Abstract
Cleft lip with or without cleft palate (CL/P) is a common congenital anomaly. Birth prevalences range from 1/500 to 1/1,000 and are consistently higher in Asian populations than in populations of European descent. Therefore, it is of interest to determine whether the CL/P etiological factors in Asian populations differ from those in white populations. A sample of 36 multiplex families were ascertained through probands with CL/P who were from Shanghai. This is the first reported genome-scan study of CL/P in any Asian population. Genotyping of Weber Screening Set 9 (387 short tandem-repeat polymorphisms with average spacing ∼9 cM [range 1–19 cM]) was performed by the Mammalian Genotyping Service of Marshfield Laboratory. Presented here are the results for the 366 autosomal markers. Linkage between each marker and CL/P was assessed by two-point and multipoint LOD scores, as well as with multipoint heterogeneity LOD scores (HLODs) plus model-free identity-by-descent statistics and the multipoint NPL statistic. In addition, association was assessed via the transmission/disequilibrium test. LOD-score and HLOD calculations were performed under a range of models of inheritance of CL/P. The following regions had positive multipoint results (HLOD ⩾1.0 and/or NPL P⩽.05): chromosomes 1 (90–110 cM), 2 (220–250 cM), 3 (130–150 cM), 4 (140–170 cM), 6 (70–100 cM), 18 (110 cM), and 21 (30–50 cM). The most significant multipoint linkage results (HLOD ⩾2.0; α=0.37) were for chromosomes 3q and 4q. Associations with P⩽.05 were found for loci on chromosomes 3, 5–7, 9, 11, 12, 16, 20, and 21. The most significant association result (P=.009) was found with D16S769 (51 cM).
Introduction
Oral-facial clefts, particularly cleft lip with or without cleft palate (CL/P), are common congenital anomalies readily observable at birth. The birth prevalence of CL/P is 1/500–1/1,000, depending on the population. Populations of either American Indian descent or Asian descent have the highest birth prevalence, populations of European descent have intermediate birth prevalence, and populations of African descent have the lowest reported birth prevalence. Given the high birth prevalence of oral-facial clefting in Asians, it is of considerable importance to investigate Asian families, to determine the population-specific etiologic factors leading to this major public-health problem. Interestingly, the first recorded surgical repair of a cleft lip, along with postoperative instructions, was described in the annals of the Chin dynasty in China, circa a.d. 390 (translated by Khoo [1966]). The subsequent political career of the affected individual was also described in detail.
The goal of the current study was to investigate genetic factors in CL/P in China. Familial factors in the etiology of oral-facial clefts have received particular attention since the 18th century. Trew (1757) was the first to report a family with several affected members. Charles Darwin (1883 [for a facsimilie of the revised second edition, see the Electronic Scholarly Publishing Web site]) highlighted a publication of “the transmission during a century of hare-lip with a cleft-palate” by Sproule (1863). Rischbieth (1910) summarized pre-1900 publications of familial cases of cleft lip (“hare-lip”) and cleft palate (see the abridged facsimile of Rischbieth [1910]), and commentary putting Rischbieth’s conclusions into historical perspective have been provided by Melnick [1997]). Rischbieth (1910) and the other members of the Galton Laboratory concluded that the inheritance of cleft lip and cleft palate was an expression of general physical and racial degeneracy, which could be traced to poor protoplasm (Melnick 1997), whereas Bateson (1909) and other proponents of Mendelism included “hare-lip” as one of a group of “dominant hereditary diseases and malformations.”
More recently, many research groups have studied the etiology of CL/P and of cleft palate (CP), with considerable success with regard to syndromic forms and limited success with regard to nonsyndromic forms. It is clear that CL/P and CP can occur as part of Mendelian syndromes, that certain chromosomal anomalies include either CL/P or CP in the phenotype, and that certain teratogens can increase the risk of having an offspring with an oral-facial cleft. However, phenotypes of known etiology comprise only a small proportion of all individuals with an oral-facial cleft (e.g., see Jones 1988; Gorlin et al. 1990; Murray 1995; Schutte and Murray 1999).
Fogh-Andersen (1942) was the first investigator to collect a systematic data set of families with nonsyndromic clefts and to evaluate the observed inheritance patterns. He concluded that the families with CL/P were consistent with segregation of alleles at a single genetic locus with variable penetrance and that families with CP were consistent with autosomal dominant inheritance with greatly reduced penetrance. In contrast to Fogh-Andersen’s hypotheses, the 1960s and 1970s saw the proposal of a specific statistical model, termed the “multifactorial threshold model” (MF/T), to explain the familial patterns of CL/P (e.g., see Carter 1976; Fraser 1976). Under the MF/T model, the occurrence of a cleft depends on a very large number of genes, each of equal, minor, and additive effect, plus environmental factors. However, tests of the MF/T in large populations were inconclusive (e.g., see Bear 1976; Melnick et al. 1980; Mendell et al. 1980; Marazita et al. 1984).
During the late 1970s and early 1980s, investigators began to apply segregation-analysis methods that allow explicit statistical tests of the MF/T versus the major-locus alternative (mixed models). In most published segregation analyses of CL/P under the mixed model, the MF/T could be rejected in favor of either a mixed model (major locus plus multifactorial background; e.g., see Marazita et al. 1984; Chung et al. 1986) or a major locus alone (e.g., see Hecht et al. 1991; Marazita et al. 1992; Nemana et al. 1992). Most such studies were conducted in populations of European descent, although a few were conducted in Asian populations (e.g., see Marazita et al. 1992). These segregation analyses of oral-facial clefts (both CL/P and CP) have consistently resulted in evidence for genes of major effect. Although such studies seem to imply a single major locus, hypotheses of multiple interacting loci or genetic heterogeneity cannot be ruled out and, indeed, have not been explicitly tested in any of the published segregation analyses to date. Analyses of recurrence-risk patterns (Farrall and Holder 1992; Mitchell and Risch 1992; Fitzpatrick and Farrall 1993; Christensen and Mitchell 1996) have been consistent with oligenic models with approximately four to seven interacting loci. With evidence that oral-facial cleft family-history patterns are consistent with involvement of one or a few loci, there are now many groups attempting to identify those genes by using the tools of linkage analysis and association analysis.
Gene-mapping studies of nonsyndromic oral-facial clefts have utilized both linkage and association methods. Candidate loci or regions on six chromosomes (chromosomes 2, 4, 6, 14, 17, and 19) have positive linkage or association results in CL/P, CP, or both (see recent reviews by Wyszynski et al. [1996] and Carinci et al. [2000]), primarily in populations of European descent. There are also a few additional loci and chromosomal regions for which only negative results have been reported in the literature and that therefore are not presented in detail in the present study.
The first published positive association with oral-facial clefts was a population-based association between CL/P and a TaqI restriction-site polymorphism in the transforming growth-factor–α locus (TGFA [Ardinger et al. 1989]). Interestingly, this locus was studied as a candidate because of its involvement in CP in the mouse. The TGFA association with CL/P has since been replicated in several studies, but several other studies have failed to confirm the association (reviewed by Wyszynski et al. [1996] and Carinci et al. [2000]). An association with TGFA has also been reported for CP, although most studies of TGFA and CP have failed to find an association.
In addition to TGFA, loci in several other chromosomal regions have shown positive results with regard to oral-facial clefting, although, like TGFA, none of these loci give consistent results across all studies. Following is a brief summary of such loci; for further details, see reviews by Wyszynski et al. (1996), Carinci et al. (2000), and Marazita (2002). For homeobox 7 (MSX1; chromosome 4p16), there have been reports of positive association, with CL/P and with CP, in individuals of European descent but reports of negative association in Filipinos. Anonymous markers on 4q31 show positive association and linkage with CL/P in individuals of European descent. For markers on chromosome 6p23 (including F13A and several anonymous markers), there have been both positive and negative reports of linkage in individuals of European descent. For transforming growth-factor–β-3 (TGFB3; 14q24), there have been one report of positive association with CP (in individuals of European descent) and two reports of negative association (one in individuals of European descent and the other in Filipinos). For retinoic acid–receptor α (RARA; 17q21), there have been multiple reports of positive associations with CL/P in individuals of European descent, but there have been only negative reports of linkage, as well as one report of negative association, with CP in individuals of European descent. For the proto-oncogene locus BCL3 (19q13), there have been both positive and negative reports of linkage and association with CL/P, as well as negative reports with CP, in individuals of European descent.
Despite the fact that Asians have the highest birth prevalence of oral-facial clefts, the majority of gene-mapping studies of CL/P have been performed in either white Europeans or white Americans. Therefore, we extended our ongoing studies of Chinese families with CL/P, to apply such gene-mapping approaches. First, we assessed linkage and association between nonsyndromic CL/P and each of the candidate regions for which there has been at least one report of positive results for individuals of European descent. As we have reported elsewhere (Marazita et al. 2002), none of those loci had positive parametric linkage results with regard to CL/P; indeed, there was significantly negative linkage evidence (LOD ⩽−2.0) for each marker when linkage homogeneity was assumed. However, with a model-free linkage method relying on identity by descent (IBD) comparisons between affected relatives, there were positive results with a marker at the APOC2 locus on chromosome 19q13. The fact that the model-free method obtained positive linkage results, whereas the parametric LOD score under homogeneity did not, may reflect the uncertainty in the underlying genetic model for CL/P, including issues such as degree of heterogeneity. In addition, significant association (P=.004) was observed for D19S49, another marker in the 19q13 region, which is ∼18 cM from APOC2, but there was no significant evidence for linkage. Evidence for association without evidence for linkage may be a characteristic of many genes predisposing to common, genetically complex disorders (Risch and Merikangas 1996).
Therefore, thus far, our results suggest that none of the candidate regions and loci that appear to play a role in CL/P in individuals of European descent have a major involvement in the Chinese population, with the possible exception of the 19q31 region. Studies of Filipinos also have ruled out the involvement of loci that are major candidates in individuals of European descent (Lidral et al. 1997). Furthermore, previous segregation analyses also have suggested that the genetic patterns are different in Chinese populations than in individuals of European descent (Marazita et al. 1986, 1992).
Given the negative results from our analyses of the candidate regions, the current study is a genome scan, at 10-cM intervals, to simultaneously search for multiple regions involved in nonsyndromic CL/P in the Chinese population. The first genome search, using 92 U.K. sib pairs with CL/P, has recently been published (Prescott et al. 1998, 2000). Initially, 11 loci with suggestive results were identified by a 10-cM mapping panel, and the results for 10 of these loci were confirmed by a 5-cM map (Prescott et al. 2000). There have not yet been any genome scans for isolated CP. The genome scan in this Chinese population will allow a comparison between genome-scan results in families of European descent that have CL/P and those in Asian families that have CL/P.
Subjects and Methods
Subjects
Families were identified as part of our ongoing studies of oral-facial clefts in China (Marazita et al. 1992, 2002; Wentzlaff et al. 1997; Cooper et al. 2000). A population-based sample of ∼2,000 nonsyndromic CL/P probands born in Shanghai formed the basis of all studies. All probands were Chinese in the Han dialect group. For the studies reported here, a subset of families each with two or more affected individuals were identified. These multiplex families had one proband, each of whom had either “CL alone” (CL) or “CL plus CP” (CLP). Affected family members were examined by Dr. Liu and coworkers, to confirm family reports and to rule out syndromic forms of clefting. For the analyses described herein, family members with either CL or CLP were all considered to be affected. Family histories as well as environmental histories were taken, and blood samples were collected for DNA extraction. Samples were obtained after the subjects had signed informed-consent forms approved by institutional review boards in both the United States and China.
Sixty multiplex families were recruited and supplied blood samples, as described by Marazita et al. (2002). Prior to the genome scan, a power study was done to determine the most informative families among the 60 total multiplex families, by the simulation method of Ploughman and Boehnke (1989) (computer program SIMLINK); simulated were flanking markers 10 cM apart and with a CL/P locus midway between them. Table 1 summarizes the results from the power studies of the 36-family subset that ultimately was used for the studies reported here. As can be seen in table 1, the power that this subset has for detection of linkage is substantial, even if heterogeneity is assumed. As further detailed below, in the “Statistical Methods” subsection, the studies presented here are the first step of a two-stage genome-scan study design. At this stage, a LOD-score threshold of 1.0 will be used as positive evidence to move to the second stage of higher-density scans. As shown in table 1, the power that a LOD ⩾1.0 would be achieved in these families was ⩾77%, even under an assumption of heterogeneity in which only 20% of the families were linked to the marker.
Table 1.
Power Estimates for 36 Multiplex Families with CL/P That Are from Shanghai[Note]
| α=1.00 | α=.80 | α=.60 | α=.40 | α=.20 | |
| Mean (SE) maximum multipoint LOD score | 23.63 (.20) | 16.04 (.23) | 10.68 (.23) | 5.56 (.16) | 2.20 (.09) |
| Probabilities: | |||||
| Maximum LOD >1.0 | 1.00 | 1.00 | 1.00 | .980 | .772 |
| Maximum LOD >2.0 | 1.00 | 1.00 | .996 | .944 | .516 |
| Maximum LOD >3.0 | 1.00 | 1.00 | .992 | .836 | .232 |
Note.— Data are based on the following assumptions: CL/P locus midway between flanking markers 10 cM apart, each with five equally frequent alleles, and autosomal recessive inheritance of CL/P with allele frequency 0.05 and 90% penetrance (for segregation analysis of CL/P in this population, see Marazita et al. 1992).
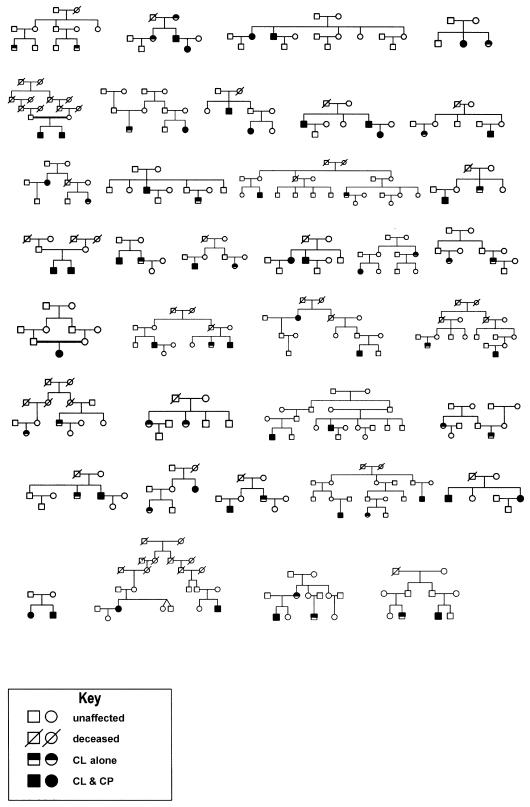
Table 2 and figure 1 summarize the families included in the present analyses. There were 36 multiplex families (466 individuals), a subset comprising the most informative families from the 60 multiplex families from Shanghai that have been reported by Marazita et al. (2002). In one family with two affected members, the proband was the only affected individual available for genotyping (see fig. 1); however, the proband’s parents were consanguineous, so the family was still informative for the linkage and association analyses presented here. The numbers of affected family members ranged from two to five and were of various degrees of relationship to the probands (including siblings, parents, cousins, aunts/uncles, grandparents, and second cousins). The mean family size of the 36 families was 12.94 (SD 5.8), ranging from a minimum of 4 family members to a maximum of 26. Figure 1 shows the family structures. Blood samples were available from 310 (80 affected individuals and 230 unaffected individuals) of the 466 family members. Table 2 summarizes the affection status of the individuals genotyped for this study.
Table 2.
Summary of Affection Status, by Sex, of 310 Genotyped Individuals from 36 Multiplex Families Ascertained in Shanghai
| Affection Status | No. ofMales | No. ofFemales | Total |
| Bilateral CL | 1 | 1 | 2 |
| Bilateral CLP | 13 | 5 | 18 |
| Unilateral CL | 15 | 14 | 29 |
| Unilateral CLP | 20 | 11 | 31 |
| Unaffected | 109 |
121 |
230 |
| Total | 158 | 152 | 310 |
Figure 1.
Pedigree diagrams of 36 families from Shanghai
DNA Analysis
DNA was extracted from blood samples, by a modified version of standard protocols described by Miller et al. (1988). Genotyping was done by the Mammalian Genotyping Service of Marshfield Laboratory, under the direction of Dr. James Weber (see the Center for Medical Genetics, Marshfield Medical Research Foundation Web site). Genotyping within the service utilizes STRPs and automated fluorescence technology. Weber Screening Set 9—a set of 387 high-quality STRPs with average spacing of ∼9 cM (range 1–19 cM)—was used; this screening set was developed at Marshfield over a period of years (Yuan et al. 1997). Lists of the Weber Screening Set 9 markers, with locus, marker, heterozygosity, centimorgan location, allele sizes, marker combinations used for multiplexing, and primer sequences, can be found at the Marshfield Lab Web site (Center for Medical Genetics, Marshfield Medical Research Foundation). The studies reported here are based on the 366 autosomal markers from this screening set.
The inheritance of each marker in all families was analyzed by PedCheck (O’Connell and Weeks 1998) to test for inconsistencies due to nonpaternity or other errors, and repeat genotyping was done as necessary. Because published allele frequencies for Weber Screening Set 9 are based on white families, allele frequencies were calculated in the founders and in the unaffected, unrelated, married-in individuals from the families. A complete list of the markers tested, with the allele sizes and calculated allele frequencies, is available at our Web site (Genetics Research Group, School of Dental Medicine, University of Pittsburgh).
Statistical Methods
Summary
Descriptive statistics were used to summarize the family data. For each genetic marker, parametric LOD scores were calculated under both dominant and recessive models, with gene frequency and penetrance as estimated by segregation analysis (Marazita et al. 1992). Model-free methods of linkage and association analysis also were performed, in particular IBD linkage tests (SimIBD) (Davis et al. 1996), and transmission/disequilibrium tests (TDTs) (Spielman et al. 1993).
Genome-scan strategy
The studies described here are the first stage of a two stage genome-scan study design. In this first stage, we screened a genomewide panel of markers spaced at 10-cM intervals. Any positive results are followed by high-resolution mapping. Under this two-stage strategy, results with either P⩽.05 or LOD scores ⩾1.0 are used to identify regions for the second-stage, high-density scan. Note that this significance level does not conclude that linkage or association exists, only that the areas identified are given high priority for the second stage, of high resolution mapping. This mapping strategy has several advantages: candidate regions can be identified even when the loci that they contain have only a moderate impact on the phenotype; the false-positive rate will be relatively small (∼6–10 false positives), whereas the majority of intervals in which a susceptibility locus is not present are excluded (see Lander and Kruglyak 1995; Hauser et al. 1996; and Kruglyak and Daly 1998).
Parameter estimates
The parametric linkage–analysis approaches required estimates of marker-allele frequencies and also of the genetic-model parameters for CL/P. Because there are no published allele-frequency estimates for the markers in Chinese populations, the frequencies were estimated in the study families, as described in the previous section.
We previously had performed population-based segregation analyses of CL/P in ∼2,000 multigenerational families from Shanghai (Marazita et al. 1992). Each family was ascertained through a surgical proband affected with nonsyndromic CL/P. Mixed-model segregation analysis of CL/P was performed under the unified model (Morton and MacLean 1974; Lalouel and Morton 1981; Lalouel et al. 1983). The best-fitting model was that of an autosomal recessive major locus, with allele frequency 0.05 (Marazita et al. 1992).
Hodge and Elston (1994) have shown that maximization of LOD scores over a range of genetic models—that is, calculation of maximized LOD scores—is a valid procedure that, without adjustment to significance levels and without need to correct for ascertainment, can be used to simultaneously evaluate linkage and determine the most likely genetic model for a data set, provided that there is a linked marker. Furthermore, if an oligogenic model pertains to CL/P or if significant heterogeneity exists, some of the loci may act in a dominant fashion whereas others may act in a recessive fashion. Therefore, we performed all the parametric tests twice, assuming the best-fitting recessive model for CL/P and also assuming the best-fitting dominant model (Marazita et al. 1992). We also calculated the SimIBD linkage statistic, which is independent of the genetic model for CL/P.
Parametric-linkage calculations: LOD scores
We calculated two-point LOD scores in the extended kindreds, using the Elston-Stewart algorithm (Elston and Stewart 1971) and employing the LINKAGE program with recent updates to speed calculations (VITESSE and FASTLINK; Cottingham et al. 1993; Terwilliger and Ott 1994; O'Connell and Weeks 1995).
Multipoint LOD-score calculations
The algorithm developed by Kruglyak et al. (1996), implemented in the computer program GENEHUNTER, was used for the multipoint LOD-score calculations and for multipoint heterogeneity LOD-score (HLOD) calculations. HLODs are based on the admixture-heterogeneity test developed by Smith (1953), in which the recombination fraction (θ) and the proportion of linked families (α) are estimated simultaneously. Recent studies have shown that, although the estimated value of α is problematic when this method is used, the HLOD remains a powerful method for detection of linkage in the presence of heterogeneity (e.g., see Greenberg and Abreu 2001; Hodge et al. 2002).
Model-free linkage tests: two-point (SimIBD)
We used the model-free IBD linkage approach developed by Davis et al. (1996) (SimIBD), which simulates marker genotypes in the affected relatives, conditional on the marker genotypes in the unaffected family members, to determine an empirical null distribution for calculation of P values. SimIBD has been shown to be robust to allele-frequency misspecification.
Model-free linkage tests: multipoint (NPL)
The model-free NPL statistics (Kruglyak et al. 1996) and associated P values were estimated by GENEHUNTER.
Allelic association: TDT method
The TDT, a powerful family-based method for detection of associations between marker and disease loci in the presence of linkage disequilibrium, was introduced by Spielman et al. (1993). Alleles at each marker were tested for association with CL/P (in all parent/affected-child triads derived from the 36 multiplex families), by standard single-allele TDT (Spielman et al. 1993—each allele vs. all others combined) and also by the multiallele TDT developed by Cleves et al. (1997) (programmed in S.A.G.E. 1999).
Results
Parametric-Linkage Results
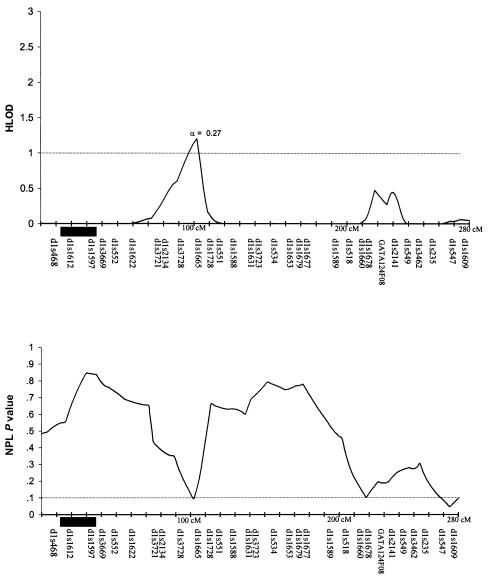
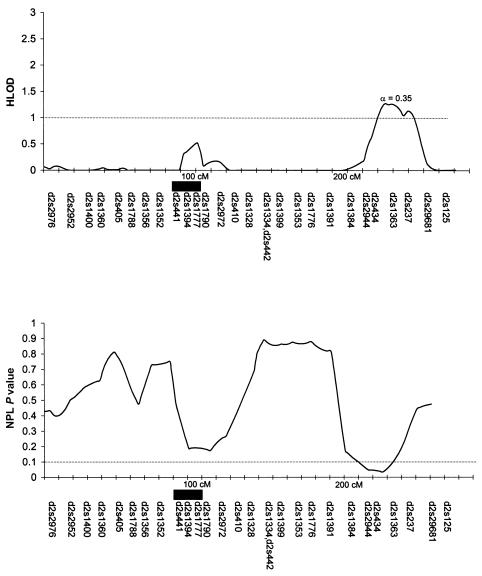
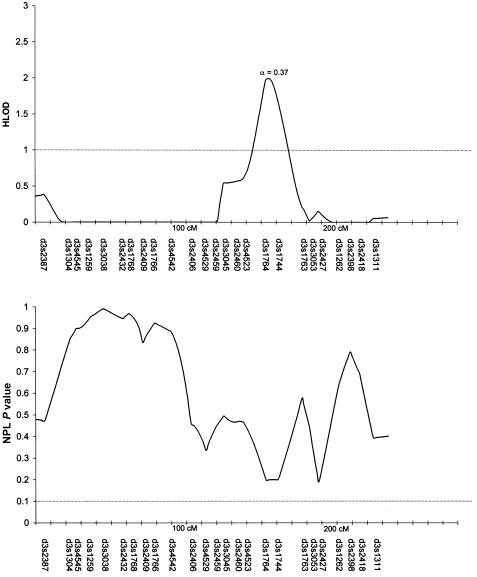
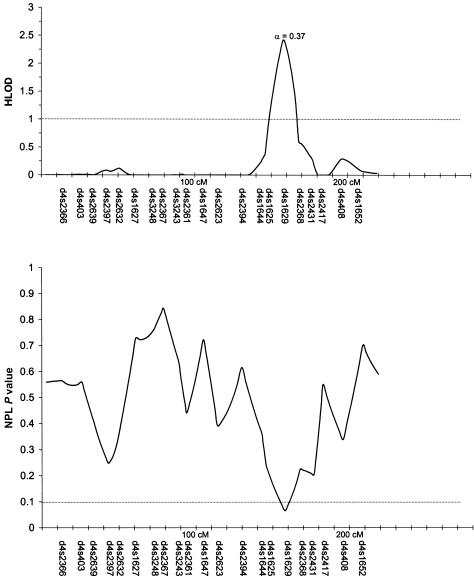
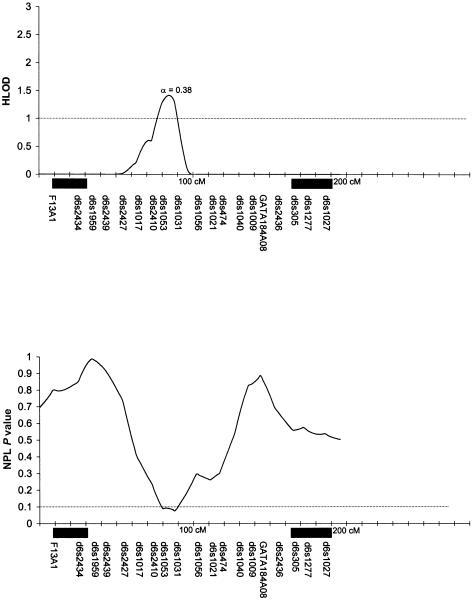
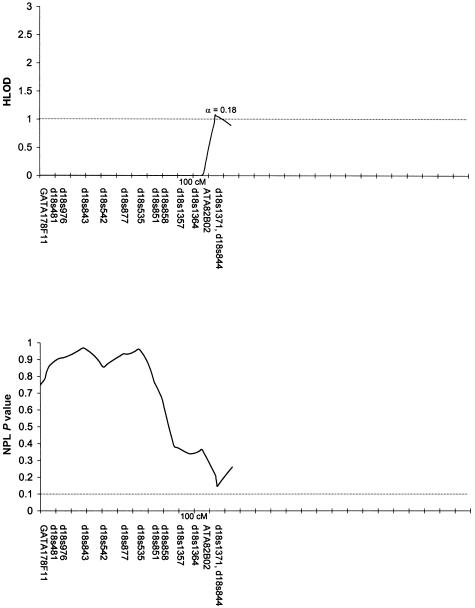
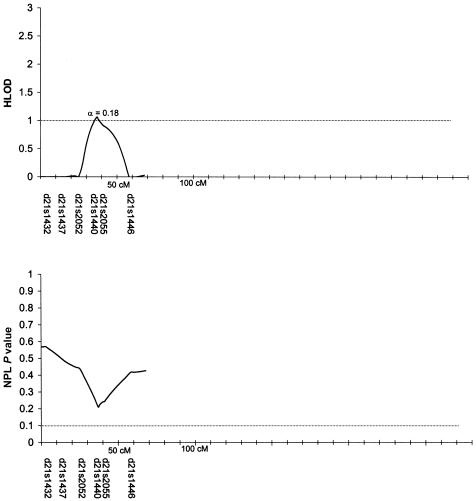
Table 3 summarizes the individual markers for which two-point LOD scores ⩾1.0 were obtained, including markers on chromosomes 1, 2, 6, 9, 10, and 14. The multipoint HLODs were uniformly negative for each chromosome. However, there were several chromosomal regions with positive multipoint HLODs. Figures 2–8 present the multipoint HLODs (with corresponding α) for those chromosomes for which one or more HLODs ⩾1.0 were obtained. Also indicated in figures 2–8 are those regions in which positive results had previously been reported for a sample of white individuals (Prescott et al. 2000). Positive multipoint HLODs were found for regions on chromosomes 1–4, 6, 18, and 21.
Table 3.
Significant Two-Point Linkage and Association Results for CL/P, with Autosomal Markers from Weber Screening Set 9
|
Parametric Linkage |
Model-Free Allelic-Association Test (TDT) |
||||
| Chromosomeand Location (Markera) | MaximumLOD Score (θ) | CLP Model | Model-FreeLinkage Test SimIBD P | χ2 (df) | P |
| 1: | |||||
| 102 cM (D1S1665) | 1.04 (.20) | Dominant | .04 | … | … |
| 2: | |||||
| 210 cM (D2S2944) | 1.45 (.10) | Recessive | … | … | … |
| 227 cM (D2S1363) | 1.91 (.20) | Recessive | .05 | … | … |
| 3: | |||||
| 26 cM (D3S4545) | … | … | … | 23.08 (10) | .010 |
| 188 cM (D3S2427) | … | … | .02 | … | … |
| 4: | |||||
| 93 cM (D4S2361) | … | … | .04 | … | … |
| 5: | |||||
| 19 cM (D5S807) | … | … | … | 14.81 (6) | .022 |
| 117 cM (D5S2501) | … | … | … | 15.66 (7) | .028 |
| 139 cM (D5S816) | … | … | … | 20.00 (7) | .006 |
| 175 cM (D5S1456) | … | … | … | 14.44 (6) | .025 |
| 6: | |||||
| 89 cM (D6S1031) | 1.41 (.20) | Dominant | … | … | … |
| 112 cM (D6S1021) | … | … | … | 13.40 (6) | .037 |
| 7: | |||||
| 7 cM (D7S3056) | … | … | … | 13.80 (7) | .055 |
| 98 cM (D7S820) | … | … | .03 | … | … |
| 114 cM (D7S1799) | … | … | … | 18.62 (10) | .045 |
| 8: | |||||
| 67 cM (D8S1110) | … | … | .01 | … | … |
| 78 cM (D8S1113) | … | … | .03 | … | … |
| 9: | |||||
| 76 cM (D9S1122) | … | … | … | 13.54 (5) | .019 |
| 104 cM (D9S910) | 1.56 (.20) | Recessive | … | … | … |
| 10: | |||||
| 125 cM (D10S1239) | 1.20 (.20) | Dominant | .05 | … | … |
| 135 cM (D10S1237) | 1.00 (.20) | Dominant | .03 | … | … |
| 165 cM (D10S1248) | 1.11 (.20) | Dominant | .05 | … | … |
| 171 cM (D10S212) | 1.07 (.20) | Dominant | … | … | … |
| 11: | |||||
| 43 cM (D11S1392) | … | … | … | 12.23 (5) | .032 |
| 12: | |||||
| 78 cM (D12S1294) | … | … | … | 13.91 (6) | .031 |
| 166 cM (D12S392) | … | … | … | 17.10 (9) | .047 |
| 14: | |||||
| 26 cM (D14S1280) | 1.09 (.20) | Recessive | … | … | … |
| 16: | |||||
| 30 cM (D16S764) | … | … | … | 10.78 (3) | .013 |
| 51 cM (D16S769) | … | … | … | 11.57 (3) | .009 |
| 19: | |||||
| 78 cM (D19S246) | … | … | .007 | … | … |
| 20: | |||||
| 39 cM (D20S470) | … | … | .0004 | 16.65 (9) | .054 |
| 21: | |||||
| 25 cM (D21S2052) | … | … | … | 15.39 (7) | .031 |
Markers shown are those for which LOD scores are ⩾1.0, SimIBD P values are ⩽.05, and/or TDT P values are ⩽.05).
Figure 2.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 1, under the dominant model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented. The black rectangles denote chromosomal regions for which positive linkage results were found, by Prescott et al. (2000), in 92 affected sib pairs from England.
Figure 3.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 2, under the dominant model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented. The black rectangles denote chromosomal regions for which positive linkage results were found, by Prescott et al. (2000), in 92 affected sib pairs from England.
Figure 4.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 3, under the dominant model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented.
Figure 5.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 4, under the recessive model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented.
Figure 6.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 6, under the dominant model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented. The black rectangles denote chromosomal regions for which positive linkage results were found, by Prescott et al. (2000), in 92 affected sib pairs from England.
Figure 7.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 18, under the recessive model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented.
Figure 8.
Results of multipoint analyses of genome-scan markers in 36 multiplex families from Shanghai, for chromosome 21, under the dominant model. Note that each multipoint analysis was performed under both dominant and recessive models of inheritance for CL/P; results under the model with the highest HLOD are presented.
Results of Model-Free Methods
Table 3 also summarizes the individual markers for which positive results (P⩽.05) were obtained under the model-free linkage (SimIBD) and association (TDT) methods. The model-free linkage results were consistent with some of the markers that had positive two-point LOD scores (i.e., markers on chromosome 1, 2, and 10), but there were additional markers that did not have positive LOD scores but that did have positive SimIBD results (markers on chromosomes 3, 4, 7, 8, 19, and 20). The model-free association results (TDT) presented are for the overall TDT (i.e., multiple allele [Cleves et al. 1997]). Interestingly, none of the markers with positive LOD scores also had a positive TDT result, although one marker (i.e., D20S470) with positive SimIBD linkage statistics also had a positive TDT. Positive TDT results were obtained for markers on chromosomes 3, 5–7, 9, 11, 12, 16, 20, and 21. Figures 2–8 present the results of the multipoint NPL statistic calculations. In every case, those regions with HLODs ⩾1.0 also had NPL P values ⩽.05.
Discussion
The most statistically significant multipoint results from this 10-cM genome scan were found for a region between a putative gene controlling risk of CL/P and regions on chromosomes 3 and 4. Note that the only previous CL/P genome scan (Prescott et al. 2000) did not find positive results for either of these chromosomes; therefore, involvement of these regions in CL/P may be unique to the Chinese population.
The 3q chromosomal region between 145 cM and 175 cM had a multipoint HLOD peak of ∼2.0 (α=0.37) with regard to CL/P in these Chinese families, under a dominant-inheritance model for CL/P. Furthermore, there were a positive IBD linkage result with D3S2427 (3q, 188 cM) and a positive-association result with D3S4545 (3p, 26 cM). There have been no previously published reports of either positive linkage or positive association between either CL/P or CP and markers on chromosome 3. There are no obvious clefting candidates on 3q, but, on 3p, there are possible candidates, including signaling molecules WNT5A and WNT7A and transcription factors RARB2, RARB4, and AP-2.
The highest multipoint HLOD peak (2.5; α=0.37) in the present study was for the 4q region near 160 cM, under a recessive-inheritance model for CL/P. Furthermore, there was a positive IBD linkage result with D4S2361 (93 cM). Anonymous markers in 4q31 (located at ∼115–130 cM) showed positive linkage and association with CL/P in white and Chilean populations (Mitchell et al. 1995). As reviewed in the “Introduction” section, there have also been positive reports for homeobox 7, MSX1 (4p16.3-16.1, 10.4 cM), although this marker did not have positive linkage or association results with CL/P in these Chinese families (Marazita et al. 2002). Other possible clefting candidates on chromosome 4q include transcription factors PITX2 and LHX7/LHX8 and growth factors/growth-factor receptors BMPR1B and FGF2.
As can be seen in figure 2, 3, and 6, chromosomes 1, 2, and 6 had positive multipoint HLOD results in both the current Chinese study and in the English genome scan (Prescott et al. 2000). However, the peak positive results were for different locations on the chromosomes. The Prescott et al. (2000) genome scan found a positive region on chromosome 1, at ∼15–25 cM; in the present study, the positive region peaked at 100 cM. The present study also obtained positive results (two-point LOD score and positive IBD linkage statistic) with D1S1665 (102 cM). Prescott et al. (2000) found positive results for chromosome 2, at 85–105 cM, whereas the present study found a peak at ∼225 cM. The present study also obtained positive results (two-point LOD scores and positive IBD linkage statistics) with D2S2944 (210 cM) and D2S163 (227 cM). As reviewed in the “Introduction” section, there have been multiple positive-association results reported for TGFA (on chromosome 2p, 71 cM), although no positive results for TGFA were found in these Chinese families (Marazita et al. 2002). The Prescott et al. (2000) genome scan found positive results for two regions on chromosome 6, at each end of the chromosome. There have also been several reports of positive results for the 6p23 region, near the p-terminus (e.g., see the review by Carinci et al. [2000]). In the present study, there was a peak HLOD (1.5; α=0.38) at ∼80 cM—that is, within the region for which Prescott et al. (2000) found positive results.
Although, for chromosomes 1, 2 and 6, the peak LOD scores found by the present study differ from those found by the Prescott et al. (2000) study, the fact that both studies found positive results for these chromosomes reinforces the possibility that CL/P susceptibility loci are present on those chromosomes. The study by Roberts et al. (1999) supports this interpretation. Roberts et al. (1999) addressed the general issue of location-estimate variation between studies that attempt to replicate gene-mapping results of complex traits. To do so, they simulated sets of two-affected-child nuclear families, with a complex trait for which the marker and disease loci were linked to each other on a simulated chromosome. The trait could be due to either of two disease loci, one of which is linked to the marker loci on the simulated chromosome and the other of which is not. To assess the precision of the location estimate for the linked disease locus, they analyzed each set of simulated families by GENEHUNTER, to obtain multipoint LOD scores every 2 cM along the simulated chromosome. Between the data sets, there was a large amount of variation in the estimate of the peak multipoint LOD scores, suggesting that, if the linkage signal for a susceptibility locus is weak because of, for example, incomplete penetrance or heterogeneity, then the location estimates may be many centimorgans from the true disease locus (Roberts et al. 1999).
In addition to the linkage results reported above, statistically significant associations (P⩽.05) were found for loci on chromosomes 3 (D3S4545, 26 cM), 5 (D5S807, 19 cM; D5S2501, 117 cM; D5S816, 139 cM; and D5S1456, 175 cM), 6 (D6S1021, 112 cM), 7 (D7S3056, 7 cM; and D7S1799, 114 cM), 9 (D9S1122, 76 cM), 11 (D11S1392, 43 cM), 12 (D12S1294, 78 cM; and D12S392, 166 cM), 16 (D16S769, 30 cM; and D16S769, 51 cM), 20 (D20S470, 39 cM), and 21 (D21S2052, 25 cM). The most significant association result (P=.009) was found for chromosome 16 (D16S769, 51 cM). A possible candidate, the transcription factor SOX 8, is near this marker on chromosome 16p (16p13.3, 19 cM). There were also two notable model-free IBD linkage results (P<.01)—D19S246 (78 cM), a marker near the only candidate marker for which positive association had been found in a previous study of these families (Marazita et al. 2002), and D20S470 (39 cM), a marker for which a positive-association result was also found in the present study. Endothelin 3 (52 cM) and BMP7 (53 cM) are possible candidates near the chromosome 20 marker. It is notable that, with the exception of the chromosome 19 region, none of the above-listed regions with positive association or IBD linkage results had previously been reported as either linked or associated with CL/P.
In summary, several regions of the genome had positive linkage and/or association with CL/P that warrant follow-up investigations, including high-resolution mapping and a search for candidate loci in these regions. On the basis of the parametric-linkage results, there appears to be heterogeneity in each of the regions for which positive results have been found, with α ranging from 0.28 to 0.37. Future studies will also focus on delineation of this heterogeneity and will also investigate gene-gene interactions. For most of the regions with positive HLODs, there were also positive results under the model-free linkage or model-free association methods, further implicating these regions in the etiology of CL/P.
Acknowledgments
We thank the families and family members who participated in these studies. We also thank Jessica Sank, for her assistance with data checking, and Roberta Giles, for assistance with database issues and pedigrees. This study was supported by National Institutes of Health grant DE09886. Some of the results in this article were obtained by the program package S.A.G.E., supported by U.S. Public Health Service Resource Grant 1-P41-RR03655 from the National Center for Research Resources. Genotyping was provided by the Mammalian Genotyping Service of Marshfield Lab, funded by the National Heart, Lung and Blood Institute.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for Weber Screening Set 9)
- Electronic Scholarly Publishing, http://www.esp.org/books/darwin/variation/facsimile/ (for facsimile of revised second edition of Darwin's The Variation of Animals and Plants under Domestication)
- Genetics Research Group, School of Dental Medicine, University of Pittsburgh, http://www.sdmgenetics.pitt.edu (for markers and allele sizes and frequencies)
References
- Ardinger HH, Buetow KH, Bell GI, Bardach J, VanDemark DR, Murray JC (1989) Association of genetic variation of the transforming growth factor-alpha gene with cleft lip and palate. Am J Hum Genet 45:348–353 [PMC free article] [PubMed] [Google Scholar]
- Bateson W (1909) Mendel's principles of hereditary. Cambridge University Press, Cambridge [Google Scholar]
- Bear JC (1976) A genetic study of facial clefting in northern England. Clin Genet 9:277–284 [DOI] [PubMed] [Google Scholar]
- Carinci F, Pezzetti F, Scapoli L, Martinelli M, Carinci P, Tognon M (2000) Genetics of nonsyndromic cleft lip and palate: a review of international studies and data regarding the Italian population. Cleft Palate Craniofac J 37:33–40 [DOI] [PubMed] [Google Scholar]
- Carter CO (1976) Genetics of common single malformations. Br Med Bull 32:21–26 [DOI] [PubMed] [Google Scholar]
- Christensen K, Mitchell LE (1996) Familial recurrence-pattern analysis of nonsyndromic isolated cleft palate—a Danish registry study. Am J Hum Genet 58:182–190 [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Bixler D, Watanabe T, Koguchi H, Fogh-Andersen P (1986) Segregation analysis of cleft lip with or without cleft palate: a comparison of Danish and Japanese data. Am J Hum Genet 39:603–611 [PMC free article] [PubMed] [Google Scholar]
- Cleves MA, Olson JM, Jacobs KB (1997) Exact transmission-disequilibrium tests with multiallelic markers. Genet Epidemiol 14:337–347 [DOI] [PubMed] [Google Scholar]
- Cooper ME, Stone RA, Liu Y-e, Hu D-n, Melnick M, Marazita ML (2000) Descriptive epidemiology of nonsyndromic cleft lip with or without cleft palate in Shanghai, China, 1980–1989. Cleft Palate Craniofac J 37:274–280 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Darwin C (1883) Inheritance. In: The variation of animals and plants under domestication, 2d ed, rev. Appleton & Co, New York, pp 445–473 [Google Scholar]
- Davis S, Schroeder M, Goldin LR, Weeks DE (1996) Nonparametric simulation-based statistics for detecting linkage in general pedigrees. Am J Hum Genet 58:867–880 [PMC free article] [PubMed] [Google Scholar]
- Elston RC, Stewart J (1971) A general model for the genetic analysis of pedigree data. Hum Hered 21:523–542 [DOI] [PubMed] [Google Scholar]
- Farrall M, Holder S (1992) Familial recurrence-pattern analysis of cleft lip with or without cleft palate. Am J Hum Genet 50:270–277 [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick D, Farrall M (1993) An estimation of the number of susceptibility loci for isolated cleft palate. J Craniofac Genet Dev Biol 13:230–235 [PubMed] [Google Scholar]
- Fogh-Andersen P (1942) Inheritance of harelip and cleft palate. Munksgaard, Copenhagen [Google Scholar]
- Fraser FC (1976) The multifactorial threshold concept-uses and misuses. Teratology 14:267–280 [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM Jr, Levin LS (1990) Syndromes of the head and neck, 3d ed. Oxford Monographs on Medical Genetics 19. Oxford University Press, New York [Google Scholar]
- Greenberg DA, Abreu P (2001) Determining trait locus position from multipoint analysis: accuracy and power of three different statistics. Genet Epidemiol 21:299–314 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M, Guo S-W, Risch N (1996) Affected-sib-pair interval mapping and exclusion for complex genetic traits: sampling considerations. Genet Epidemiol 13:117–137 [DOI] [PubMed] [Google Scholar]
- Hecht JT, Yang P, Michels VV, Buetow KH (1991) Complex segregation analysis of nonsyndromic cleft lip and palate. Am J Hum Genet 49:674–681 [PMC free article] [PubMed] [Google Scholar]
- Hodge SE, Elston RC (1994) Lods, wrods, and mods: the interpretation of lod scores calculated under different models. Genet Epidemiol 11:329–342 [DOI] [PubMed] [Google Scholar]
- Hodge SE, Vieland VJ, Greenberg DA (2002) HLODs remain powerful tools for detection of linkage in the presence of genetic heterogeneity. Am J Hum Genet 70:556–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MC (1988) Etiology of facial clefts: prospective evaluation of 428 patients. Cleft Palate J 25:16–20 [PubMed] [Google Scholar]
- Khoo B-c (1966) An ancient Chinese text on a cleft lip. Plast Reconstr Surg 38:89–91 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ (1998) Linkage thresholds for two-stage genome scans. Am J Hum Genet 62:994–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lalouel JM, Morton NE (1981) Complex segregation analysis with pointers. Hum Hered 31:312–321 [DOI] [PubMed] [Google Scholar]
- Lalouel JM, Rao DC, Morton NE, Elston RC (1983) A unified model for complex segregation analysis. Am J Hum Genet 35:816–826 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lidral AC, Murray JC, Buetow KH, Basart AM, Schearer H, Shiang R, Naval A, Layda E, Magee K, Magee W (1997) Studies of the candidate genes TGFB2, MSX1, TGFA, and TGFB3 in the etiology of cleft lip and palate in the Philippines. Cleft Palate Craniofac J 34:1–6 [DOI] [PubMed] [Google Scholar]
- Marazita ML (2002) Genetic etiologies of facial clefting. In: Mooney MP, Siegel MI (eds) Understanding craniofacial anomalies: the etiopathogenesis of craniosynostoses and facial clefting. John Wiley & Sons, New York, pp 147–161 [Google Scholar]
- Marazita ML, Field LL, Cooper ME, Tobais R, Maher BS, Peanchitlertkajorn S, Liu Y-e (2002) Non-syndromic cleft lip with or without cleft palate in China: assessment of candidate regions. Cleft Palate Craniofac J 39:149–156 [DOI] [PubMed] [Google Scholar]
- Marazita ML, Hu DN, Spence MA, Liu YE, Melnick M (1992) Cleft lip with or without cleft palate in Shanghai, China: evidence for an autosomal major locus. Am J Hum Genet 51:648–653 [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Spence MA, Melnick M (1984) Genetic analysis of cleft lip with or without cleft palate in Danish kindreds. Am J Med Genet 19:9–18 [DOI] [PubMed] [Google Scholar]
- ——— (1986) Major gene determination of liability to cleft lip with or without cleft palate: a multi-racial view. J Craniofac Genet Dev Biol Suppl 2:89–97 [PubMed] [Google Scholar]
- Melnick M (1997) Cleft lip and palate etiology and its meaning in early 20th century England: Galton/Pearson vs. Bateson; polygenically poor protoplasm vs. Mendelism. J Craniofac Genet Dev Biol 17:65–79 [PubMed] [Google Scholar]
- Melnick M, Bixler D, Fogh-Andersen P, Conneally PM (1980) Cleft lip ± cleft palate: an overview of the literature and an analysis of Danish cases born between 1941 and 1968. Am J Med Genet 6:83–97 [DOI] [PubMed] [Google Scholar]
- Mendell NR, Spence MA, Gladstien K, Brunette J, Stevens A, Clifford E, Georgiade N, Pickrell KL, Serafin D, Quinn GW (1980) Multifactorial/threshold models and their application to cleft lip and cleft palate. In: Melnick M, Bixler D, Shields ED (eds) Etiology of cleft lip and cleft palate. Alan R Liss, New York, pp 387–406 [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE, Healey SC, Chenevix-Trench G (1995) Evidence for an association between nonsyndromic cleft lip with or without cleft palate and a gene located on the long arm of chromosome 4. Am J Hum Genet 57:1130–1136 [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE, Risch N (1992) Mode of inheritance of nonsyndromic cleft lip with or without cleft palate: a reanalysis. Am J Hum Genet 51:323–332 [PMC free article] [PubMed] [Google Scholar]
- Morton NE, MacLean CJ (1974) Analysis of family resemblance. III. Complex segregation of quantitative traits. Am J Hum Genet 26:489–503 [PMC free article] [PubMed] [Google Scholar]
- Murray JC (1995) Face facts: genes, environment, and clefts. Am J Hum Genet 57:227–232 [PMC free article] [PubMed] [Google Scholar]
- Nemana LJ, Marazita ML, Melnick M (1992) Genetic analysis of cleft lip with or without cleft palate in Madras, India. Am J Med Genet 42:5–9 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- ——— (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploughman LM, Boehnke M (1989) Estimating the power of a proposed linkage study for a complex genetic trait. Am J Hum Genet 44:543–551 [PMC free article] [PubMed] [Google Scholar]
- Prescott N, Lees M, Malcolm S. Winter R (1998) Non-syndromic cleft lip and palate: a genome-wide sib-pair analysis. Am J Hum Genet Suppl 63:A1764 [Google Scholar]
- Prescott NJ, Lees MM, Winter RM, Malcolm S (2000) Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib pairs. Hum Genet 106:345–50 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Rischbieth H (1910) Hare-lip and cleft palate. Part 4 in: Pearson K (ed) Treasury of human inheritance.. Dulau, London, pp 79–123 [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAGE (1999) Statistical analysis for genetic epidemiology, release 4.0. Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland [Google Scholar]
- Schutte BC, Murray JC (1999) The many faces and factors of orofacial clefts. Hum Mol Genet 8:1853–1859 [DOI] [PubMed] [Google Scholar]
- Smith CAB (1963) Testing for heterogeneity of recombination fraction values in human genetics. Ann Hum Genet 27:175–182 [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Sproule J (1863) Hereditary nature of hare-lip. Br Med J 1:412 [Google Scholar]
- Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. Johns Hopkins University Press, Baltimore [Google Scholar]
- Trew CJ (1757) Sistens plura exempla palati deficientis. Nova Acta Phys Med Acad Caesarae Leopoldion-Carolinae 1:445–447 [Google Scholar]
- Wentzlaff KA, Cooper ME, Yang P, Aston CP, Liu Y-e, Melnick M, Marazita ML (1997) Association between non-right-handedness and cleft lip with or without cleft palate in a Chinese population. J Craniofacial Genet Dev Biol 17:141–147 [PubMed] [Google Scholar]
- Wyszynski DF, Beaty TH, Maestri NE (1996) Genetics of nonsyndromic oral clefts revisited. Cleft Palate Craniofac J 33:406–417 [DOI] [PubMed] [Google Scholar]
- Yuan B, Vaske D, Weber JL, Beck J, Sheffield VC (1997) Improved set of short-tandem-repeat polymorphisms for screening the human genome. Am J Hum Genet 60:459–460 [PMC free article] [PubMed] [Google Scholar]