98 | 23.23 | .00 | 1.12 | 1.55 | .38 | |
| D6S299 | D6S299 | 28.08 | 26.78 | −.01 | .24 | .68 | .59 | |
| D6S276 | D6S276 | 29.18 | 28.26 | −.01 | .16 | .57 | .51 | |
| MFD61 | D6S105 | 29.28 | 31.42 | .01 | .21 | .65 | .48 | |
| D6S273 | D6S273 | 32.58 | 35.32 | .07 | .21 | .78 | .56 | |
| D6S291 | D6S291 | 34.08 | 43.02 | .04 | .13 | .52 | .32 | |
Note.— The two primary peaks are boxed.
Chromosomal location in UCSC August 2001 draft assembly of the genome (see the UCSC Human Genome Project Working Draft Web site).
On the basis of these previously published linkage results, we investigated region 6p22 in further detail, by sequencing to find SNPs, genotyping, and analysis using both case-control and family-based TDT designs. We first concentrated on the 250-kb interval between D6S1676 and the CAG repeat, in gene SCA1. We tested a total of 34 SNPs in subsets of individuals (n=47–300), in an “internal case-control” design. The most highly selected group of cases (one per family) consisted of affected individuals in categories D1–D2 who were selected from those families that, during those separate GENEHUNTER runs (results not shown), yielded positive H-LOD/NPL scores with the 16 SSLP markers in 6p23-22 (D6S1653–D6S422) but did not do so with the adjacent set of 15 SSLP markers in 6p25-22 (D6S477–D6S443). The control group comprised unaffected individuals who were unrelated (i.e., married-in) to the affected individuals in their families and were chosen from families that produced negative results across the entire region. For these 34 SNPs, no sizable or regionally consistent differences in allele frequencies were observed (data not shown).
On the basis of these negative results, we then focused on the 670-kb interval between D6S260 and D6S1676, the two SSLP markers located atop the 6p22 multipoint-linkage peak. We identified SNPs by database searches and by sequencing of pools of the highly selected cases.
Table 2 shows the map locations and marker characteristics for the 3 SSLPs and 17 SNPs in 6p22 that were tested in the entire sample. The markers are presented in chromosomal order, with D6S260 telomeric. The first three markers in the table are in the gene Jumonji, the next 12 SNPs are in the gene DTNBP1 (i.e., the dysbindin gene), and the next 5 markers are proximal to the dysbindin gene. Markers P1655, P1635, and P1765, as well many others that we identified but did not test in the entire sample, were first detected by sequencing of pools of affected individuals, as described above. All SNPs tested in dysbindin are intronic, except P1328, which is in the 3′ UTR of transcript AL136637. Analysis using multiple splice-site prediction programs (Mount 2000) showed that none of the SNPs are predicted to affect splicing, but this must be tested experimentally.
The pairwise TRANSMIT results are shown in table 3. When all individuals are included in the analysis, the pattern of significance varies somewhat across diagnostic categories, with maximal evidence for a given marker usually occurring when either categories D1–D5 or categories D1–D8 are used. One clear exception to this pattern, however, is SNP P1320. Except for the most centromeric marker, the SSLP AFM189YE3, the strongest evidence was never found for the very-broad definition (i.e., categories D1–D9), which includes psychiatric disorders not considered to be within the schizophrenia spectrum. When the analysis was restricted to one affected offspring per nuclear family per extended pedigree, the median P value of 20 runs was not significant for markers P1333, P1655, P1325, markers that reached significance when all individuals were included. None of the markers were positive when categories D1–D9 were used. All of the markers in the entire sample were in Hardy-Weinberg equilibrium. To test the possibility that the positive TDT results were artifactual, due, in some way, to transmission distortion, for P1635 we recoded the affected individuals as unknown and the unaffected individuals as affected. No transmission distortion was observed.
Table3.
Pairwise TRANSMIT Results
|
Transmit P Valuea |
|||||||||
| All Individuals |
One Affected Offspring perNuclear Family per Extended Pedigreeb |
||||||||
| Gene and Marker (NCBI Number) | OvertransmittedAllele (Frequency) | D1–D2 | D1–D5 | D1–D8 | D1–D9 | D1–D2 | D1–D5 | D1–D8 | D1–D9 |
| Jumonji: | |||||||||
| D6S260 | .193 | .096 | .018 | .077 | .579 | .333 | .341 | .355 | |
| P1730 (rs1474588) | .403 | .743 | .487 | .187 | .499 | .242 | .635 | .605 | |
| P1283 (rs760659) | .536 | .797 | .831 | .411 | .616 | .535 | .552 | .626 | |
| Dysbindin: | |||||||||
| P1328 (rs742106) | .127 | .125 | .275 | .256 | .502 | .332 | .467 | .436 | |
| P1333 (rs742105) | T (.480) | .0099 | .0036 | .0027 | .019 | .533 | .507 | .504 | .559 |
| P1287 (rs760666) | .769 | .603 | .597 | .954 | .717 | .638 | .615 | .578 | |
| P1655 (rs2619539) | G (.463) | .0059 | .001 | .0008 | .014 | .558 | .504 | .465 | .544 |
| P1635 | A (.102) | .00009 | .00008 | .00004 | .024 | .006 | .0066 | .027 | .114 |
| P1325 (rs1011313) | C (.088) | .174 | .0377 | .0104 | .024 | .562 | .399 | .512 | .628 |
| P1765 (rs2619528) | G (.167) | .109 | .036 | .095 | .738 | .0466 | .171 | .158 | .561 |
| P1757 (rs2005976) | G (.167) | .091 | .037 | .065 | .960 | .0078 | .0366 | .039 | .157 |
| P1320 (rs760761) | T (.178) | .0004 | .020 | .172 | .929 | .0029 | .107 | .127 | .526 |
| P1763 (rs2619522) | A (.155) | .202 | .101 | .130 | .604 | .0465 | .229 | .392 | .677 |
| P1578 (rs1018381) | .762 | .738 | .488 | .176 | .473 | .824 | .553 | .705 | |
| P1583 (rs909706) | .969 | .308 | .299 | .244 | .282 | .558 | .645 | .469 | |
| Unknown: | |||||||||
| P1586 (rs885773) | .478 | .378 | .226 | .065 | .293 | .489 | .495 | .501 | |
| P1294 (rs441539) | .018 | .049 | .051 | .103 | .071 | .117 | .146 | .414 | |
| P1140 (rs1000117) | .316 | .137 | .193 | .078 | .436 | .604 | .364 | .604 | |
| D6S1676 | .617 | .359 | .371 | .354 | .166 | .218 | .195 | .090 | |
| AFM189YE3 | .644 | .086 | .544 | .036 | .580 | .695 | .696 | .623 | |
Generated by simulation.
Median of 20 runs. Values <.05 are in boldface italic.
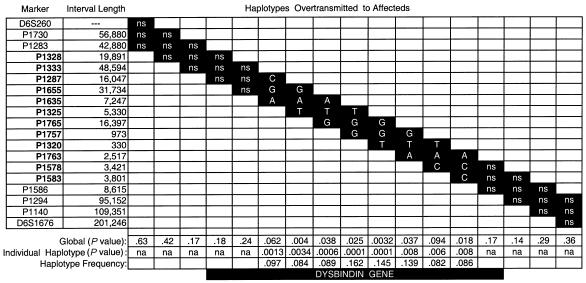
We also analyzed two-, three-, and four-marker haplotypes for the 17 SNPs and for the 2 flanking SSLPs. Figure 1 shows the results, when diagnostic categories D1–D2 were used, for all of the three-marker haplotypes tested. The “global” P value represents the overall significance when the observed versus expected transmissions of all of the haplotypes are considered together. Also shown are the most significant of the overtransmitted haplotypes from that median run, the haplotype frequency, and the associated P value. All of the adjacent, overtransmitted three-marker haplotypes are consistent with each other in terms of the SNP alleles represented. Results with the two- and four-marker haplotypes (data not shown) were also in quite good agreement with the results for the pairwise and three-marker haplotypes. One difference between the pairwise results and the haplotype results occurred with marker P1325, for which the C allele was slightly overtransmitted in all pairwise runs but for which the T allele was contained in all three overtransmitted three-marker haplotypes. The frequency of the most positive haplotypes varied between 0.082 and 0.162, which is consistent with our previous estimates based on linkage results—that <30% of the families segregate susceptibility alleles on 6p.
Figure 1.
TRANSMIT results with three-marker haplotypes. When diagnostic categories D1–D2 and the condition of one affected offspring per nuclear family per extended pedigree are used, a sliding window of three markers was tested, at two-marker overlaps. Below each (most significant) overtransmitted haplotype is the P value for that haplotype, the haplotype frequency, and the global P value (median value of 20 runs), derived by evaluation of the transmissions of all haplotypes at once. SNPs within the dysbindin gene are in boldface and are offset, and the location of the gene is shown below the haplotype frequencies. Interval Length = marker-to-marker distance (in bp); na = instances in which no individual haplotype (either over- or undertransmitted) resulted in P<.05.
Table 4 shows the pairwise marker-to-marker LD results; below the diagonal is the standard Lewontin D′ statistic, and above the diagonal is the d2 statistic. Note the extremely strong LD (a) between P1333 and the 60-kb region bounded by P1287 and P1325 and (b) with the region bounded by P1578 and P1586, but not with the markers between these two regions. In contrast, the more proximal marker P1287 is in strong LD with the entire 72-kb region bounded by P1655 and P1583. In addition, P1325 is in strong LD with P1765 and P1757 (17 kb), is not at all in LD with P1320, and is “again” in LD with P1763–P1294 (131 kb). With this complex pattern of LD, neither the number of susceptibility alleles nor their specific location(s) is readily apparent.
Table 4.
Pairwise (Marker-to-Marker) LD Results
|
Pairwise LD Resulta |
|||||||||||||||||
| P1730 | P1283 | P1328 | P1333 | P1287 | P1655 | P1635 | P1325 | P1765 | P1757 | P1320 | P1763 | P1578 | P1583 | P1586 | P1294 | P1140 | |
| P1730 | … | .00 | .16 | .04 | .10 | .04 | .02 | .04 | .03 | .03 | .01 | .02 | .14 | .07 | .08 | .00 | .02 |
| P1283 | .15 | … | .02 | .00 | .02 | .00 | .01 | .13 | .00 | .00 | .00 | .01 | .03 | .00 | .06 | .00 | .00 |
| P1328 | .45 | .38 | … | .22 | .05 | .19 | .00 | .00 | .02 | .01 | .00 | .02 | .01 | .21 | .01 | .01 | .00 |
| P1333 | .26 | .00 | .57 | … | .28 | .81 | .11 | .09 | .00 | .00 | .02 | .00 | .09 | .54 | .05 | .05 | .01 |
| P1287 | .79 | .54 | .49 | .95 | … | .14 | .38 | .38 | .36 | .06 | .06 | .05 | .03 | .15 | .01 | .09 | .00 |
| P1655 | .25 | .07 | .52 | .93 | 1.00 | … | .10 | .08 | .01 | .01 | .02 | .00 | .05 | .49 | .03 | .05 | .00 |
| P1635 | .61 | .53 | .04 | .96 | 1.00 | .86 | … | .00 | .36 | .38 | .46 | .36 | .12 | .02 | .00 | .02 | .01 |
| P1325 | .93 | .58 | .19 | .94 | .99 | .94 | .08 | … | .30 | .28 | .00 | .30 | .15 | .14 | .01 | .05 | .00 |
| P1765 | .28 | .08 | .37 | .15 | 1.00 | .17 | .79 | .99 | … | .92 | .66 | .87 | .35 | .09 | .26 | .03 | .00 |
| P1757 | .29 | .07 | .34 | .14 | .94 | .15 | .82 | .94 | .96 | … | .70 | .94 | .38 | .13 | .23 | .03 | .00 |
| P1320 | .14 | .06 | .10 | .28 | .95 | .26 | .93 | .00 | .85 | .87 | … | .69 | .26 | .04 | .14 | .04 | .00 |
| P1763 | .25 | .11 | .38 | .14 | .99 | .14 | .76 | 1.00 | .96 | .99 | .90 | … | .41 | .49 | .25 | .04 | .00 |
| P1578 | .93 | .32 | .45 | .90 | .95 | .83 | .87 | 1.00 | .93 | .97 | .82 | .97 | … | .07 | .60 | .01 | .01 |
| P1583 | .29 | .09 | .47 | .89 | .86 | .83 | .51 | .94 | .83 | .82 | .54 | .99 | 1.00 | … | .03 | .02 | .02 |
| P1586 | .61 | .39 | .37 | .73 | .52 | .60 | .22 | .75 | .71 | .68 | .56 | .68 | .85 | .74 | … | .01 | .01 |
| P1294 | .12 | .04 | .17 | .30 | .42 | .30 | .56 | .90 | .48 | .53 | .52 | .62 | .50 | .20 | .43 | … | .02 |
| P1140 | .17 | .00 | .07 | .14 | .16 | .09 | .38 | .20 | .01 | .01 | .07 | .01 | .22 | .16 | .22 | .16 | … |
For each pair of markers, the standardized D′ is shown below the diagonal, and d2 is shown above the diagonal. D′ values >0.8 are in boldface italic.
Discussion
The picture emerging from detailed studies of the limited number of genes that have been identified in complex disorders is one of subtle, rather than striking, relationships between genetic variation, gene function, and increased risk (Risch 2000; Reich and Lander 2001). For example, the role of specific SNPs in the calpain-10 gene that are associated with type 2 diabetes is still unclear, despite extensive efforts directed toward both SNP detection and genotyping in multiple, large samples (Altshuler et al. 2000a; Horikawa et al. 2000; Weiss and Terwilliger 2000; Cox 2001; Evans et al. 2001). Another example from type 2 diabetes is the frequent PPAR-γ Pro12Ala polymorphism, for which a meta-analysis of >3,000 individuals was necessary to resolve apparently conflicting studies (Altshuler et al. 2000b). Likewise, despite extensive efforts, the relationship between the sequence diversity in the APOE gene (Nickerson et al. 2000) and the contribution of this variation to the risk of Alzheimer disease remains incompletely understood (Daly 1998; Martin et al. 2000). Given these well-documented difficulties, the results presented here represent only an initial step toward an understanding of the possible etiologic role that dysbindin plays in schizophrenia. The complex pattern of positive versus negative TDT results that we observed with individual markers may indicate multiple susceptibility alleles (haplotypes), although other explanations are possible; if true, this would again support the expectation of a high degree of allelic heterogeneity in most complex phenotypes (Weiss and Terwilliger 2000). Nevertheless, the central location of the positive SNPs within the 140-kb dysbindin gene strongly implicates this gene as a source of the positive linkage signal in this chromosomal region. Our results suggest that the susceptibility haplotypes probably do not extend distally to the neuronally expressed gene Jumonji, which directly abuts the dysbindin gene but is transcribed in the opposite direction. Proximally, there are no ESTs or consistently predicted exons for the next few hundred kilobases, but this certainly does not rule out genes being present. It remains a formal possibility that dysbindin itself is not aberrant in schizophrenia but, rather, that variations in it affect expression of nearby genes; and this can be tested.
In view of the recent data showing the punctate, usually 10–100-kb haplotype block structure of the human genome (Daly et al. 2001; Johnson et al. 2001; Reich et al. 2001), confirmation that the dysbindin gene is the sole susceptibility gene in this region may be relatively straightforward, since the most strongly associated markers and haplotypes reside within this gene. It does appear that marker P1635, which we found by sequencing affected individuals from 6p22-linked families but which other groups have not yet deposited into the public databases, is an important boundary. In the pairwise analysis, P1635 is strongly associated with schizophrenia, whereas the marker that is 5.3 kb proximal (i.e., marker P1325) is not. This boundary effect is also reflected in both the haplotype and the marker-to-marker LD results. Although higher marker densities achieved by use of the available (i.e., common) SNPs may be of some utility in finer mapping of the risk alleles, the sobering findings from the haplotype-tagging study by Todd and colleagues (Johnson et al. 2001) suggest that the publicly available SNPs will be inadequate for “mutation” detection, and so saturation resequencing in affected individuals, starting near P1635, is necessary. The SNPs tested are intronic, and, although none is located in a canonical splice site, very little is known about the function that specific intronic sequences have with regard to hnRNA secondary structure, protein binding, stability, and splicing efficiency. Thus, any of these associated SNPs may affect expression, and this possibility has to be investigated directly by testing for differences in transcript structures, quantity, and spatial and developmental distribution between patients and control subjects.
Dysbindin is evolutionarily conserved and is predicted to have a coiled-coil secondary structure (Burkhard et al. 2001) similar to that of the mouse ortholog (Dtnbp1) (Benson et al. 2001), with which it exhibits a high degree of protein-sequence identity. On the basis of EST alignments alone, there appear to be at least five alternative transcripts.
In mouse brain, the dysbindin protein is found in multiple anatomical locations, including axon fibers in the corpus callosum, mossy-fiber terminal fields in the hippocampus and cerebellum, and neuropil areas of the neocortex, hippocampus, and substantia nigra (Benson et al. 2001). Using semiquantitative PCR, we have found that human dysbindin transcripts are expressed in all 24 tissues (e.g., brain, heart, lung, etc.) tested, as well as in all 12 brain regions (frontal lobe, temporal lobe, cerebellum, hippocampus, substantia nigra, caudate nucleus, amygdala, thalamus, hypothalamus, pons, medulla, and spinal cord) tested (data not shown).
Dysbindin binds to β-dystrobrevin and thus is likely to also be a component of the dystrophin protein complex (DPC) (Mehler 2000; Roberts 2001) found in postsynaptic densities (PSD) (Blake et al. 1999) and elsewhere in brain. In addition to its structural role in neuromuscular synapse formation and maintenance (Grady et al. 2000), the DPC appears to be involved in signal transduction—for example, by regulating nicotinic receptor clustering and by recruiting specific signaling molecules such as neuronal nitric oxide synthase (Bredt 1999; Grady et al. 1999), an enzyme that also interacts with PSD proteins (e.g., PSD-93 and PSD-95) that are involved in N-methyl-d-aspartate–receptor clustering. The composition of the DPC in muscle and brain (e.g., content of dystrophin isoforms Dp 427, Dp71, and Dp140) differs and is also heterogeneous within the brain (Moukhles and Carbonetto 2001), but it is almost certain both to exert effects on synaptic function in brain as well and to modulate other receptors. For example, dystrophin is extensively colocalized with postsynaptic gamma-aminobutyric acid (GABA-A)–receptor subtypes in hippocampus, cortex, and cerebellum (Kneusel et al. 1999). Also, Kneusel et al. (1999) have found that, in the mdx (dystrophin deficient) mouse cerebellum and hippocampus, there was a marked reduction in the number (but not in the size) of synaptic GABA-A–receptor clusters, whereas this was not true for the striatum, which does not normally contain dystrophin.
The relationship between genomic variation in dysbindin and the biological processes involved in schizophrenia is entirely speculative. Nevertheless, if dysbindin plays a role in synaptic signaling and plasticity, it would be well positioned to play a pathogenic role. There is abundant neuropathological evidence that synaptic changes are associated with schizophrenia; for example, both a reduced density of dendritic spines on the excitatory pyramidal neurons and reductions in multiple proteins found in afferent terminals in frontal cortex and hippocampus have been reported (Weinberger 1999; Glantz and Lewis 2001). Such changes suggest reduced cortical connectivity and plasticity, which are likely to be primary to the illness. Numerous as they may be, genes coding for synaptic proteins, whether functioning on dendritic spines presynaptically or postsynaptically, are all tenable functional candidate genes (Mirnics et al. 2001). In addition to the still rather diffuse dopaminergic and developmental “hypotheses” of schizophrenia, both the glutamatergic and GABA-ergic systems have been implicated (Goldman-Rakic and Selemon 1997; Benes 2000; Pearlson 2000), and these four areas of investigation already overlap considerably. It is conceivable that defects in dysbindin function could affect more than one of these systems, during development, adulthood, or both.
The intermediate (schizophrenia) phenotypes, of moderately diminished premorbid IQ and related cognitive measures (Elvevag and Goldberg 2000; Weickert et al. 2000), may be an interesting connection between the function of the DPC in synaptic transmission and the observed association between dysbindin and schizophrenia. The nonprogressive, cognitive deficits present in Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD) are reflected in significantly lower IQ scores—approximately one-quarter of boys with DMD (whose dystrophin is either missing entirely or functionally mutant) are mildly mentally retarded (Blake and Kroger 2000; Anderson et al. 2002). The impairment is found to be greater on verbal than on nonverbal tests (Moizard et al. 1998) and may be due to disruption of DPC function during embryogenesis, although, as in the case of schizophrenia, the majority of patients with DMD exhibit neither gross nor histological brain abnormalities. Since mutations in dysbindin may also compromise DPC function, this may explain, at least in some small measure, the well-known observation that the average IQ of schizophrenics is lower than that in control subjects. Overall, there appears to be little evidence of comorbidity between schizophrenia and the muscular dystrophies, but this alone would not preclude some overlap in molecular mechanisms. Early reports had described (a) a family in which four of five adult patients with BMD also had schizophrenia or related disorders (Zatz et al. 1993) and, separately, (b) an association between schizophrenia and DMD (Melo et al. 1993), but these results may be anecdotal. Southern blotting of DNA from 94 schizophrenics was used to screen for deletions in exons 1–59 of dystrophin, but no deletions were identified (Lindor et al. 1994). No SNP association studies have been reported for dystrophin or other components of the DPC, such as syntrophins, sarcoglycans, and dystroglycans.
A considerable amount of work will be required in order to confirm and extend our findings of an association between dysbindin and schizophrenia. At present, the possibility that this is a false-positive result, due to any of several possible artifacts, cannot, of course, be formally ruled out. However, given the internal consistency and strength of the results, we think that genetic variation in dysbindin influences the risk of schizophrenia and related schizophrenia-spectrum disorders and, thus, that this gene should be the subject of further inquiry.
Acknowledgments
This project was largely supported by National Institute of Mental Health grants MH41953, MH52537, and MH45390. Data were collected under the supervision of S. Humphries, M. Healy, and A. Finnerty. Additional interviews were conducted by J. Burke, B. Murphy, F. Duke, R. Shinkwin, M. Ni Nuallain, F. McMahon, J. Downing, T. Hebron, B. Hanratty, E. Crowe, M. Doherty, J. Bray, and L. Lowry. This project would not have been possible without the cooperation of the families themselves and of the staffs of the many psychiatric hospitals and units in Ireland and Northern Ireland, and their efforts are gratefully acknowledged. We also wish to acknowledge the assistance of R. McClelland.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- NCBI Single Nucleotide Polymorphism, http://www.ncbi.nlm.nih.gov/SNP/ (for reference identification numbers for SNPs)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCZD3 [MIM 600511])
- UCSC Human Genome Project Working Draft, http://genome.ucsc.edu/ (for marker and SNP locations)
References
- Altshuler D, Daly M, Kruglyak L (2000a) Guilt by association. Nat Genet 26:135–137 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl M, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000b) The common PPAR Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Anderson JL, Head SI, Rae C, Morley JW (2002) Brain function in Duchenne muscular dystrophy. Brain 125 (Pt 1): 4–13 [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Blouin JL, Pulver AE, Wolyniec P, Lasseter VK, Nestadt G, Kasch L, Babb R, Kazazian HH, Dombroski B, Kimberland M, Ott J, Housman D, Karayiorgou M, MacLean CJ (1995) Schizophrenia susceptibility and chromosome 6p24-22. Nat Genet 11:235–236 [DOI] [PubMed] [Google Scholar]
- Arolt V, Lencer R, Nolte A, Müller-Myhsok B, Purmann S, Schürmann M, Leutelt J, Pinnow M, Schwinger E (1996) Eye tracking dysfunction is a putative phenotypic susceptibility marker of schizophrenia and maps to a locus on chromosome 6p in families with multiple occurrence of the disease. Am J Med Genet 67:564–579 [DOI] [PubMed] [Google Scholar]
- Bailer U, Leisch F, Meszaros K, Lenzinger E, Willinger U, Strobl R, Gebhardt C, Gerhard E, Fuchs K, Sieghart W, Kasper S, Hornik K, Aschauer HN (2000) Genome scan for susceptibility loci for schizophrenia. Neuropsychobiology 42:175–182 [DOI] [PubMed] [Google Scholar]
- Baron M (2001) Genetics of schizophrenia and the new millennium: progress and pitfalls. Am J Hum Genet 68:299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM (2000) Emerging principles of altered neural circuitry in schizophrenia. Brain Res Brain Res Rev 31:251–269 [DOI] [PubMed] [Google Scholar]
- Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ (2001) Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem 276:24232–24241 [DOI] [PubMed] [Google Scholar]
- Blake DJ, Hawkes R, Benson MA, Beesley PW (1999) Different dystrophin-like complexes are expressed in neurons and glia. J Cell Biol 147:645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DJ, Kroger S (2000) The neurobiology of Duchenne muscular dystrophy: learning lessons from muscle? Trends Neurosci 23:92–99 [DOI] [PubMed] [Google Scholar]
- Bray NJ, Owen MJ (2001) Searching for schizophrenia genes. Trends Mol Med 7:169–174 [DOI] [PubMed] [Google Scholar]
- Bredt DS (1999) Knocking signalling out of the dystrophin complex. Nat Cell Biol 1:E89–E91 [DOI] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11:82–88 [DOI] [PubMed] [Google Scholar]
- Cervino ACL, Hill AVS (2000) Comparison of tests for association and linkage in incomplete families. Am J Hum Genet 67:120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kwok PY (1999) Homogeneous genotyping assays for single nucleotide polymorphisms with fluorescence resonance energy transfer detection. Genet Anal 14:157–163 [DOI] [PubMed] [Google Scholar]
- Chen X, Levine L, Kwok PY (1999) Fluorescence polarization in homogenous nucleic acid analysis. Genome Res 9:492–498 [PMC free article] [PubMed] [Google Scholar]
- Clayton D (1999) A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet 65:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D, Jones H (1999) Transmission/disequilibrium tests for extended marker haplotypes. Am J Hum Genet 65:1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ (2001) Challenges in identifying genetic variation affecting susceptibility to type 2 diabetes: examples from studies of the calpain-10 gene. Hum Mol Genet 10:2301–2305 [DOI] [PubMed] [Google Scholar]
- Daly MJ (1998) Untangling the genetics of a complex disease. JAMA 280:652–653 [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Koeleman BP, Todd JA, Clayton DG (2000) Unbiased application of the transmission/disequilibrium test to multilocus haplotypes. Am J Hum Genet 66:2009–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvevag B, Goldberg TE (2000) Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol 14:1–21 [PubMed] [Google Scholar]
- Evans JC, Frayling TM, Cassell PG, Saker PJ, Hitman GA, Walker M, Levy JC, et al (2001) Studies of association between the gene for calpain-10 and type 2 diabetes mellitus in the United Kingdom. Am J Hum Genet 69:544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA (2001) Dendritic spine density in schizophrenia and depression. Arch Gen Psychiatry 58:203 [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS, Selemon LD (1997) Functional and anatomical aspects of prefrontal pathology in schizophrenia. Schizophr Bull 23:437–458 [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J (1982) Schizophrenia: the epigenetic puzzle. Cambridge University Press, New York [Google Scholar]
- Grady RM, Grange RW, Lau KS, Maimone MM, Nichol MC, Stull JT, Sanes JR (1999) Role for alpha-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat Cell Biol 1:215–220 [DOI] [PubMed] [Google Scholar]
- Grady RM, Zhou H, Cunningham JM, Henry MD, Campbell KP, Sanes JR (2000) Maturation and maintenance of the neuromuscular synapse: genetic evidence for roles of the dystrophin-glycoprotein complex. Neuron 25:279–293 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Hovatta I, Lichtermann D, Juvonen H, Suvisaari J, Terwilliger JD, Arajarvi R, Kokko-Sahin M-L, Ekelund J, Lonnqvist J, Peltonen L (1998) Linkage analysis of putative schizophrenia gene candidate regions on chromosomes 3p, 5q, 6p, 8p, 20p, and 22q in a population-based sampled Finnish family set. Mol Psychiatry 3:452–457 [DOI] [PubMed] [Google Scholar]
- Hwu HG, Lin MW, Lee PC, Lee SF, Ou-Yang WC, Liu CM (2000) Evaluation of linkage of markers on chromosome 6p with schizophrenia in Taiwanese families. Am J Med Genet 96:74–78 [PubMed] [Google Scholar]
- Imai K, Harada S, Kawanishi Y, Tachikawa H, Okubo T, Suzuki T (2001) The (CTG)n polymorphism in the NOTCH4 gene is not associated with schizophrenia in Japanese individuals. BMC Psychiatry 1:1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 [DOI] [PubMed] [Google Scholar]
- Joo EJ, Lee JH, Cannon TD, Price RA (1999) Possible association between schizophrenia and a CAG repeat polymorphism in the spinocerebellar ataxia type 1 (SCA1) gene on human chromosome 6p23. Psychiatr Genet 9:7–11 [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Gogos JA (1997) Dissecting the genetic complexity of schizophrenia. Mol Psychiatry 2:211–223 [DOI] [PubMed] [Google Scholar]
- Kendler KS (2000) The genetics of schizophrenia. In: Sadock BJ, Sadock VA (eds) Comprehensive textbook of psychiatry, 7th ed. Williams & Wilkins, New York, pp 1147–1159 [Google Scholar]
- Kendler KS, Diehl SR (1993) The genetics of schizophrenia: a current, genetic-epidemiologic perspective. Schizophr Bull 19:261–285 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Lieberman JA, Walsh D (1989) The Structured Interview for Schizotypy (SIS): a preliminary report. Schizophr Bull 15:559–571 [DOI] [PubMed] [Google Scholar]
- Kendler KS, McGuire M, Gruenberg AM, O'Hare A, Spellman M, Walsh D (1993a) The Roscommon family study. I. Methods, diagnosis of probands, and risk of schizophrenia in relatives. Arch Gen Psychiatry 50:527–540 [DOI] [PubMed] [Google Scholar]
- ——— (1993b) The Roscommon Family Study. II. The risk of nonschizophrenic nonaffective psychoses in relatives. Arch Gen Psychiatry 50:645–652 [DOI] [PubMed] [Google Scholar]
- ——— (1993c) The Roscommon family study. III. Schizophrenia-related personality disorders in relatives. Arch Gen Psychiatry 50:781–788 [DOI] [PubMed] [Google Scholar]
- Kendler KS, McGuire M, Gruenberg AM, Walsh D (1994) An epidemiologic, clinical, and family study of simple schizophrenia in County Roscommon, Ireland. Am J Psychiatry 151:27–34 [DOI] [PubMed] [Google Scholar]
- Kendler KS, O'Neill FA, Burke J, Murphy B, Duke F, Straub RE, Shinkwin R, Nuallain MN, MacLean CJ, Walsh D (1996) Irish study of high-density schizophrenia families: field methods and power to detect linkage. Am J Med Genet 67:179–190 [DOI] [PubMed] [Google Scholar]
- Kneusel I, Mastrocola M, Zuellig RA, Bornhauser B, Schaub MC, Fritschy J-M (1999) Altered synaptic clustering of GABA-A receptors in mice lacking dystrophin (mdx mice). Eur J Neurosci 11:4457–4462 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1988) On measures of gametic disequilibrium. Genetics 120:849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm E, Ekholm B, Balciuniene J, Johansson G, Castensson A, Koisti M, Nylander PO, Pettersson U, Adolfsson R, Jazin E (1999) Linkage analysis of a large Swedish kindred provides further support for a susceptibility locus for schizophrenia on chromosome 6p23. Am J Med Genet 88:369–377 [PubMed] [Google Scholar]
- Lindor NM, Sobell JL, Heston LL, Thibodeau SN, Sommer SS (1994) Screening the dystrophin gene suggests a high rate of polymorphism in general but no exonic deletions in schizophrenics. Am J Med Genet 54:1–4 [DOI] [PubMed] [Google Scholar]
- Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari AJ , Riley J, Finch KL, Stevens JF, Livak KJ, Slotterbeck BD, Slifer SH, Warren LL, Conneally PM, Schmechel DE, Purvis I, Pericak-Vance MA, Roses AD, Vance JM (2000) SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet 67:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziade M, Bissonnette L, Rouillard E, Martinez M, Turgeon M, Charron L, Pouliot V, Boutin P, Cliche D, Dion C, Fournier JP, Garneau Y, Lavallee JC, Montgrain N, Nicole L, Pires A, Ponton AM, Potvin A, Wallot H, Roy MA, Merette C (1997) 6p24-22 region and major psychoses in the eastern Quebec population: Le Groupe IREP. Am J Med Genet 74:311–318 [PubMed] [Google Scholar]
- Maziade M, Roy MA, Rouillard E, Bissonnette L, Fournier JP, Roy A, Garneau Y, Montgrain N, Potvin A, Cliche D, Dion C, Wallot H, Fournier A, Nicole L, Lavallee JC, Merette C (2001) A search for specific and common susceptibility loci for schizophrenia and bipolar disorder: a linkage study in 13 target chromosomes. Mol Psychiatry 6:684–693 [DOI] [PubMed] [Google Scholar]
- McGinnis RE, Fox H, Yates P, Cameron LA, Barnes MR, Gray IC, Spurr NK, Hurko O, St Clair D (2001) Failure to confirm NOTCH4 association with schizophrenia in a large population-based sample from Scotland. Nat Genet 28:128–129 [DOI] [PubMed] [Google Scholar]
- Mehler MF (2000) Brain dystrophin, neurogenetics and mental retardation. Brain Res Brain Res Rev 32:277–307 [DOI] [PubMed] [Google Scholar]
- Melo M, Vieira AHG, Passos-Bueno MR, Zatz M (1993) Association of schizophrenia and Duchenne muscular dystrophy. Br J Psychiatry 162:711–712 [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Lewis DA, Levitt P (2001) Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci 24:479–486 [DOI] [PubMed] [Google Scholar]
- Moizard M-P, Billard C, Toutain A, Berret F, Marmin N, Moraine C (1998) Are Dp71 and Dp140 brain dystrophin isoforms related to cognitive impairment in Duchenne muscular dystrophy? Am J Med Genet 80:32–41 [DOI] [PubMed] [Google Scholar]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62:690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukhles H and Carbonetto S (2001) Dystroglycan contributes to the formation of multiple dystrophin-like complexes in brain. J Neurochem 78:824–834 [DOI] [PubMed] [Google Scholar]
- Mount S (2000) Genomic sequence, splicing, and gene annotation. Am J Hum Genet 67:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE (1999) Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am J Hum Genet Suppl 65:A436 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Taylor S, Fullerton SM, Weiss KM, Clark A, Stengard J, Salomaa V, Boerwinkle E, Sing CF (2000) Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res 10:1532–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlson GD (2000) Neurobiology of schizophrenia. Ann Neurol 48:556–566 [PubMed] [Google Scholar]
- Pujana MA, Martorell L, Volpini V, Valero J, Labad A , Vilella E, Estivill X (1997) Analysis of amino-acid and nucleotide variants in the spinocerebellar ataxia type 1 (SCA1) gene in schizophrenic patients. Hum Genet 99:772–775 [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES (2001) On the allelic spectrum of human disease. Trends Genet 17:502–510 [DOI] [PubMed] [Google Scholar]
- Riley BP, McGuffin P (2000) Linkage and associated studies of schizophrenia. Am J Med Genet 97:23–44 [DOI] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Roberts RG (2001) Dystrophins and dystrobrevins. Genome Biol 2:REVIEWS3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6 and 8 (1996) Additional support for schizophrenia linkage on chromosomes 6 and 8: a multicenter study. Am J Med Genet 67:580–594 [DOI] [PubMed] [Google Scholar]
- Schwab SG, Albus M, Hallmayer J, Hönig S, Borrmann M, Lichtermann D, Ebstein RP, Ackenheil M, Lerer B, Risch N, Maier W, Wildenauer DB (1995) Evaluation of a susceptibility gene for schizophrenia on chromosome 6p by multipoint affected sib-pair linkage analysis. Nat Genet 11:325–327 [DOI] [PubMed] [Google Scholar]
- Schwab SG, Hallmayer J, Albus M, Lerer B, Eckstein GN, Borrmann M, Segman RH, Hanses C, Freymann J, Yakir A, Trixler M, Falkai P, Rietschel M, Maier W, Wildenauer DB (2000) A genome-wide autosomal screen for schizophrenia susceptibility loci in 71 families with affected siblings: support for loci on chromosome 10p and 6. Mol Psychiatry 5:638–649 [DOI] [PubMed] [Google Scholar]
- Sklar P, Schwab SG, Williams NM, Daly M, Schaffner S, Maier W, Albus M, Trixler M, Eichhammer P, Lerer B, Hallmayer J, Norton N, Williams H, Zammit S, Cardno AG, Jones S, McCarthy G, Milanova V, Kirov G, O'Donovan MC, Lander ES, Owen MJ, Wildenauer DB (2001) Association analysis of NOTCH4 loci in schizophrenia using family and population-based controls. Nat Genet 28:126–128 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Spitzer RL, Williams JBW, Gibbon M, First MB (1992) The structured clinical interview for DSM-III-R (SCID). I. History, rationale, and description. Arch Gen Psychiatry 49:624–629 [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Kendler KS (1996) The putative schizophrenia locus on chromosome 6p: a brief overview of the linkage studies. Mol Psychiatry 1:89–92 [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O'Neill FA, Walsh D, Kendler KS (2002) Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and followup of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry 7:542–559 [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, O'Neill FA, Burke J, Murphy B , Duke F, Shinkwin R, Webb BT, Zhang J, Walsh D, Kendler KS (1995) A potential vulnerability locus for schizophrenia on chromosome 6p24-22: evidence for genetic heterogeneity. Nat Genet 11:287–293 [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Jiang Y, Neale MC, Kendler KS, Straub RE (2001) Association of the tryptophan hydroxylase gene with smoking initiation but not progression to nicotine dependence. Am J Med Genet 105:479–484 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Stone WS, Faraone SV (1999) Schizophrenia: a review of genetic studies. Harv Rev Psychiatry 7:185–207 [PubMed] [Google Scholar]
- Turecki G, Rouleau GA, Joober R, Mari J, Morgan K (1997) Schizophrenia and chromosome 6p. Am J Med Genet 74:195–198 [PubMed] [Google Scholar]
- Ujike H, Takehisa Y, Takaki M, Tanaka Y, Nakata K, Takeda T, Kodama M, Fujiwara Y, Yamamoto A, Kuroda S (2001) NOTCH4 gene polymorphism and susceptibility to schizophrenia and schizoaffective disorder. Neurosci Lett 301:41–44 [DOI] [PubMed] [Google Scholar]
- Vieland VJ (2001) The replication requirement. Nat Genet 29:244–245 [DOI] [PubMed] [Google Scholar]
- Wang S, Detera-Wadleigh SD, Coon H, Sun C, Goldin LR, Duffy DL, Byerley WF, Gershon ES, Diehl SR (1996) Evidence of linkage disequilibrium between schizophrenia and the SCA1 CAG repeat on chromosome 6p23. Am J Hum Genet 59:731–736 [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun C, Walczak CA, Ziegle JS, Kipps BR, Goldin LR, Diehl SR (1995) Evidence for a susceptibility locus for schizophrenia on chromosome 6pter-6p22. Nat Genet 10:41–46 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Sobel E, O'Connell JR, Lange K (1995) Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet 56:1506–1507 [PMC free article] [PubMed] [Google Scholar]
- Wei J, Hemmings GP (2000) The NOTCH4 locus is associated with susceptibility to schizophrenia. Nat Genet 25:376–377 [DOI] [PubMed] [Google Scholar]
- Weickert TW, Goldberg TE, Gold JM, Bigelow LB, Egan MF, Weinberger DR (2000) Cognitive impairments in patients with schizophrenia displaying preserved and compromised intellect. Arch Gen Psychiatry 57:907–913 [DOI] [PubMed] [Google Scholar]
- Weinberger DR (1999) Cell biology of the hippocampal formation in schizophrenia. Biol Psychiatry 45:395–402 [DOI] [PubMed] [Google Scholar]
- Weiss KM, Terwilliger JD (2000) How many diseases does it take to map a gene with SNPs? Nat Genet 26:151–157 [DOI] [PubMed] [Google Scholar]
- Wright P, Nimgaonkar VL, Donaldson PT, Murray RM (2001) Schizophrenia and HLA: a review. Schizophr Res 47:1–12 [DOI] [PubMed] [Google Scholar]
- Zatz M, Vallada H, Melo MS, Passos-Bueno MR, Vieira AHG, Vainzof M, Gill M, Gentil V (1993) Cosegregation of schizophrenia with Becker muscular dystrophy: susceptibility locus for schizophrenia at Xp21 or an effect of the dystrophin gene in the brain. J Med Genet 30:131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]