Abstract
Duplications and deletions are known to cause a number of genetic disorders, yet technical difficulties and financial considerations mean that screening for these mutations, especially duplications, is often not performed. We have adapted multiplex amplifiable probe hybridization (MAPH) for the screening of the DMD gene, mutations in which cause Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy. MAPH involves the quantitative recovery of specifically designed probes following hybridization to immobilized genomic DNA. We have engineered probes for each of the 79 exons of the DMD gene, and we analyzed them by using a 96-capillary sequencer. We screened 24 control individuals, 102 patients, and 23 potential carriers and detected a large number of novel rearrangements, especially small, one- and two-exon duplications. A duplication of exon 2 alone was the most frequently occurring mutation identified. Our analysis indicates that duplications occur in 6% of patients with DMD. The MAPH technique as modified here is simple, quick, and accurate; furthermore, it is based on existing technology (i.e., hybridization, PCR, and electrophoresis) and should not require new equipment. Together, these features should allow easy implementation in routine diagnostic laboratories. Furthermore, the methodology should be applicable to any genetic disease, it should be easily expandable to cover >200 probes, and its characteristics should facilitate high-throughput screening.
Introduction
Most techniques currently applied to reveal disease-causing mutations are PCR based and do not readily produce quantitative data. Consequently, although copy-number changes (i.e., deletions and duplications) are frequently involved, they will go undetected unless specific techniques are applied (Petrij-Bosch et al. 1997; Wijnen et al. 1998; Morgan et al. 1999). The major reason behind this failure is economical: obtaining quantitative data is feasible but is technically demanding, labor intensive, and, thus, costly. When specific precautions are taken, Southern blotting and quantitative PCR are able to detect deletions and/or duplications, but they are both laborious and difficult to implement on a routine basis.
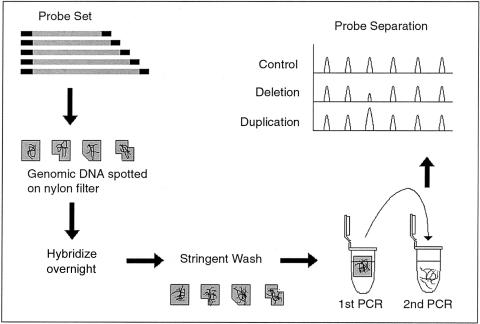
A technique that might fill this gap has recently been described—namely, multiplex amplifiable probe hybridization (MAPH) (fig. 1) (Armour et al. 2000). MAPH is based on the quantitative recovery of probes, after their hybridization to immobilized DNA. Each probe is engineered to allow simultaneous amplification with only one set of primers. This overcomes one of the most difficult elements of quantitative multiplex PCR—namely, differences, between primers, in annealing efficiency. In the original MAPH protocol, after the hybridization step, the PCR products were radioactively labeled during PCR amplification and were analyzed on a polyacrylamide gel. To facilitate a more-automated, higher-throughput application of MAPH, we have chosen to label the products fluorescently and to separate them on a 96-capillary sequencer. Each product is identifiable on the basis of length, with the size of the resultant peak being directly proportional to the copy number of the relevant probe. Changes in peak heights will therefore reflect deletions and duplications in genomic DNA.
Figure 1.
Outline of the MAPH technique. Probes are prepared such that all can be amplified with one primer pair. After overnight hybridization to immobilized genomic DNA, unbound probes are removed by stringent washing. Bound probes are then released and amplified in a quantitative manner. By fluorescent labeling and capillary electrophoresis, it is possible to both discriminate and quantify each probe. Changes in peak heights correspond to copy-number changes (i.e., deletions and duplications).
We have applied the modified MAPH protocol to scan for copy-number changes in patients with Duchenne muscular dystrophy (DMD) (MIM #310200). DMD is the most commonly inherited neuromuscular disease, affecting 1 in 3,500 male individuals (Worton and Thompson 1988). It is an X-linked disorder that is caused by mutations in the DMD gene. This gene is the largest known, covering 2.4 Mb (den Dunnen et al. 1989; Boyce et al. 1991) and containing 79 exons that encode a 14-kb mRNA (Koenig et al. 1987). Translation-truncating mutations in DMD lead to the lethal phenotype of DMD, whereas mutations that retain the reading frame generally cause the less severe phenotype of Becker muscular dystrophy (BMD) (Monaco et al. 1988; Koenig et al. 1989). An accurate molecular diagnosis is therefore essential both to confirm the clinical diagnosis and to distinguish the two allelic forms.
In approximately two-thirds of cases, the mutation is a deletion or duplication of one or more of the exons, clustered in two hotspot regions (Forrest et al. 1987; Koenig et al. 1987; Darras et al. 1988; den Dunnen et al. 1989; Gillard et al. 1989). In affected male patients, deletion detection is relatively simple. For multiplex PCR, two nine-exon sets (the Chamberlain et al. [1988] and Beggs et al. [1990] sets) have been designed around these hotspots, which together detect 90%–95% of the deletions in male patients. Alternative methods must be applied to determine the exact boundaries of the deletion, as well as to detect duplications and carrier status in female individuals. The size (2.4 Mb) and complexity (79 exons) of DMD, however, make this a daunting task. Quantitative Southern blotting has been the most commonly used technique (den Dunnen et al. 1989; Hu et al. 1990; Yamagishi et al. 1996). By the comparison of band intensities between test samples and control samples, it is possible to detect copy-number changes in the individual exons. The preparation of high-quality blots is technically demanding, and six to eight hybridizations are required in order to scan all the exons. In addition, duplication detection is difficult, especially in female carriers and when the duplications are small (i.e., covering only one or two exons). Quantitative PCR is another, more recently applied technique (Ioannou et al. 1992; Mansfield et al. 1993; Yau et al. 1996), in which a multiplex PCR is performed that has a limited number of cycles, ensuring that quantitative products are yielded. Again, the technique is technically demanding, and the incomplete coverage of the exons means that mutations outside the hotspots will be missed. Not surprisingly, therefore, mutation-analysis reports differ considerably in the frequency of duplications detected, ranging between 0 and 6%, depending on the techniques applied for analysis (Koenig et al. 1987; den Dunnen et al. 1989; Hu et al. 1990; Mendell et al. 2001).
Here we describe a MAPH-based method that scans all 79 DMD exons for deletions and duplications. We have been able to detect and define a large series of new mutations—in particular, duplications—with several not detected by Southern blotting and/or quantitative PCR. The simplicity of this technique should allow its easy implementation in diagnostic laboratories, and its utility means that it can be readily adapted for the screening of duplications and deletions in any genetic disease.
Methods
Probe Generation
All probes were based on individual exons. Some DMD probes were created using primers from the Chamberlain et al. (1988) and Beggs et al. (1990) kits. The remainder were based on sequences obtained from the Leiden Muscular Dystrophy Pages. To facilitate analysis, we prepared control probes by using genomic sequence obtained from GenBank. For each product, the presence of duplicated and/or repetitive sequences was excluded using the BLAST program. The sequences were checked against the nr (nonredundant) and htgs (high-throughput genomic sequences) databases. No probe showed an intraspecies homology >90% for a stretch of ⩾30 nt (expected value >e−11).
Products were amplified from genomic DNA by PCR and were cloned into the pGEM-T easy vector (Promega). The correct insert was confirmed by sequencing through use of the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). This was performed at the Leiden Genome Technology Center, where reactions were analyzed on the ABI 3700 Sequencer (Applied Biosystems).
Each probe was amplified from the vector by use of the primers MAPH-F1 (GGCCGCGGGAATTCGATT) and MAPH-R1 (GCCGCGAATTCACTAGTG). Products were purified with the Qiagen PCR cleanup kit (Qiagen) and were then added to the appropriate probe mixture, which had a final concentration of 100–500 pg/μl. Probe sets A and B were prepared containing 40 and 39 DMD exons, respectively. Nine-probe control mixtures were made specifically for use with each probe set.
MAPH
MAPH was performed using a protocol adapted from the original MAPH protocol (Armour et al. 2000), as follows (for detailed protocol, see the Leiden Muscular Dystrophy Pages). At least 1 μg genomic DNA was denatured in 1 μl 1N NaOH and spotted on a small nylon filter, followed by UV cross-linking. Up to 16 filters were hybridized together in one tube; the filters were prehybridized in 1 ml prehybridization solution (0.5 M sodium phosphate [pH 7.2], 1 mM EDTA, 7% SDS, and 100 ng/μl Herring Sperm DNA [Gibco BRL]) for ∼2 h at 60°C; this solution was replaced with 200 μl prehybridization solution that contained 2 μg denatured Cot1 DNA (Gibco BRL) and was incubated at 60°C for 30 min. Probe mixture (1 μl combined probes, 1 μl Cot1 DNA [1 μg/μl], 1 μl Herring Sperm DNA [10 μg/μl; Gibco BRL], 1 μl blocking mixture [blocking primers that each had a final concentration of 20 μM], and 3 μl H2O) was denatured by the addition of 2 μl 1N NaOH and incubated at 37°C for 1 min. After cooling on ice, 3 μl 1M NaH2PO4 was added, and the mixture was added to the tube that contained the filters. Hybridization was performed overnight at 60°C. Washing was performed the next day with five times in 25 ml salt-sodium citrate (SSC) and 1% SDS, followed by five washes in 25 ml 0.1 × SSC and 0.1% SDS, all at 60°C. Each filter was transferred to a PCR tube, and a five-cycle PCR amplification was performed under the following conditions: 94°C for 5 min; five cycles of 94°C for 45 s, 57°C for 1 min, and 68°C for 1 min; and 68°C for 10 min.
Two and one-half microliters of this mixture was transferred to a second PCR, which was performed under the same conditions as the first reaction except that one of the primers was fluorescently labeled and the reaction was for 23 cycles. Two microliters of this product was added to 10 μl (Hi Di) Formamide (Applied Biosystems) and 0.15 μl ROX-500 size standard (Applied Biosystems) in a 96-well plate. This was heated at 95°C for 5 min, followed by immediate cooling on ice. The samples were analyzed on an ABI 3700 capillary sequencer (Applied Biosystems).
Data Analysis
Data were analyzed using the programs Gene Scan (Applied Biosystems) and Excel (Microsoft). Peaks were considered unreliable if they were outside predefined thresholds (upper and lower limits of 12,000 and 150 units, respectively).
Male samples were initially visually assessed, to detect any deletions. Presence of a peak that corresponds to a Y chromosome–specific probe confirmed the sex of the sample. Absence of one or more DMD peaks was taken to be a deletion, and no calculations were performed.
Samples were analyzed using a combination of methods described for analysis of MAPH (Armour et al. 2000; Sismani et al. 2001) and array-based comparative genomic hybridization (Hodgson et al. 2001) experiments. In the original MAPH publication (Armour et al. 2000), each peak was compared with the two nearest peaks, for normalization. Since each DMD exon could potentially be altered in copy number, we also added probes from exons of autosomal genes unrelated to DMD. For each dystrophin exon, the peak height was divided by the sum of the peak heights of the two nearest unlinked probes, to give a ratio. Within one hybridization, the median of the ratio for each exon was calculated and was used as a reference value against which all exons were compared. Each exon was divided by this number, thereby normalizing all unaffected exons to 1.0. For each sample, initial estimates for deletions or duplications were performed visually, by setting arbitrary thresholds on the basis of expected ratios (Hodgson et al. 2001). Wild-type exons were expected to fall in the ranges of 1.0±0.5 for male patients and 1.0±0.25 for female carriers. The median and SD of the exons that fell within this range were calculated, and each exon was divided by the median to correct for variations between samples. Any exon that was outside 3 SDs of the “normal” exons was assumed to be altered in copy number. Samples that showed an SD >15% over the unaffected exons or that appeared to show noncontiguous deletions or duplications were deemed to be unreliable.
Results
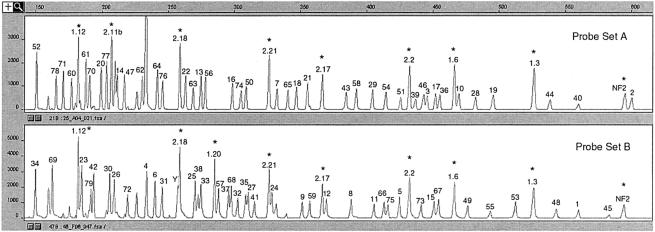
Probes were initially tested by hybridization to control DNA from 24 healthy individuals, as well as to DNA from a patient with a deletion encompassing the entire DMD gene. From the control samples, all the probes could be recovered (fig. 2), whereas only control probes from outside DMD were recovered from the patient sample (data not shown). Thus, none of the probes hybridize to other regions in the genome, which would lead to false-positive signals.
Figure 2.
Example of trace patterns obtained from an unaffected male individual. The numbers refer to DMD exon numbers: “1.x” and “2.x” (where x is the exon number) refer to BRCA1 and BRCA2, respectively, and “NF2” denotes a probe homologous to the first exon of the NF2 gene. Asterisks (*) indicate control peaks, and unlabeled peaks indicate noise. Probes range in size from 151 bp (DMD exon 34) to 602 bp (DMD exon 2).
A total of 125 samples were screened in a semiblind manner (table 1table 1 and figs. 3 and 4). These were a mixture of fully and partially characterized cases, as well as samples from cases in which no mutation had been found. In several cases, the DNA was from a potential carrier in whom the mutation sought was already known. With a threshold of 3 SDs and the assumption that the unaffected ratios are normally distributed, a false-positive result should only occur ∼0.3% of the time, which is the equivalent of one exon per four DMD genes tested. This is approximately the ratio of false-positive results seen among samples that were not excluded for other reasons (e.g., peak height being outside the boundaries or SD being >15%). Therefore, all samples that showed a single-exon rearrangement were tested at least twice. Following these criteria, we found no sample that showed evidence of more than one mutational event.
Table 1.
Samples Screened
|
Mutation Foundb by |
|||
| Sample (Sex) | Method(s)a | OtherMethods | MAPH |
| D1 (M) | 1,3 | Del 5-7 | Del 5-7 |
| D2 (F) | Dup 52-55 | ||
| D3 (M) | 3 | Del 50 | Del 50 |
| D4 (F) | Dup 50-55 | ||
| D5 (M) | 1,3 | nm | Dup 50-55 |
| D6 (M) | 3 | Del 8-44 | Del 8-44 |
| D7 (M) | 1,3 | nm | nm |
| D8 (M) | 1,3 | Dup 43 | Dup 43 |
| D9 (F) | Dup 58-63 | ||
| D10 (M) | 1,3 | ?c | Dup 58-63 |
| D11 (M) | 3 | Del 49-52 | Del 49-52 |
| D12 (M) | 3 | Del 48-50 | Del 48-50 |
| D13 (F) | Del 45 | ||
| D14 (M) | 3 | Del 1m | Del 1m |
| D15 (M) | 3 | nm | Del 53 |
| D16 (M) | 1,3 | nm | nm |
| D17 (M) | 3 | Del (3)-(10) | Del 4-12 |
| D18 (M) | 3 | Del 14-60 | Del 14-60 |
| D19 (M) | 1,3 | Del 69 | Del 64-67 |
| D20 (F) | 3 | Del (45)-(50) | Del 49-54 |
| D21 (M) | 3 | Del 2-(33) | Del 2-30 |
| D22 (M) | 3 | nm | nm |
| D23 (M) | 3 | Dup 12 | Dup 12 |
| D24 (F) | Dup 2 | ||
| D25 (M) | 2,3 | nm | Dup 2 |
| D26 (M) | 1,3 | nm | nm |
| D27 (F) | Dup 12-13 | ||
| D28 (F) | Dup 44 | ||
| D29 (M) | 3 | Dup 2-9 | Dup 2-9 |
| D30 (F) | nm | ||
| D31 (M) | 3 | Del 1-79 | Del 1-79 |
| D32 (F) | 3 | Dup 44-57 | Dup 44-57 |
| D33 (F) | Dup 2-7 | ||
| D34 (M) | 1,3 | Dup 2-7 | Dup 2-7 |
| D35 (M) | 1,3 | Dup 2-7 | Dup 2-7 |
| D36 (F) | 3 | Del 10-(?) | Del 10-46 |
| D37 (M) | 3 | Del 3-(?) | Del 3-19 |
| D38 (F) | 3 | Del (50) | Del 48-50 |
| D39 (M) | nm | ||
| D40 (M) | 1,3 | Del 46-51 | Del 46-51 |
| D41 (M) | 3 | Del XJ10d | Del 4-13 |
| D42 (M) | 2,3 | Del 3-16 | Del 3-16 |
| D43 (M) | 1,3 | nm | nm |
| D44 (F) | 2,3 | Dup 2e | Dup 2 |
| D45 (M) | Dup 2 | ||
| D46 (F) | 3 | nm | nm |
| D47 (M) | 1,3 | nm | nm |
| D48 (M) | 1,3 | nm | nm |
| D49 (M) | 3 | Del 8-(16) | Del 8-39 |
| D50 (M) | 1,3 | nm | nm |
| D51 (F) | 2,3 | nm | nm |
| D52 (M) | 1,2,3 | Dup 51 | Dup 51 |
| D53 (M) | 1,3 | nm | nm |
| D54 (M) | 1,3 | nm | nm |
| D55 (M) | 1,3 | nm | nm |
| D56 (M) | 1,3 | nm | nm |
| D57 (M) | Dup 6 | ||
| D58 (M) | 1,3 | nm | nm |
| D59 (M) | 1,2,3 | nm | nm |
| D60 (M) | 1,3 | nm | nm |
| D61 (M) | 1,3 | Del 2-7 | Del 3-6 |
| D62 (M) | 1,2,3 | Del 20-29 | Del 20-29 |
| D63 (F) | Dup 2-(7) | Dup 3-7 | |
| D64 (M) | Dup 17 | Dup 17 | |
| D65 (M) | 1,2,3 | nm | nm |
| D66 (F) | nm | ||
| D67 (M) | 1,2,3 | Del 19-43 | Del 21-43 |
| D68 (M) | 1,2,3 | Del 19-43 | Del 21-43 |
| D69 (M) | 1,3 | nm | nm |
| D70 (F) | Dup 3-7 | Dup 3-7 | |
| D71 (M) | Dup 3 | Dup 3 | |
| D72 (F) | Dup 3 | ||
| D73 (M) | 1,2,3 | nm | nm |
| D74 (M) | Dup 51-55 | Dup 51-55 | |
| D75 (F) | 2,3 | Dup 51-55 | Dup 51-55 |
| G1 (M) | 1,4 | nm | Del 21 |
| G2 (M) | 1,4 | nm | Dup 10-11 |
| G3 (M) | 1,4 | nm | Dup 18-23 |
| G4 (M) | 1,4 | nm | nm |
| G5 (M) | 1,4 | nm | Del 48 |
| G6 (M) | 1,4 | nm | Dup 6-7 |
| G7 (M) | 1,4 | nm | Del 66 |
| G8 (M) | 1,4 | nm | nm |
| G9 (M) | 1,4 | nm | nm |
| G10 (F) | 1,4 | nm | Del 48-50 |
| G11 (M) | 1,4 | nm | nm |
| H1 (M) | 1 | nm | nm |
| H2 (M) | 1 | nm | nm |
| H3 (M) | 1 | nm | nm |
| H4 (F) | 1 | nm | nm |
| H5 (M) | 1 | nm | Del 45-50 |
| H6 (M) | 1 | nm | Dup 44 |
| H7 (M) | 1 | nm | nm |
| H8 (M) | 1 | nm | nm |
| H9 (M) | 1 | nm | nm |
| H10 (M) | 1 | nm | Dup 3-4 |
| L1 (M) | 1,4 | nm | Dup 8-13 |
| L2 (M) | 1,4 | nm | Del 18 |
| L3 (M) | 1,4 | nm | nm |
| L4 (M) | 1,4 | nm | nm |
| L5 (M) | 1,4 | nm | Dup 5-6 |
| L6 (M) | 1,4 | nm | Dup 54 |
| L7 (M) | 1,4 | nm | Dup 8-9 |
| L8 (M) | 1,4 | nm | nm |
| L9 (M) | 1,4 | nm | Dup 45-52 |
| L10 (M) | 1,4 | nm | Dup 2 |
| L11 (M) | 1,4 | nm | Dup 8-13 |
| L12 (M) | 1,4 | nm | Dup 53-55 |
| L13 (M) | 1,4 | nm | Dup 61-64 |
| L14 (M) | 1,4 | nm | nm |
| L15 (M) | 1,4 | nm | Dup 51-57 |
| L16 (M) | 1,4 | nm | Dup 8-9 |
| L17 (M) | 1,4 | nm | Dup 2 |
| L18 (M) | 1,4 | nm | Dup 3-30 |
| L19 (M) | 1,4 | nm | Dup 20 |
| L20 (M) | 1,4 | nm | Dup 14-21 |
| L21 (M) | 1,4 | nm | nm |
| L22 (M) | 1,4 | nm | Dup 2 |
| L23 (M) | 1,4 | nm | Dup 8-9 |
| L24 (M) | 1,4 | nm | Dup 42-43 |
| L25 (M) | 1,4 | nm | Dup 6-7 |
| L26 (M) | 1,4 | nm | Del 56 |
| L27 (M) | 1,4 | nm | nm |
| M1 (M) | 1,4 | nm | Dup 18-32 |
| M2 (M) | 1,4 | nm | Dup 20-27 |
1 = PCR by use of the Chamberlain et al. (1988) and Beggs et al. (1990) sets; 2 = quantitative Southern blotting; 3 = quantitative multiplex PCR; 4 = point-mutation detection.
Del = deletion; Dup = duplication; nm = no mutation found. Numbers denote exons; those in parentheses indicate an uncertain breakpoint.
Duplication of 30–50 kb around exon 60 detected using pulsed-field gel electrophoresis.
Probe located in intron 7.
Detectable by quantitative PCR, not evident with quantitative Southern blotting.
Table 1.
Samples Screened
|
Mutation Foundb by |
|||
| Sample (Sex) | Method(s)a | OtherMethods | MAPH |
| D1 (M) | 1,3 | Del 5-7 | Del 5-7 |
| D2 (F) | Dup 52-55 | ||
| D3 (M) | 3 | Del 50 | Del 50 |
| D4 (F) | Dup 50-55 | ||
| D5 (M) | 1,3 | nm | Dup 50-55 |
| D6 (M) | 3 | Del 8-44 | Del 8-44 |
| D7 (M) | 1,3 | nm | nm |
| D8 (M) | 1,3 | Dup 43 | Dup 43 |
| D9 (F) | Dup 58-63 | ||
| D10 (M) | 1,3 | ?c | Dup 58-63 |
| D11 (M) | 3 | Del 49-52 | Del 49-52 |
| D12 (M) | 3 | Del 48-50 | Del 48-50 |
| D13 (F) | Del 45 | ||
| D14 (M) | 3 | Del 1m | Del 1m |
| D15 (M) | 3 | nm | Del 53 |
| D16 (M) | 1,3 | nm | nm |
| D17 (M) | 3 | Del (3)-(10) | Del 4-12 |
| D18 (M) | 3 | Del 14-60 | Del 14-60 |
| D19 (M) | 1,3 | Del 69 | Del 64-67 |
| D20 (F) | 3 | Del (45)-(50) | Del 49-54 |
| D21 (M) | 3 | Del 2-(33) | Del 2-30 |
| D22 (M) | 3 | nm | nm |
| D23 (M) | 3 | Dup 12 | Dup 12 |
| D24 (F) | Dup 2 | ||
| D25 (M) | 2,3 | nm | Dup 2 |
| D26 (M) | 1,3 | nm | nm |
| D27 (F) | Dup 12-13 | ||
| D28 (F) | Dup 44 | ||
| D29 (M) | 3 | Dup 2-9 | Dup 2-9 |
| D30 (F) | nm | ||
| D31 (M) | 3 | Del 1-79 | Del 1-79 |
| D32 (F) | 3 | Dup 44-57 | Dup 44-57 |
| D33 (F) | Dup 2-7 | ||
| D34 (M) | 1,3 | Dup 2-7 | Dup 2-7 |
| D35 (M) | 1,3 | Dup 2-7 | Dup 2-7 |
| D36 (F) | 3 | Del 10-(?) | Del 10-46 |
| D37 (M) | 3 | Del 3-(?) | Del 3-19 |
| D38 (F) | 3 | Del (50) | Del 48-50 |
| D39 (M) | nm | ||
| D40 (M) | 1,3 | Del 46-51 | Del 46-51 |
| D41 (M) | 3 | Del XJ10d | Del 4-13 |
| D42 (M) | 2,3 | Del 3-16 | Del 3-16 |
| D43 (M) | 1,3 | nm | nm |
| D44 (F) | 2,3 | Dup 2e | Dup 2 |
| D45 (M) | Dup 2 | ||
| D46 (F) | 3 | nm | nm |
| D47 (M) | 1,3 | nm | nm |
| D48 (M) | 1,3 | nm | nm |
| D49 (M) | 3 | Del 8-(16) | Del 8-39 |
| D50 (M) | 1,3 | nm | nm |
| D51 (F) | 2,3 | nm | nm |
| D52 (M) | 1,2,3 | Dup 51 | Dup 51 |
| D53 (M) | 1,3 | nm | nm |
| D54 (M) | 1,3 | nm | nm |
| D55 (M) | 1,3 | nm | nm |
| D56 (M) | 1,3 | nm | nm |
| D57 (M) | Dup 6 | ||
| D58 (M) | 1,3 | nm | nm |
| D59 (M) | 1,2,3 | nm | nm |
| D60 (M) | 1,3 | nm | nm |
| D61 (M) | 1,3 | Del 2-7 | Del 3-6 |
| D62 (M) | 1,2,3 | Del 20-29 | Del 20-29 |
| D63 (F) | Dup 2-(7) | Dup 3-7 | |
| D64 (M) | Dup 17 | Dup 17 | |
| D65 (M) | 1,2,3 | nm | nm |
| D66 (F) | nm | ||
| D67 (M) | 1,2,3 | Del 19-43 | Del 21-43 |
| D68 (M) | 1,2,3 | Del 19-43 | Del 21-43 |
| (continued) | |||
Table 1 (continued).
|
Mutation Foundb by |
|||
| Sample (Sex) | Method(s)a | OtherMethods | MAPH |
| D69 (M) | 1,3 | nm | nm |
| D70 (F) | Dup 3-7 | Dup 3-7 | |
| D71 (M) | Dup 3 | Dup 3 | |
| D72 (F) | Dup 3 | ||
| D73 (M) | 1,2,3 | nm | nm |
| D74 (M) | Dup 51-55 | Dup 51-55 | |
| D75 (F) | 2,3 | Dup 51-55 | Dup 51-55 |
| G1 (M) | 1,4 | nm | Del 21 |
| G2 (M) | 1,4 | nm | Dup 10-11 |
| G3 (M) | 1,4 | nm | Dup 18-23 |
| G4 (M) | 1,4 | nm | nm |
| G5 (M) | 1,4 | nm | Del 48 |
| G6 (M) | 1,4 | nm | Dup 6-7 |
| G7 (M) | 1,4 | nm | Del 66 |
| G8 (M) | 1,4 | nm | nm |
| G9 (M) | 1,4 | nm | nm |
| G10 (F) | 1,4 | nm | Del 48-50 |
| G11 (M) | 1,4 | nm | nm |
| H1 (M) | 1 | nm | nm |
| H2 (M) | 1 | nm | nm |
| H3 (M) | 1 | nm | nm |
| H4 (F) | 1 | nm | nm |
| H5 (M) | 1 | nm | Del 45-50 |
| H6 (M) | 1 | nm | Dup 44 |
| H7 (M) | 1 | nm | nm |
| H8 (M) | 1 | nm | nm |
| H9 (M) | 1 | nm | nm |
| H10 (M) | 1 | nm | Dup 3-4 |
| L1 (M) | 1,4 | nm | Dup 8-13 |
| L2 (M) | 1,4 | nm | Del 18 |
| L3 (M) | 1,4 | nm | nm |
| L4 (M) | 1,4 | nm | nm |
| L5 (M) | 1,4 | nm | Dup 5-6 |
| L6 (M) | 1,4 | nm | Dup 54 |
| L7 (M) | 1,4 | nm | Dup 8-9 |
| L8 (M) | 1,4 | nm | nm |
| L9 (M) | 1,4 | nm | Dup 45-52 |
| L10 (M) | 1,4 | nm | Dup 2 |
| L11 (M) | 1,4 | nm | Dup 8-13 |
| L12 (M) | 1,4 | nm | Dup 53-55 |
| L13 (M) | 1,4 | nm | Dup 61-64 |
| L14 (M) | 1,4 | nm | nm |
| L15 (M) | 1,4 | nm | Dup 51-57 |
| L16 (M) | 1,4 | nm | Dup 8-9 |
| L17 (M) | 1,4 | nm | Dup 2 |
| L18 (M) | 1,4 | nm | Dup 3-30 |
| L19 (M) | 1,4 | nm | Dup 20 |
| L20 (M) | 1,4 | nm | Dup 14-21 |
| L21 (M) | 1,4 | nm | nm |
| L22 (M) | 1,4 | nm | Dup 2 |
| L23 (M) | 1,4 | nm | Dup 8-9 |
| L24 (M) | 1,4 | nm | Dup 42-43 |
| L25 (M) | 1,4 | nm | Dup 6-7 |
| L26 (M) | 1,4 | nm | Del 56 |
| L27 (M) | 1,4 | nm | nm |
| M1 (M) | 1,4 | nm | Dup 18-32 |
| M2 (M) | 1,4 | nm | Dup 20-27 |
1 = PCR by use of the Chamberlain et al. (1988) and Beggs et al. (1990) sets; 2 = quantitative Southern blotting; 3 = quantitative multiplex PCR; 4 = point-mutation detection.
Del = deletion; Dup = duplication; nm = no mutation found. Numbers denote exons; those in parentheses indicate an uncertain breakpoint.
Duplication of 30–50 kb around exon 60 detected using pulsed-field gel electrophoresis.
Probe located in intron 7.
Detectable by quantitative PCR, not evident with quantitative Southern blotting.
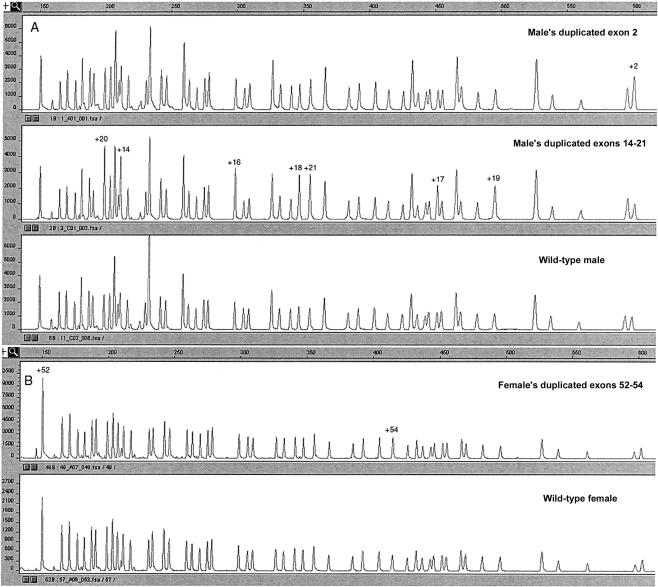
Figure 3.
Patterns obtained from analysis of patients by use of probe set A. A, Male patient's duplicated exon 2, male patient's duplicated exons 14–21, and male control individual. B, Female carrier's duplicated exons 52–54 and female control individual.
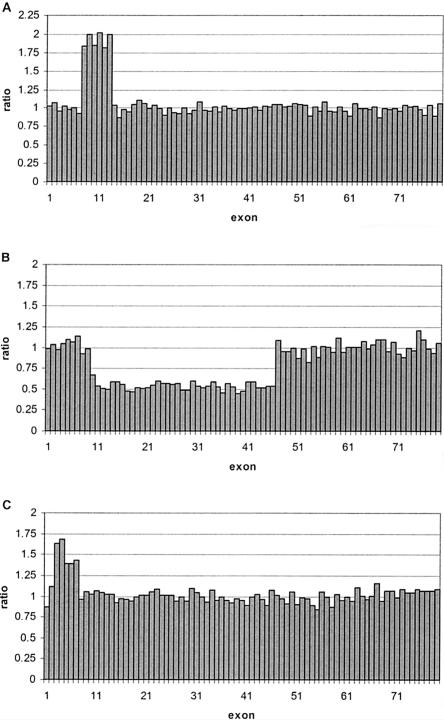
Figure 4.
Analysis of different patient samples. A, Male patient L1's duplicated exons 8–13, with SD 0.05. B, Female carrier D36's deleted exons 10–46, with SD 0.05 C, Female carrier D70's duplicated exons 3–7, with SD 0.06.
False-negative results are more difficult to assess. An estimate can be made by looking at patient samples in which more than two exons are deleted or duplicated. A result would be considered to be false negative when one or more exons within a mutated series was found to be normal. In no patient sample was this seen. Although this does not exclude the possibility that false-negative calls will occur, it does suggest that they will happen very rarely.
The exon 75 probe was the probe that showed the highest variation among the 79 DMD probes used. This appeared to be due to slight variations in PCR/washing conditions, rather than variations in a polymorphic sequence, since no sample consistently showed a duplication of exon 75. For any hybridization in which exon 75 showed such variation, the results for that exon were ignored, and the exon was retested in a subsequent experiment. In no cases could an exon 75 duplication be confirmed.
Initially, some probes could not be recovered. Close examination of the sequences revealed that all had a relatively low GC content (<40%). In some cases—for example, exon 2—it was not possible to raise this percentage, since the entire region was extremely AT rich. To solve this problem, we made the probes longer. In this manner, we were able to use a 602-bp exon 2 probe with a GC content as low as 30%.
Discussion
Of the 24 mutations previously characterized in our laboratory, all were detected using the MAPH technique (table 1table 1). In one case, the breakpoints did not match exactly. This was in a male patient (D61) that, with MAPH, was seen as having a deletion of exons 3–6 but had previously been diagnosed as having a deletion of exons 3–7. Southern blot analysis showed a junction fragment for exon 7, suggesting that the breakpoint may be within the exon. In a hybridization-based technique, a breakpoint may be misdiagnosed if the deletion occurs within the sequence to which it is bound by the probe and if there is enough of the exon remaining for the probe to hybridize; this is likely to occur rarely however, since it has been calculated that, in ∼99% of cases of DMD and/or BMD, the breakpoints are outside the coding exon (den Dunnen et al. 1989). By contrast, PCR from genomic DNA may lead to false-negative results if there is a polymorphism within the priming site (Abbs et al. 1991).
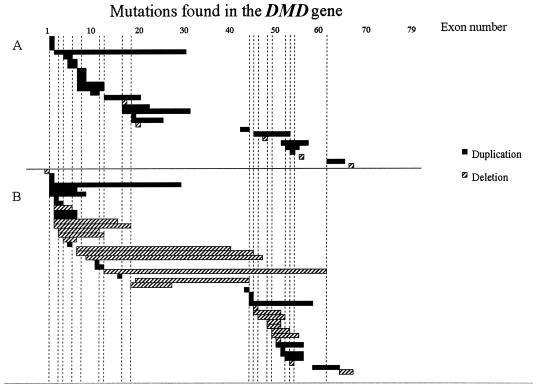
Of the 72 male samples in which no mutation had previously been found, 37 (51%) had mutations that were detectable by use of MAPH (table 1table 1). When only those samples that had been checked for deletions and point mutations were included, the frequency was 74% (29/39). These were composed of five deletions (all of one exon) and, strikingly, but not unexpectedly, 24 duplications. To present an unbiased view of duplication distribution, we have depicted those mutations detected in samples that were also screened for point mutations and deletions (fig. 5A) separately from an overview of all the mutations detected (fig. 5B).
Figure 5.
Independent mutations detected during the present study. Vertical bars represent the 18 exons tested using the Chamberlain et al. (1988) and Beggs et al. (1990) kits. A, Mutations detected in samples in which point mutations and deletions had been excluded, mainly by multiplex PCR. B, All other mutations detected.
Samples LA1–LA27 (table 1table 1) were from a point-mutation screening by use of the DOVAM-S (detection of virtually all mutations–SSCP) technique (Mendell et al. 2001). A total of 141 samples were tested, and 108 point mutations were found. We analyzed 27 of the remaining 33 samples, finding two deletions and 20 duplications. Samples M1 and M2 (table 1table 1) remained from a study, using denaturing high-performance liquid chromatography, that screened eight patients with DMD for point mutations (Bennett et al. 2001) but found no obvious pathological mutations in two of these samples. By use of MAPH, both samples showed a duplication, thereby completing the mutation study.
A total of 23 female potential or proven carriers were tested. Of the 15 samples in which no mutation had previously been found, two deletions and eight duplications were found. Analysis of potential carriers was facilitated when it was known what mutation to expect.
Newly found mutations could often be confirmed using other methods. Small duplications, such as that in sample D52 (with an exon 51 duplication), could be confirmed by retrospective examination of Southern blots that had been previously prepared and analyzed in our laboratory. The DNA in sample D45 showed an exon 2 duplication by use of MAPH, as did DNA from the mother (sample D44). The result from sample D44 was confirmed by quantitative PCR, yet was not evident on a Southern blot.
The exon 2 (samples D25 and D120.7) and exons 58–63 (samples D10 and DL33.2) duplications, which have been described elsewhere (den Dunnen et al. 1989), are interesting cases. Pulsed-field gel electrophoresis analysis indicated that there were rearrangements of ∼150 kb, at the 5′ end of the gene, and 30–50 kb, around exon 60. Despite a focused analysis of both regions, no duplications could be detected using Southern blotting.
Duplication of exon 2 alone is extremely difficult to detect by Southern blotting, since the band is very weak. This may be due to the very low GC content (∼30%) of exon 2 and its surrounding region, leading, under stringent conditions, to weak hybridization. Given the extremely large size (190 and 170 kb) of the introns flanking exon 2, it is not surprising that a deletion or duplication of exon 2 by itself is a mutation that has been found more than once. In fact, it was the single most common duplication found, occurring five times. Interestingly, however, no deletion of exon 2 alone has so far been reported (Leiden Muscular Dystrophy Pages).
Our results show that, even when the DMD gene is screened for deletions, duplications, and point mutations (DOVAM-S or denaturing gradient gel electrophoresis), a small number of samples remain in which no disease-causing mutation can be detected. There are several possible explanations why no mutation was found in these samples. When RNA has not yet been analyzed in a patient, mutations that affect splicing are the most plausible candidates. Indeed, RNA-based techniques, such as the protein-truncation test, detect mutations that would be missed using DNA-based techniques (Roest et al. 1996; Whittock et al. 1997). It is also possible that the disease was misdiagnosed and that the mutation lies in a gene responsible for other muscular disorders. Germline mosaicism has been reported elsewhere (Bakker et al. 1987; Wood and McGillivray 1988) and would not necessarily be detectable by use of the methods described herein. Another, less likely reason is mutations in a gene that is involved in the regulation of dystrophin expression.
Although mutation detection obviously is critical for diagnosis, it may also be important for future therapeutic purposes. Recent reports have showed the potential use of read-through protein synthesis (Gentamycin) (Barton-Davis et al. 1999) and exon skipping (with antisense oligoribonucleotides) (van Deutekom et al. 2001) in the restoration of the reading frame of the dystrophin transcript. In particular, single-exon duplications, as detected in 12 cases in this study, would make an ideal target for exon skipping. The presence of two targets not only would double the efficiency but also should produce a normal transcript, leading to a wild-type protein.
The MAPH approach's primary advantages over Southern blotting and quantitative PCR are the relative simplicity, speed, and completeness of coverage of all 79 exons. Although 90%–95% of the deletions can be detected using multiplex PCR, the breakpoints are often not determined, and rare mutations outside the hotspots will be missed. In previously published reports on MAPH (Armour et al. 2000; Sismani et al. 2001), recovered probes were radioactively labeled and were separated on a polyacrylamide gel. For speed and convenience, we chose to use a combination of fluorescent labeling and capillary electrophoresis. Capillary electrophoresis is becoming more widely used in mutation detection, since it provides greater sensitivity and has high-throughput capabilities (Bosserhoff et al. 2000). We used the ABI 3700 (Applied Biosystems), which allows the simultaneous analysis of 96 samples. One run of 96 samples takes ∼4 h, with the data analyzed by software provided with the machine.
There are several ways in which the current system can be further enhanced. In the present study, only two (blue [FAM sample] and red [ROX size standard]) of the four available colors were used. By use of up to three sets of primers, each labeled with a different fluorophore, it should be possible to expand the potential number of probes by threefold. Hybridizing the PCR products to a microarray composed of each individual probe could further increase the number of probes tested, with the additional advantage that they would no longer need to be differentiated in length.
In contrast to many other methods, this technique should be easy to implement in a standard diagnostic laboratory, since no new technology needs to be introduced. The critical techniques are hybridization and PCR, and the products can be analyzed on any apparatus that is used for sequence analysis. Furthermore, it can easily be applied to any disease gene of interest, and the resolution provided and the potential of array implementation may even allow future genomewide screening.
Acknowledgments
We would like to thank R. Hofstra, E. Hoffman, S. Sommer, and R. Bennett for providing DNA samples. This work was supported by grants from ZorgOnderzoek Nederland (9607.031.1) and the Prinses Beatrix Fonds.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for nr and htgs databases)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
- Leiden Muscular Dystrophy Pages, http://www.dmd.nl/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DMD [MIM #310200])
References
- Abbs SJ, Yau SC, Mathew CG, Bobrow M (1991) A convenient multiplex PCR system for the detection of dystrophin gene deletions: a comparative analysis with cDNA hybridisation shows mistypings by both methods. J Med Genet 28:304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour JA, Sismani C, Patsalis PC, Cross G (2000) Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res 28:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E, Van Broeckhoven C, Bonten EJ, Van De Vooren MJ, Veenema H, Van Hul W, van Ommen GJB, Vandenberghe A, Pearson PL (1987) Germline mosaicism and Duchenne muscular dystrophy mutations. Nature 329:554–556 [DOI] [PubMed] [Google Scholar]
- Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL (1999) Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest 104:375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs AH, Koenig M, Boyce FM, Kunkel LM (1990) Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 86:45–48 [DOI] [PubMed] [Google Scholar]
- Bennett RR, Dunnen J, O'Brien KF, Darras BT, Kunkel LM (2001) Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genet 2:17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosserhoff AK, Buettner R, Hellerbrand C (2000) Use of capillary electrophoresis for high throughput screening in biomedical applications: a minireview. Comb Chem High Throughput Screen 3:455–466 [DOI] [PubMed] [Google Scholar]
- Boyce FM, Beggs AH, Feener CA, Kunkel LM (1991) Dystrophin is transcribed in brain from a distant upstream promoter. Proc Natl Acad Sci USA 88:1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT (1988) Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 16:11141–11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras BT, Blattner P, Harper JF, Spiro AJ, Alter S, Francke U (1988) Intragenic deletions in 21 Duchenne muscular dystrophy (DMD)/Becker muscular dystrophy (BMD) families studied with the dystrophin cDNA: location of breakpoints on HindIII and BglII exon-containing fragment maps, meiotic and mitotic origin of the mutations. Am J Hum Genet 43:620–629 [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Grootscholten PM, Bakker E, Blonden LAJ, Ginjaar HB, Wapenaar MC, van Paassen HMB, van Broeckhoven C, Pearson PL, van Ommen GJB (1989) Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet 45:835–847 [PMC free article] [PubMed] [Google Scholar]
- Forrest SM, Cross GS, Speer A, Gardner-Medwin D, Burn J, Davies KE (1987) Preferential deletion of exons in Duchenne and Becker muscular dystrophies. Nature 329:638–640 [DOI] [PubMed] [Google Scholar]
- Gillard EF, Chamberlain JS, Murphy EG, Duff C, Smith B, Burghes AHM, Thompson MW, Sutherland J, Oss I, Bodrug SE, Klamut HJ, Ray PN, Worton RG (1989) Molecular and phenotypic analysis of patients with deletions within the deletion-rich region of the Duchenne muscular dystrophy (DMD) gene. Am J Hum Genet 45:507–520 [PMC free article] [PubMed] [Google Scholar]
- Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Albertson DG, Pinkel D, Collins C, Hanahan D, Gray JW (2001) Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet 29:459–464 [DOI] [PubMed] [Google Scholar]
- Hu X, Ray PN, Murphy E, Thompson MW, Worton RG (1990) Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin and phenotype/genotype correlation. Am J Hum Genet 46:682–695 [PMC free article] [PubMed] [Google Scholar]
- Ioannou P, Christopoulos G, Panayides K, Kleanthous M, Middleton L (1992) Detection of Duchenne and Becker muscular dystrophy carriers by quantitative multiplex polymerase chain reaction analysis. Neurology 42:1783–1790 [DOI] [PubMed] [Google Scholar]
- Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, Meng G, et al (1989) The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 45:498–506 [PMC free article] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener CA, Kunkel LM (1987) Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50:509–517 [DOI] [PubMed] [Google Scholar]
- Mansfield ES, Robertson JM, Lebo RV, Lucero MY, Mayrand PE, Rappaport E, Parrella T, Sartore M, Surrey S, Fortina P (1993) Duchenne/Becker muscular dystrophy carrier detection using quantitative PCR and fluorescence-based strategies. Am J Med Genet 48:200–208 [DOI] [PubMed] [Google Scholar]
- Mendell JR, Buzin CH, Feng J, Yan J, Serrano C, Sangani DS, Wall C, Prior TW, Sommer SS (2001) Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurology 57:645–650 [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM (1988) An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2:90–95 [DOI] [PubMed] [Google Scholar]
- Morgan NV, Tipping AJ, Joenje H, Mathew CG (1999) High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am J Hum Genet 65:1330–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FBL, Hageman S, Arts PJ, Ligtenberg MJ, Meijers-Heijboer H, Klijn JG, Vasen HF, Cornelisse CJ, van 't Veer LJ, Bakker E, van Ommen GJB, Devilee P (1997) BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 17:341–345 [DOI] [PubMed] [Google Scholar]
- Roest PAM, Bout M, Van Der Tuijn AC, Ginjaar HB, Bakker E, Hogervorst FBL, van Ommen GJB, den Dunnen JT (1996) Splicing mutations in DMD/BMD detected by RT-PCR/PTT: detection of a 19AA insertion in the cysteine-rich domain of dystrophin compatible with BMD. J Med Genet 33:935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sismani C, Armour JA, Flint J, Girgalli C, Regan R, Patsalis PC (2001) Screening for subtelomeric chromosome abnormalities in children with idiopathic mental retardation using multiprobe telomeric FISH and the new MAPH telomeric assay. Eur J Hum Genet 9:527–532 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AAM, Ginjaar HB, Baas F, den Dunnen JT, van Ommen GJB (2001) Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet 10:1547–1554 [DOI] [PubMed] [Google Scholar]
- Whittock NV, Roberts RG, Mathew CG, Abbs SJ (1997) Dystrophin point mutation screening using a multiplexed protein truncation test. Genet Test 1:115–123 [DOI] [PubMed] [Google Scholar]
- Wijnen J, van der Klift H, Vasen H, Khan PM, Menko F, Tops C, Meijers HH, Lindhout D, Moller P, Fodde R (1998) MSH2 genomic deletions are a frequent cause of HNPCC. Nat Genet 20:326–328 [DOI] [PubMed] [Google Scholar]
- Wood S, McGillivray BC (1988) Germinal mosaicism in Duchenne muscular dystrophy. Hum Genet 78:282–284 [DOI] [PubMed] [Google Scholar]
- Worton RG, Thompson MW (1988) Genetics of Duchenne muscular dystrophy. Annu Rev Genet 22:601–629 [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Kato S, Hiraishi Y, Ishihara T, Hata J, Matsuo N, Takano T (1996) Identification of carriers of Duchenne/Becker muscular dystrophy by a novel method based on detection of junction fragments in the dystrophin gene. J Med Genet 33:1027–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau SC, Bobrow M, Mathew CG, Abbs SJ (1996) Accurate diagnosis of carriers of deletions and duplications in Duchenne/Becker muscular dystrophy by fluorescent dosage analysis. J Med Genet 33:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]