Abstract
Cyclin D1 is a component of the core cell-cycle machinery and is frequently overexpressed in breast cancer. It physically interacts with the tumor suppressor Dmp1 that attenuates the oncogenic signals from Ras and HER2 by inducing Arf/p53-dependent cell-cycle arrest. Currently, the biological significance of Dmp1–cyclin D1 interplay in breast cancer has not been determined. Here, we show that cyclin D1 bound to Dmp1 to activate both Arf and Ink4a promoters and, consequently, induced apoptosis or G2/M cell-cycle delay in normal cells to protect them from neoplastic transformation. The cyclin D1–induced Ink4a/Arf gene expression was dependent on Dmp1 because the induction was not detected in Dmp1-deficient or DMP1-depleted cells. Arf/Ink4a expression was increased in pre-malignant mammary glands from Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice but significantly down-regulated in those from Dmp1-deficient mice. Selective Dmp1 deletion was found in 21% of the MMTV-D1 and D1T286A mammary carcinomas, and the Dmp1 heterozygous status significantly accelerated mouse mammary tumorigenesis with reduced apoptosis and increased metastasis. Overall, our study reveals a pivotal role of combined Dmp1 loss and cyclin D1 overexpression in breast cancer.
As a key sensor and integrator of extracellular signals in early-to-mid G1 phase, cyclin D1 deregulation, including chromosomal translocation, gene amplification, or reduced degradation, results in tumorigenesis and has been linked to breast cancer and other human cancers.1,2 Cyclin D1 drives cells to enter S phase by binding and activating the Cdk4/6 kinase. The cyclin D1/Cdk4 complex phosphorylates the retinoblastoma protein (pRb), which releases E2F transcriptional factors from pRb constraint. E2Fs then activate the transcription of genes required for cells to enter S phase.3 Apart from its catalytic function, cyclin D1 also exerts a transcription regulation without the participation of Cdks, even though it is not a transcriptional factor. Cyclin D1 executes the transcriptional regulation through its interaction with transcriptional factors and chromatin modifiers.4,5 For instance, it directly binds and activates estrogen receptor α (ERα) in breast cancer cell lines in a CDK-independent fashion.6 By contrast, cyclin D1 interacts and inhibits the transcriptional activity of androgen receptor (AR), Myb-related protein B (B-Myb), E1A-binding protein p300, and NF-κB.7–10 Overall, these studies suggested that cyclin D1 regulates cell proliferation, growth, and differentiation, and its transcriptional regulatory activity has great potential significance in human cancer progression.5,11
Among known cyclin D1–associated transcriptional factors, cyclin D–interacting Myb-like Protein 1 (Dmp1; alias Dmtf1) is a bona fide tumor suppressor.12,13 Dmp1 directly binds and activates Arf, thereby inducing Arf-p53–dependent cell-cycle arrest.14 Dmp1-deficient cells can be transformed by oncogenic Ras alone without altering the status of Arf and p53 genes, suggesting that the Arf-p53 pathway is significantly attenuated in Dmp1-deficient cells.13 Ectopically expressed Dmp1 inhibits the growth of breast cancer cells with wild-type p53, indicating that p53 is a critical target for Dmp1 to exhibit its biological activity.15 Both Dmp1-null and Dmp1-heterozygous mice are prone to develop tumors, and a wild-type Dmp1 allele is often retained in tumors from Dmp1-heterozygous mice. This suggests that Dmp1 is haploinsufficient as a tumor suppressor.12,13,16 Our recent study in MMTV-neu mice showed that mammary carcinogenesis was significantly accelerated in both Dmp1+/− and Dmp1−/− backgrounds, with no difference between groups that lacked one or both Dmp1 alleles. Hemizygous Dmp1 deletion was found in >50% of neu mammary carcinomas accompanied with significant down-regulation of p19Arf and p21Cip1/WAF1, indicating that Dmp1 is a physiological regulator of the Arf-p53 pathway in vivo.17 Consistent with the mouse model studies, the loss of heterozygosity of DMP1 was found in nearly 45% of human breast carcinomas and mutually exclusive with that of INK4a/ARF and P53, suggesting primary involvement of DMP1 in human mammary carcinogenesis.15
Cyclin D1 physically interacts with Dmp1, and antagonizes its ability of activating the CD13/aminopeptidase N promoters in the absence of functional Cdks.18 However, by an unknown mechanism, Dmp1 and cyclin D1 collaborate to activate the Arf promoter, which suggests that whether cyclin D1 acts as a corepressor or coactivator of Dmp1 is dependent on the context of a target gene.19 Although Dmp1 and cyclin D1 play vital roles in breast cancer prevention and development, respectively, the biological functions and significance of Dmp1–cyclin D1 interaction in breast cancer remain to be determined. We conducted the current study to elucidate the cooperative role of cyclin D1 and Dmp1 in breast cancer development. We showed that Dmp1 modulated the activation of the Arf/Ink4a tumor suppressor pathways induced by cyclin D1 overexpression. We crossed the MMTV-cyclin D1 and MMTV-D1T286A mice with Dmp1-deficient mice and found that Dmp1 heterozygosity significantly accelerated cyclin D1–induced mammary carcinogenesis. We also observed the metastasis of cyclin D1–induced mammary tumors in a Dmp1-deficient background.
Materials and Methods
Cell Culture and Luciferase Reporter Assays
Wild-type and Dmp1-null murine embryonic fibroblasts (MEFs) were established from 13.5-day-old embryos as previously described.12 MEF, NIH 3T3, and human mammary epithelial cells were cultured as described previously.12,15,17 To study the responsiveness of the Arf/Ink4a promoters to cyclin D1 and D1T286A, 2 × 105 cells were seeded into 60-mm-diameter culture dishes 24 hours before transfection, and then transfected with 4 μg of luciferase reporter DNA with or without increasing amount of pFLEX1-cyclin D1, pFLEX1 D1T286A,20 or pFLEX1-cyclin D1Δ21 and 4 μg of a control plasmid with β-actin promoter–driven secreted alkaline phosphatase (SEAP).18,19,21 GeneJuice transfection reagent (Novagen; EMD Millipore, Billerica, MA) was used in all transfections.
Chromatin Immunoprecipitation
Tissue and cell chromatin immunoprecipitations (ChIP) were performed as described previously.19,22–24 Briefly, in tissue ChIP, lysates from Dmp1+/+;MMTV-neu tumors were precipitated with antibodies against cyclin D1 (SP4; NeoMarkers/Lab Vision, Fremont, CA, and sc-753; Santa Cruz Biotechnology, Santa Cruz, CA) and incubated at 4°C overnight. After reverse cross-linking, the immunoprecipitated DNA was amplified by PCR after being mixed with 1 μCi of [α-32P]dATP (PerkinElmer, Waltham, MA) and separated on a 10% nondenaturing polyacrylamide gel. In cell ChIP, wild-type and Dmp1-null MEFs were infected with lentivirus expressing hemagglutinin (HA)-cyclin D1. Forty-eight hours after infection, the cell lysates were precipitated with anti-HA affinity gel (E6779; Sigma-Aldrich, St. Louis, MO). Mouse IgG-agarose (A0919; Sigma-Aldrich) was used as a negative control. For the detection of cyclin D1 on the Arf promoter, a sense primer 5′-ACTCGGAGCAAGGGAAACCT-3′ and an anti-sense primer 5′-TAGCAGTAGCTGCGCCCTTT-3′ were used. For the detection of cyclin D1 on the Ink4a promoter, a sense primer 5′-GCAAATAGCGCCACCTATGG-3′ and an anti-sense primer 5′-CTGCTCCAGATGGCTCTCCT-3′ were used.
Retrovirus and Lentivirus Production
For retrovirus production, 293T cells were transfected with pSRαMSV-tkneo vector, pSRαMSV-cyclin D1-tkneo, pSRαMSV-D1T286A-tkneo, pSR vector, or pSR-1131 (DMP1 shRNA)16 together with a helper retrovirus plasmid. Viruses were harvested and used to infect cells as previously described.19 For lentivirus production, 293T cells were transfected with pSL4-HA, pSL4-HA-cyclin D1, or pSL4-HA-cyclin D1T286A with helper lentivirus plasmids including VSVG, RSV-REV, and PMDL g/p RRE. Viruses were harvested 48 hours after transfection. For preparation of doxycycline-inducible cyclin D1, the cyclin D1 cDNA was PCR amplified by Pfu DNA polymerase and then subcloned downstream of a 6xTRE minimal promoter in an all-in-one doxycycline-inducible vector that expresses rtTA-advanced protein, generated by our colleagues.
Real-Time PCR
Analysis of Dmp1 gene copy number and quantification of Dmp1, p14ARF, p19Arf, p21Cip1/WAF1, p16Ink4a, and CYCLIN D1 mRNAs were conducted by real-time PCR TaqMan assays on an Applied Biosystems ABI 7500 Real-Time PCR machine (Life Technologies, Foster City, CA). β-Actin was used as internal control.22–24
Western Blot Analysis
Proteins were extracted from cell lysates in ice-cold EBC buffer with proteinase inhibitors.25 After being separated by gel electrophoresis and transferred to nitrocellulose membranes, proteins were detected by immunoblotting with affinity-purified polyclonal antibodies for Dmp1 [Rabbit Antibody D (RAD)],22 cyclin D1 (sc-753; Santa Cruz Biotechnology), p53 (sc-6243G; Santa Cruz Biotechnology), Mdm2, or HDM2 (ab16896 [2A10]; Abcam, Cambridge, UK), p19Arf (sc-32748; Santa Cruz Biotechnology), p14ARF (sc-53639; Santa Cruz Biotechnology), cleaved poly(ADP-ribose) polymerase (PARP) (#AF-600-NA; R&D Systems, Minneapolis, MN), cleaved caspase 3 (#9661; Cell Signaling Technology, Danvers, MA), p16Ink4a (sc-74401; Santa Cruz Biotechnology), p21CIP1/WAF1 (sc-397G; Santa Cruz Biotechnology), or β-actin (sc-1615, sc-47778; Santa Cruz Biotechnology), followed by incubation with horseradish peroxidase–conjugated secondary antibodies, and visualization using an enhanced chemiluminescence detection kit (PerkinElmer).
Dmp1+/−; and Dmp1−/−; MMTV-cyclin D1 and MMTV-D1T286A Compound Mice
The mouse model studies were performed according to a protocol approved by the Institutional Animal Care and Use Committee of Wake Forest University School of Medicine. Dmp1-heterozygous females were backcrossed to the same FVB/NJ males (The Jackson Laboratory, Bar Harbor, ME) for more than eight generations to obtain Dmp1+/− mice with >99% FVB/NJ background overall. A male MMTV-cyclin D1 mouse (provided by Dr. E.V. Schmidt, Harvard Medical School)26 or MMTV-D1T286A mouse (provided by Dr. J.A. Diehl, University of Pennsylvania)27 in a pure FVB/NJ background was crossed with a Dmp1+/− female to obtain Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice. Then Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A compound transgenic mice were further crossed with Dmp1+/− mice to obtain >15 mice with each genetic background. Littermate wild-type mice were used as controls.
Cell-Cycle Analysis
Wild-type and Dmp1-null MEFs were infected with lentivirus carrying empty vector or expressing cyclin D1 or D1T286A. Forty-eight hours after infection, cells were trypsinized, washed with PBS, fixed in 70% of ethanol, treated with propidium iodide and ribonuclease A, and then run through a flow cytometer (Accuri; Becton Dickinson, Franklin Lakes, NJ). Cell-cycle distributions were analyzed by ModFit LT software version 3.0 (Verity Software House, Topsham, ME).
Immunohistochemical Staining
Immunohistochemical staining of tissues and tumors were conducted as described previously.15 The following antibodies were used for immunohistochemistry with formalin-fixed, paraffin-embedded sections: Ki-67 (SP6; NeoMarkers/Lab Vision), cyclin D1 (SP4; NeoMarkers/Lab Vision), cleaved caspase-3 (#9661; Cell Signaling Technology), cytokeratin 8 (ab59400; Abcam), cytokeratin 14 (ab7800; Abcam), and ERα (sc-542; Santa Cruz Biotechnology).
In Vitro Mutagenesis
The murine Ink4a promoter Dmp1-binding site deletion/point mutants were generated by use of an in vitro mutagenesis kit (Stratagene; Agilent Technologies, Santa Clara, CA). For deletion mutagenesis, the sense primer 5′-CCATCCCTTTCCCCTCCCGTGGGGGGAACAGCAGTG-3′ and its reverse complementary sequence were used. For point mutagenesis, the sense primer 5′-CCTTTCCCCTCCCCCTTCCGGAGGTGGGGGGAA-3′ and its reverse complementary sequence were used.
Electrophoretic Mobility Shift Assay
Recombinant DMP1 protein was prepared in Sf9 cells.25 Electrophoretic mobility shift assays were performed using 32P-labeled oligonucleotide probe covering the DMP1/Ets site on murine or human INK4a promoter obtained by annealing oligonucleotide 5′-GGGATCCCTTTCCCCTCCCCCATCCGGAGGTGGGGGGAACAGCA-3′ (mouse sense strand) or 5′-GGGCTGGGATCAGCTCTCAGCATCCGGAAGCCTTTGCCTACTAG-3′ (30911 human sense strand) or 5′-GGGATAGACGTGAGCCACCGCATCCGGACTTTCCTTTTATGTAA-3′ (33549 human sense strand) (the DMP1/Ets consensus sequence is underlined) with a complementary antisense strand. The probe containing the DMP1/Ets site on murine Arf promoter has been described.14 For competition experiments, a 50-fold excess of unlabeled oligonucleotides were added to reaction mixtures before probe incubation.
Statistical Analysis
Statistical differences of survival in Dmp1+/+;MMTV-cyclin D1, Dmp1+/−;MMTV-cyclin D1, Dmp1+/+;MMTV-D1T286A, and Dmp1+/−;MMTV-D1T286A mice were analyzed by Medcalc software version 12.7.0 (Mariakerke, Belgium). Statistical analyses for all experiments were conducted using unpaired Student’s t-tests.
Results
Dmp1 Is Essential to Cyclin D1–Mediated Activation of the Arf and Ink4a Promoters
We previously demonstrated that Dmp1 directly binds and activates the Arf promoter.14,28 To determine whether Dmp1 also regulates Ink4a gene expression, we analyzed the murine Ink4a promoter and found a candidate Dmp1/Ets consensus sequence (TCCGGATGG: −178 to −166 relative to the transcription initiation site; two mismatches from the originally reported consensus sequences25 at both ends; the DMP1/Ets consensus sequence is underlined) (Supplemental Figure S1A). In electrophoretic mobility shift assay studies, recombinant Dmp1 directly associated with the oligonucleotide with this sequence at a relatively low affinity compared to its binding to the oligonucleotide with the Dmp1 consensus in the Arf promoter (Supplemental Figure S1B). In reporter assays, Dmp1 activated the Ink4a promoter by sixfold to eightfold over a control vector, but this effect was not observed in reporter constructs with the Dmp1/Ets site mutated or deleted (Supplemental Figure S1C), suggesting that this consensus was necessary for Dmp1-mediated Ink4a transactivation. We also found two consensus sequences of Dmp1 in the human INK4a promoter, which was bound and activated by Dmp1 up to threefold over a control vector (Supplemental Figure S1, D and E). Thus, our data support that Dmp1 regulates Ink4a expression in both humans and mice.
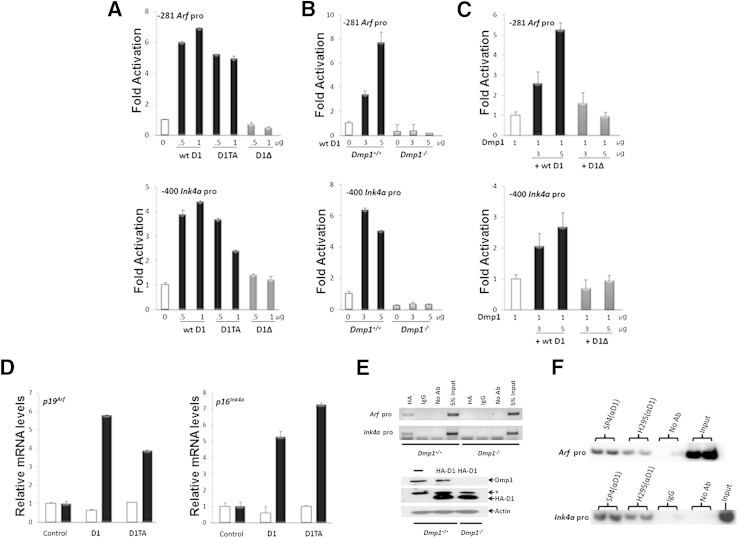
Cyclin D1 induces cell cycle arrest, senescence, and apoptosis when overexpressed.29 Thus, we investigated the potential effect of cyclin D1 on the Arf and Ink4a promoters. Both wild-type cyclin D1 and its constitutively active mutant D1T286A activated the Arf and Ink4a promoters, although the former showed better response than the latter (sevenfold versus fourfold) (Figure 1A). Cyclin D1–mediated activation of Arf and Ink4a was also observed in Dmp1+/+ MEFs, but this effect was abrogated in Dmp1−/− MEFs (Figure 1B). However, ectopically expressed Dmp1 in Dmp1−/− MEFs restored cyclin D1–induced luciferase activity mediated by the Arf and Ink4a promoters, suggesting that Dmp1 is indispensable for cyclin D1–activated Arf and Ink4a expression (Figure 1C). Moreover, a cyclin D1 mutant, D1Δ142–253 that is deficient in Dmp1 binding,21 failed in activating either the Arf or Ink4a promoter, suggesting that the effect of cyclin D1 on these promoters depended on cyclin D1–Dmp1 interaction (Figure 1, A and C). Additionally, the transcription of endogenous p19Arf and p16Ink4a was promoted by ectopic cyclin D1 or D1T286A in wild-type MEFs, but this effect was diminished in Dmp1-null MEFs (Figure 1D).
Figure 1.
Both p19Arf and p16Ink4a promoters are activated by cyclin D1 and the cyclin D1T286A mutant. A: The Arf (−281) or Ink4a (−400) promoter luciferase construct was cotransfected with indicated amounts of cyclin D1 [wild-type (wt)], D1T286A (D1TA), or D1Δ142–253 (D1Δ)21 expression vectors in NIH 3T3 cells. The numbers show the fold activation of the luciferase reporter normalized by internal controls of SEAP. B: The Arf (−281) or Ink4a (−400) promoter luciferase construct was cotransfected with increasing amounts of cyclin D1 expression vector in Dmp1+/+ or Dmp1−/− MEFs. The numbers show the fold activation of the luciferase reporter normalized by internal controls of SEAP. C: Luciferase reporter construct encoding Arf (−281) promoter or Ink4a (−400) promoter was cotransfected with 1 μg of pFLEX1-Dmp1 together with either 3 or 5 μg of pFLEX1-cyclin D1 or pFLEX1-D1Δ142–253 into Dmp1-null MEFs. The numbers show the fold activation of the luciferase reporter normalized by internal controls of SEAP. D: Primary Dmp1+/+ (black bars) or Dmp1−/− (white bars) MEFs were infected with retrovirus carrying an empty vector or vector expressing cyclin D1 or D1T286A protein. After neomycin selection, cells were harvested for the analysis of Arf and Ink4a mRNA levels by real-time PCR using β-actin as a control. E: ChIP analysis of cyclin D1 binding to the Arf and Ink4a promoters in Dmp1+/+ and Dmp1−/− MEFs infected with lentivirus expressing HA-cyclin D1. Lysates derived from Dmp1+/+ and Dmp1−/− MEFs were immunoblotted for Dmp1 and HA. β-Actin was used as a loading control. The asterisk indicates a nonspecific band. F: ChIP analysis for the binding of endogenous cyclin D1 to the Arf and Ink4a promoters in MMTV-neu tumors. Tissue ChIP was conducted with formalin-fixed tumors from the MMTV-neu mice with wild-type Dmp1. The cyclin D1 protein was precipitated by two different antibodies to cyclin D1 (SP4 and H295). Data are means ± SD from n = 3 individual experiments (A–D). Ab, antibody; pro, promoter.
Consistently, we observed the recruitment of cyclin D1 on the Arf and Ink4a promoters in ChIP assays in wild-type MEFs infected with lentivirus expressing HA-tagged cyclin D1, but this binding was not detected in Dmp1-null MEFs (Figure 1E). The association of endogenous cyclin D1 with the Arf and Ink4a promoters was confirmed by ChIP assays using lysates from MMTV-neu mammary tumors that expressed high levels of both Dmp1 and cyclin D1 (Figure 1F).17,30 Collectively, these data suggest that cyclin D1 binds and activates both the Arf and Ink4a promoters, and this regulation depends on the presence of Dmp1.
Cyclin D1 Induces G2/M Cell-Cycle Delay and Apoptosis Mediated by Dmp1
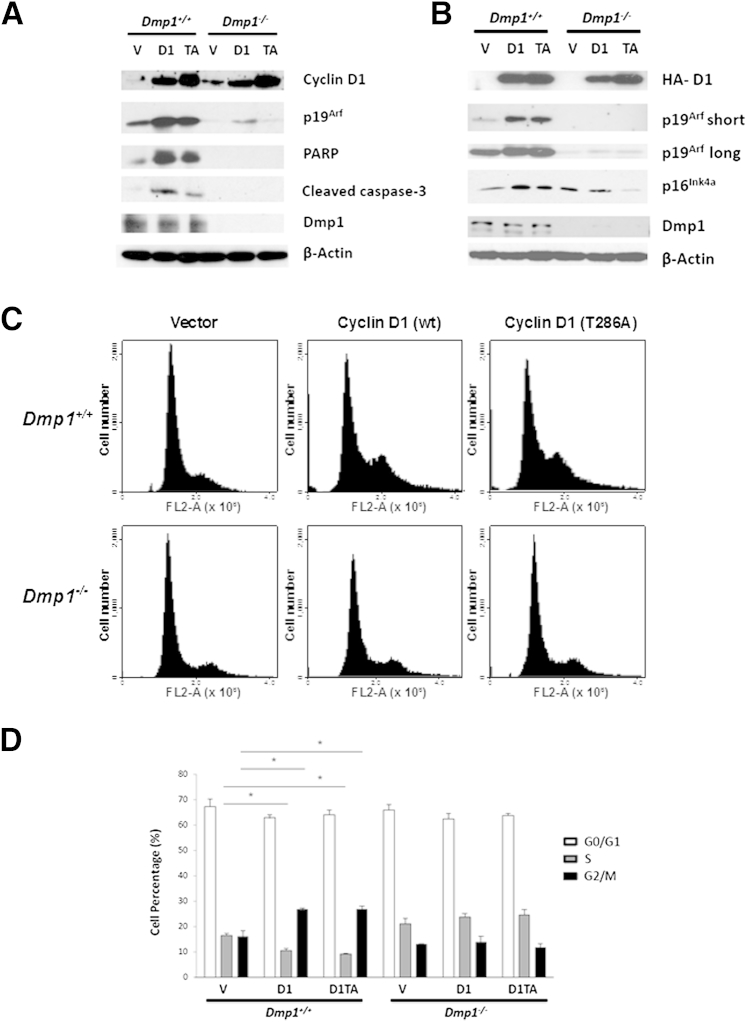
Both cyclin D1 and D1T286A increased p19Arf protein level when wild-type MEFs were starved in serum-free medium for 48 hours. Those cells underwent apoptosis as indicated by increased levels of cleaved PARP and caspase-3. However, there was no increase of p19Arf or induction of cleaved PARP and caspase-3 in Dmp1−/− MEFs on cyclin D1 or D1T286A overexpression (Figure 2A). We also stained the cells with annexin V and determined apoptotic cells by flow cytometry. Ectopic expression of cyclin D1 or D1T286A resulted in 20% increase of apoptosis in Dmp1+/+ MEFs, but this effect was not observed in Dmp1−/− MEFs (data not shown). Consistent with the aforementioned data, ectopic expression of cyclin D1 and D1T286A in wild-type MEFs by lentiviral infection promoted p19Arf expression. We also detected increased expression of p16Ink4a protein in those cells, although neither p19Arf nor p16Ink4a showed this response toward cyclin D1 in Dmp1−/− MEFs (Figure 2B).
Figure 2.
Overexpression of cyclin D1 or D1T286A induces apoptosis and G2/M phase delay in MEFs. A: Primary Dmp1+/+ and Dmp1−/− MEFs were infected with control retrovirus (V) or retrovirus expressing cyclin D1 (D1) or D1T286A (TA) protein. Cells were starved for 48 hours in serum-free medium and then harvested for immunoblot analysis of cyclin D1, p19Arf, PARP, cleaved caspase-3, and Dmp1. β-Actin was used as a loading control. B: Primary Dmp1+/+ and Dmp1−/− MEFs were infected with control lentivirus or lentivirus expressing HA-tagged cyclin D1 or D1T286A protein. Cells were harvested after 48 hours for immunoblot analysis of HA-cyclin D1, p19Arf, p16Ink4a, and Dmp1. β-Actin was used as a loading control. C: Representative images of cell-cycle profiles of Dmp1+/+ and Dmp1−/− MEFs infected by lentivirus carrying an empty vector or expressing cyclin D1 or D1T286A (T286A) and stained with propidium iodide 48 hours after infection. D: Quantification of flow cytometric analyses was from three independent experiments. Error bars indicate means ± SD. FL-2A, fluorescent pulse-area; wt, wild-type. ∗P < 0.05.
We further investigated the potential effect of cyclin D1 on cell-cycle profile. Ectopic expression of cyclin D1 and D1T286A through lentiviral infection led to G2/M phase delay of wild-type MEFs, but they only slightly increased the S phase fraction of Dmp1−/− MEFs (Figure 2, C and D), indicating that cyclin D1–mediated G2/M phase delay of the cell cycle depends on Dmp1.
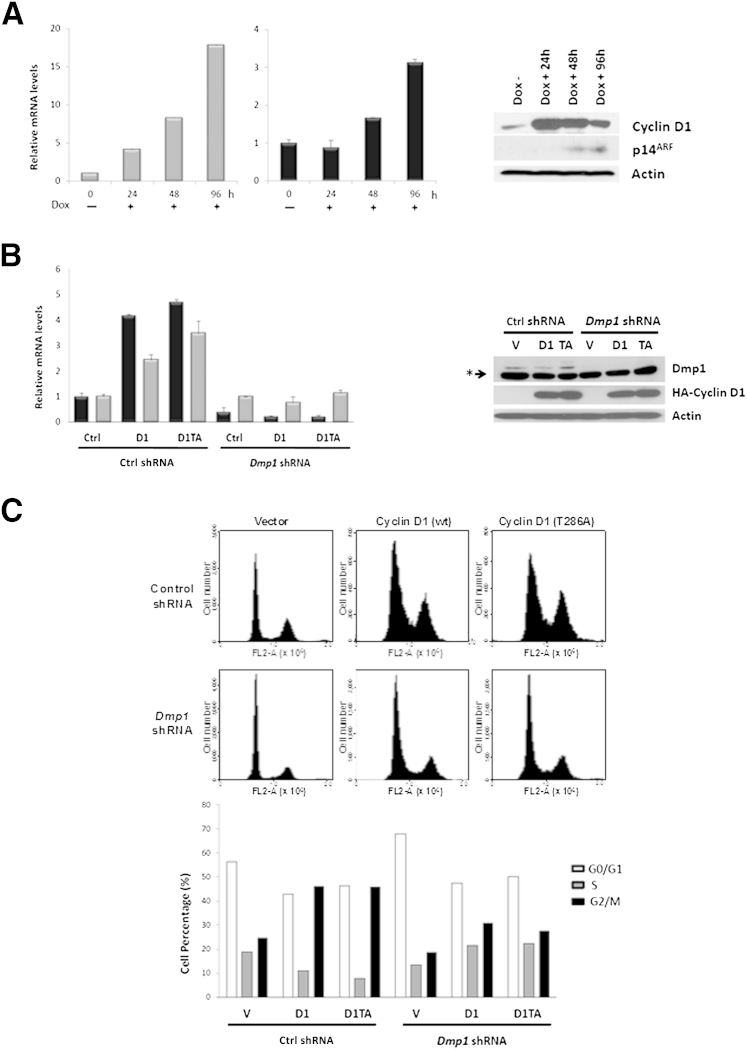
Similar to the findings in MEFs, both p14ARF and p16INK4a mRNA levels increased in human mammary epithelial cells when ectopic cyclin D1 was expressed by a doxycycline-inducible system (Figure 3A). The protein level of p14ARF was increased accordingly, whereas the p16INK4a level was too low to be detected (Figure 3A). With DMP1 knockdown by its shRNA15,16 in human mammary epithelial cells, ectopic cyclin D1 or D1T286A failed in altering p14ARF or p16INK4a transcription (Figure 3B), although the two cyclin D1 proteins were expressed at comparable levels despite Dmp1 expression (Figure 3B). Moreover, human mammary epithelial cells underwent G2/M phase delay on cyclin D1 and D1T286A overexpression, whereas DMP1 silencing abolished this effect (Figure 3C). Taken together, our data indicate that elevated cyclin D1 expression activates Arf/Ink4a genes in both mice and humans, and induces G2/M phase delay or apoptosis in a Dmp1-dependent fashion.
Figure 3.
Overexpression of cyclin D1 activates both p14ARF and p16INK4a in human mammary epithelial cells (HMECs). A: HMECs were infected with lentivirus expressing doxycycline (Dox)-induced cyclin D1. After puromycin selection, 0.5 μg/mL Dox was added, and cells were harvested after 0, 24, 48, and 96 hours, respectively. p14ARF (light gray bars) and p16INK4a (dark gray bars) mRNA levels measured by real-time PCR using β-actin as a control. Cyclin D1 and p14ARFexpression were analyzed by immunoblots with β-actin as a loading control. B: HMECs were manipulated to coexpress control or DMP1 shRNA (Ctrl) and cyclin D1 (D1) or D1T286A (D1TA) as indicated. Real-time PCR analysis of p14ARF (light gray bars) and p16INK4a (dark gray bars) mRNA levels after normalized against β-actin. Immunoblot analysis of Dmp1 and HA-tagged cyclin D1 expression. β-Actin was used as loading control. The asterisk indicates nonspecific bands. C: HMEC cells were treated as in B and stained with propidium iodide followed by flow cytometric analysis (top panel). Quantification of cell-cycle distribution is shown in the bottom panel.
Dmp1 Is Required for the Increased p19Arf/p16Ink4a Expression in Mammary Tumors of MMTV-D1 and D1T286A Mice
Because Dmp1 deletion was frequently detected (approximately 50%) in mammary tumors of MMTV-neu transgenic mice, whereas the Ink4a/Arf or p53 locus deletion was not observed,17 we next focused on the Dmp1 gene copy number study in mammary tumors of MMTV-D1 and MMTV-D1T286A mice. We found one Dmp1 allele deletion in two of nine mammary tumors from MMTV-D1 mice and 2 of 10 mammary tumors from MMTV-D1T286A mice (4 of 19, or 21%) (Supplemental Figure S2A). Significant induction of the p19Arf and p16Ink4a (2-fold to 18-fold) gene transcription was detected in more than half of Dmp1 wild-type mammary tumors compared to those in normal mammary glands. Markedly, among four tumors with one Dmp1 allele lost, two tumors showed significant down-regulation of p19Arf (Supplemental Figure S2B), and three exhibited very low p16Ink4a expression (Supplemental Figure S2C). A previous study discovered that the p53 pathway can be inactivated by mutations in MMTV-D1 and D1T286A tumors.27 To determine whether the increased p19Arf expression was due to p53 inactivation,28 we sequenced the region of the p53 gene encoding its DNA-binding domain. One MMTV-D1 tumor and one MMTV-D1T286A tumor had p53 mutations, whereas others with elevated p19Arf expression maintained wild-type p53. As expected, tumors with p53 mutations showed a dramatically increased level of p19Arf (Supplemental Figure S2B). In summary, these observations indicate that Dmp1 is essential for cyclin D1–activated p19Arf/p16Ink4a expression in mammary tumors of Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice.
Cyclin D1– and D1T286A-Induced Mammary Carcinogenesis Is Accelerated in Dmp1+/− Mice
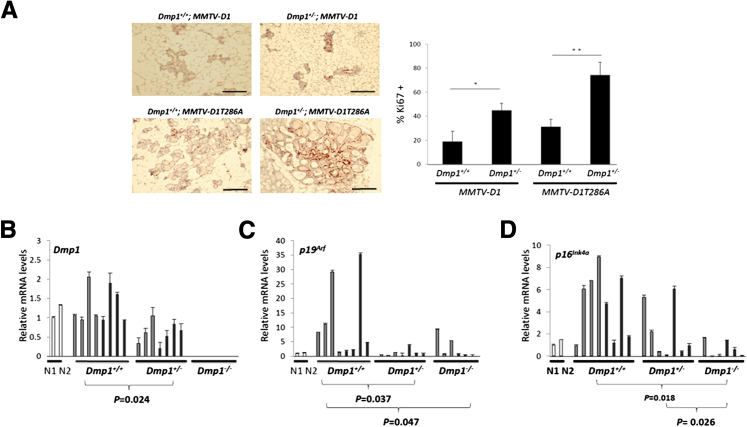
To study whether there is any cooperation between Dmp1 loss and cyclin D1 overexpression in vivo, we crossed MMTV-cyclin D1 and MMTV-D1T286A mice with Dmp1-null mice to generate the compound mice (Supplemental Figure S3). Consistent with a previous study,27 both cyclin D1 and D1T286A were highly expressed in mammary glands of the transgenic mice, and D1T286A showed increased nuclear intensity in mammary epithelial cells relative to these in the mice with wild-type cyclin D1 (Supplemental Figure S4A). We did not detect cyclin D1 or D1T286A expression in other organs. Dmp1 showed substantially reduced expression in the mammary glands of Dmp1+/− mice compared to these in wild-type mice (Supplemental Figure S4B). We next assessed the proliferation and p19Arf/p16Ink4a expression in pre-malignant mammary glands from female mice between the ages of 8 and 10 months. The percentages of Ki-67–positive cells were significantly increased in the mammary glands of Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice over these in Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice (Figure 4A). Consistent with our observation in mammary tumors (Supplemental Figure S2, B and C), both p19Arf and p16Ink4a mRNA levels were significantly increased (5-fold to 35-fold for p19Arf, 4-fold to 9-fold for p16Ink4a) in mammary glands of Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice compared to their counterparts without the CYCLIN D1 transgene. On the other hand, the p19Arf mRNA levels were significantly decreased in mammary glands from both Dmp1+/− and Dmp1−/− mice compared to those from Dmp1+/+ mice (Figure 4, B and C), whereas p16Ink4a showed marked reduction only in mammary glands from Dmp1−/− mice (Figure 4D). Taken together, these findings suggest that cyclin D1 overexpression in mammary glands up-regulates both p19Arf and p16Ink4a in Dmp1+/+ mice, but this regulation is compromised in Dmp1-deficient backgrounds.
Figure 4.
Mammary glands from Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice show lower proliferation and increased Arf/Ink4a expression. A: Mammary tissues derived from MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice showed increased proliferation at Dmp1 heterozygous background. Representative images of Ki-67 immunostaining (red) in mammary glands from Dmp1+/+; and Dmp1+/−; MMTV-cyclin D1, and MMTV-D1T286A mice. Nuclei were counterstained with hematoxylin (blue). Scale bars: 100 μm. Quantification of Ki-67–positive cells per mammary gland section is shown. Error bars indicate means ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01. B–D: Real-time PCR analysis of Dmp1, p19Arf, and p16Ink4a mRNA levels in mammary glands from MMTV-cyclin D1 and MMTV-D1T286A mice in Dmp1+/+, Dmp1+/−, and Dmp1−/− backgrounds. Data were normalized against β-actin mRNA levels. Gray columns represent the samples from MMTV-cyclin D1 mice and black columns represent the samples from MMTV-cyclin D1T286A mice. N, normal mammary gland from nontransgenic mice.
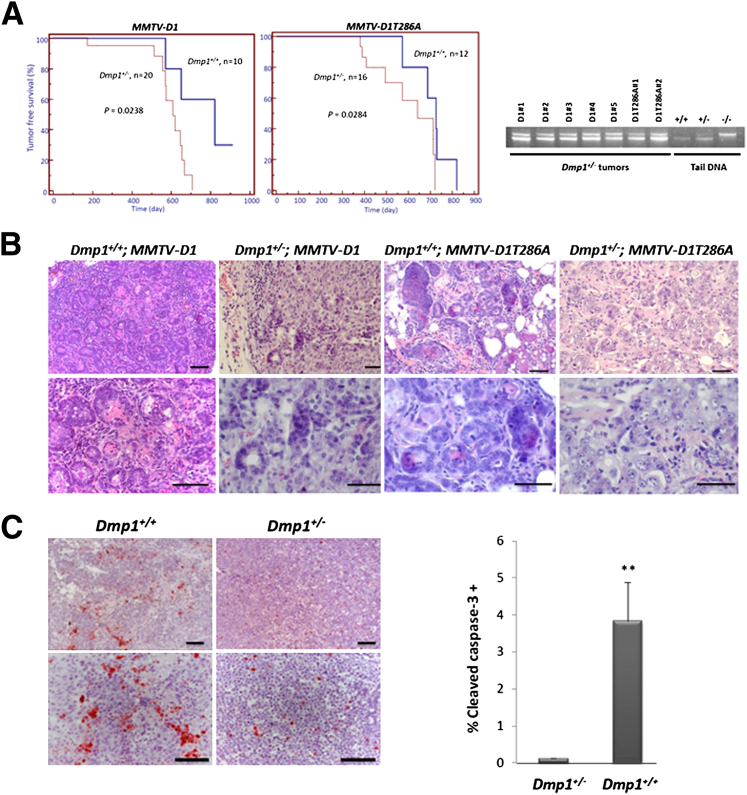
We established long-term murine cohorts to assess the effect of cyclin D1 overexpression and Dmp1 loss on tumor formation in these transgenic mice. We focused on mammary tumor development in Dmp1+/− mice because the Dmp1−/−;MMTV-cyclin D1 and Dmp1−/−;MMTV-D1T286A mice frequently developed lung carcinomas or had high incidence of deaths in the early months after birth for unknown reasons before they developed mammary tumors. This design was also clinically relevant, because human breast cancers showed hemizygous deletion of human DMP1 (hDMP1) in nearly half of the cases, whereas biallelic deletion of hDMP1 was rare.15 The cyclin D1– and D1T286A-induced mammary tumor development was significantly accelerated in Dmp1+/− mice, with an estimated median disease-free survival from 810 to 600 days (P = 0.0238) for wild-type cyclin D1, and from 730 to 645 days in D1T286A (P = 0.0284) (Figure 5A). There were higher percentages of Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice developing mammary tumors, with shorter survival times than those of the Dmp1 wild-type cohorts (Table 1). In genomic DNA analyses, mammary tumors of Dmp1+/− mice retained a wild-type Dmp1 allele in all of the seven tumors examined (Figure 5A), confirming the haploinsufficiency of Dmp1 in suppressing cyclin D1–driven tumor formation. Dmp1+/+; and Dmp1+/−; MMTV-cyclin D1 and MMTV-D1T286A mice developed mammary ductal adenocarcinomas. The mammary tumors from Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice showed squamous metaplasia and microinvasion as indicated by blurred epithelial–stromal interface in some glands and a corresponding tissue desmoplasia, whereas the tumors from Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice yielded a more aggressive phenotype as measured by an increased nuclear/cytoplasmic ratio, nuclear pleomorphism, and layers of disorganized epithelium (Figure 5B). Moreover, mammary tumors from Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mice displayed a significantly higher rate of apoptosis compared to those from Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice (Figure 5C), as indicated by cleaved caspase-3 staining (percentage of cleaved caspase-3–positive cells: 3.83% in Dmp1+/+ versus 0.11% in Dmp1+/−, P < 0.01).
Figure 5.
Cyclin D1– and D1T286A-induced mammary carcinogenesis is accelerated in Dmp1+/− mice. A: Tumor-free survival of Dmp1+/+;MMTV-cyclin D1 (blue) and Dmp1+/−;MMTV-cyclin D1 (red) compound transgenic mice (left panel) and Dmp1+/+;MMTV-DT286A (blue) and Dmp1+/−;MMTV-DT286A (red) compound transgenic mice (center panel). Cyclin D1–mediated-tumor development was significantly more accelerated in Dmp1+/− than in Dmp1+/+ genetic background. PCR confirmed retention of the wild-type Dmp1 locus in mammary carcinomas from Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice (right panel). MMTVD1, MMTV-cyclin D1. B: Representative images of mammary adenocarcinoma from Dmp1+/+;MMTV-cyclin D1 and Dmp1+/−;MMTV-cyclin D1 mice and Dmp1+/+;MMTV-D1T286A and Dmp1+/−;MMTV-D1T286A mice. Histological sections were stained with H&E. C: Representative images of immunostaining for cleaved caspase-3 (red) in mammary tumors from Dmp1+/+;MMTV-cyclin D1, Dmp1+/+;MMTV-D1T286A, Dmp1+/−;MMTV-cyclin D1, and Dmp1+/−;MMTV-D1T286A mice. Nuclei were counterstained with hematoxylin (blue). Scale bars: 100 μm. Quantification of caspase-3–positive area in the tumor sections. For each section, three independent areas were scanned and quantified using Image-Pro Plus software version 6.3.0. Error bars indicate means ± SD. ∗∗P < 0.01.
Table 1.
Tumor Characterization in Dmp1+/+; and Dmp1+/−; MMTV-D1 and MMTV-TA Mice
|
Dmp1+/+;MMTV-D1 |
Dmp1+/−;MMTV-D1 |
|
| Percentage of tumor-bearing mice | 40% (4/10) | 57.9% (11/19) |
| Mean survival time (months) | 21 | 19 |
| Tumor spectrum | ||
| Adenocarcinoma of breast | 3/4 | 9/11∗ |
| Lymphoma | 1/4 | 1/11 |
| Sarcoma |
0/4 |
1/11 |
|
Dmp1+/+;MMTV-TA |
Dmp1+/−;MMTV-TA |
|
| Percentage of tumor-bearing mice | 58.3% (7/12) | 62.6% (10/16) |
| Mean survival time (months) | 22.71 | 19.38 |
| Tumor spectrum | ||
| Adenocarcinoma of breast | 6/7 | 7/10† |
| Sarcoma | 1/7 | 1/10 |
| Lung carcinoma | 0/7 | 2/10 |
MMTV-D1, MMTV-cyclin D1; MMTV-TA, MMTV-D1T286A.
The tumors metastasized to other organs, including liver, ovary, and uterus, in four of nine Dmp1+/−;MMTV-cyclin D1 mice.
The tumors metastasized to other organs, including liver, ovary, and uterus, in one of seven Dmp1+/−;MMTV-D1T286A mice with mammary adenocarcinomas.
Most human breast carcinomas with high cyclin D1 levels also express ER, and a large portion of mammary tumors from MMTV-cyclin D1 and MMTV-D1T286A mice with intact Dmp1 are ER-positive.27 Thus, we wanted to determine whether cyclin D1–induced mammary tumors on the Dmp1+/− background retained ERα expression. Our immunohistochemical analyses revealed >50% of mammary tumors (12 of 21) from Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice were still ERα-positive (Supplemental Figure S5), suggesting that the loss of Dmp1 did not change the estrogen dependence of cyclin D1–induced mammary tumors.
Frequent Metastasis of Mammary Tumors in Dmp1+/−;MMTV-cyclin D1 Mice
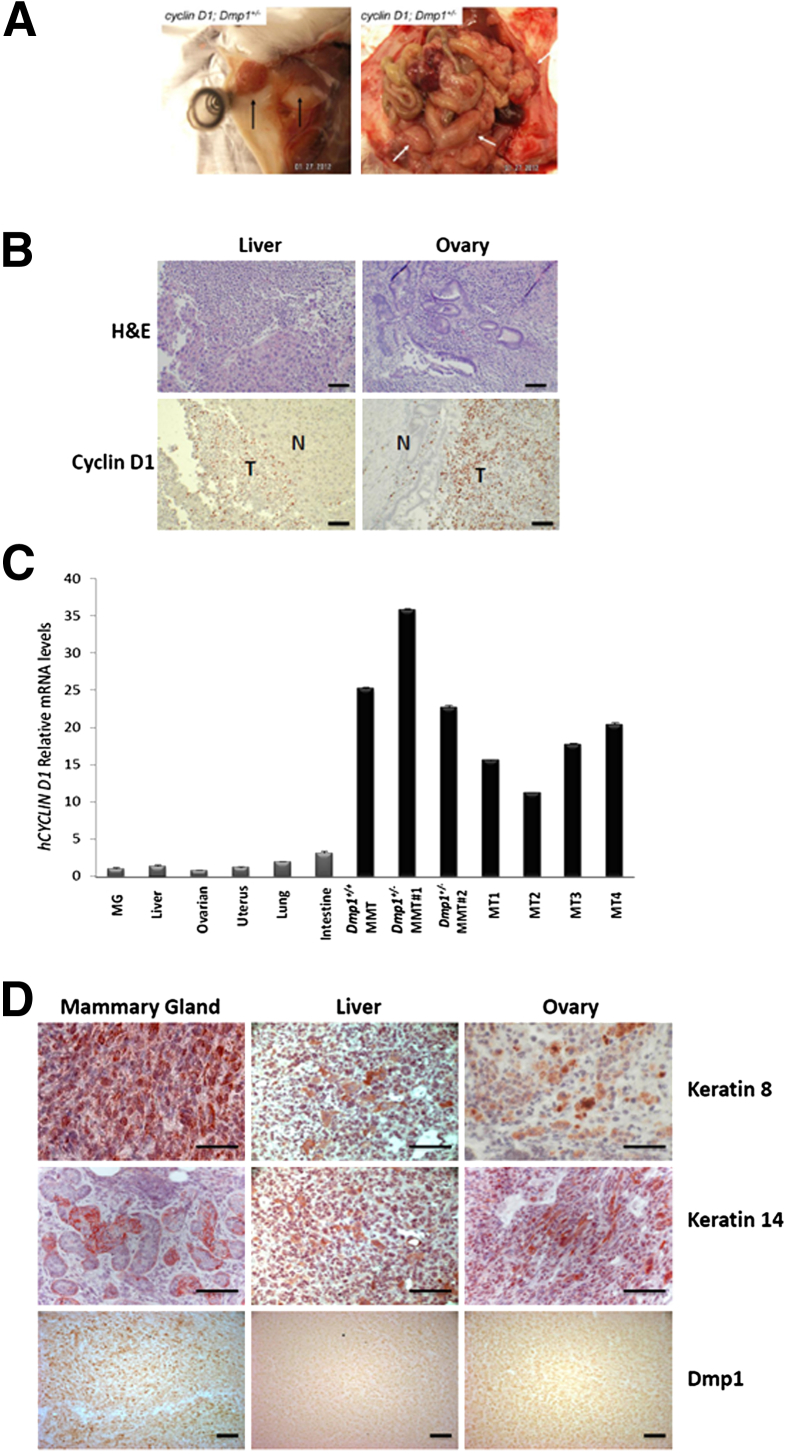
Apart from mammary adenocarcinomas, a small number of other tumor types, including lymphoma and sarcoma, were also found in both Dmp1+/+ and Dmp1+/− cohorts (Table 1). Of note, lung carcinomas were only found in Dmp1+/− mice, confirming the haploinsufficiency of Dmp1 in lung cancer suppression.16,31 Importantly, four of nine mammary tumors from Dmp1+/−;MMTV-cyclin D1 (multiparous) mice and one of seven tumors from Dmp1+/−;MMTV-D1T286A (nulliparous) mice metastasized to other organs, including liver, ovary, and uterus (Table 1 and Figure 6A). Intense cyclin D1 staining was detected in the metastatic tumors but not in adjacent normal tissues (Figure 6B). The high expression of human CYCLIN D1 transgene in both primary mammary and metastatic tumors was confirmed by real-time PCR studies (Figure 6C). Furthermore, both primary and metastatic tumors expressed cytokeratin 8 and 14 (Figure 6D), indicating that they derived from the same epithelial origin. Finally, we still detected low levels of Dmp1 in the metastatic tumors, as in primary tumors, suggesting that they still retained the one Dmp1 allele (Figure 6D). In summary, these results suggest that Dmp1 reduction due to its heterozygosity promotes the metastasis of cyclin D1– and D1T286A-initiated mammary tumors.
Figure 6.
Mammary tumors induced by cyclin D1 and D1T286A metastasize in Dmp1+/− mice. A: Photomicrographs of mammary and metastatic tumors from one Dmp1+/−;MMTV-cyclin D1 mouse; tumor cells disseminated throughout the body to other organs, including the uterus and ovaries. Left panel: primary mammary tumor (arrows). Right panel: metastasis of tumor cells (arrows) throughout the abdomen. B: Representative images of immunostaining for H&E and cyclin D1 (red) in metastatic tumors in liver and ovary from Dmp1+/−;MMTV-cyclin D1 mice. High cyclin D1 expression was detected only in the tumor region but not adjacent normal tissues. Scale bars: 100 μm. N, normal; T, tumor. C: Real-time PCR analyses of human cyclin D1 transgene mRNA levels in normal organs of nontransgenic mice and mammary/metastatic tumors in Dmp1+/+;MMTV-cyclin D1 and Dmp1+/−;MMTV-cyclin D1 mice. Data were normalized against that of β-actin. MG, mammary gland; MMT, mouse mammary tumor; MT, metastatic tumor. D: Representative images of immunostaining for keratin 8 (red), keratin 14 (red) and Dmp1 (red) in primary mammary tumors and metastatic tumors from liver and ovary, respectively, of Dmp1+/−;MMTV-cyclin D1 mice. Nuclei were counterstained with hematoxylin after keratin 8 and keratin 14 staining. Scale bars: 100 μm.
Discussion
It has been shown that Dmp1 is a physiological activator of the Arf promoter.13,31 The Arf promoter is transactivated by a variety of oncogenic signaling to prevent incipient cells from undergoing full transformation to cancers. It has been reported that Ets transcription factors play essential roles in activating the p16INK4a promoter through an Ets consensus in response to oncogenic Ras-Raf-MEK kinase signaling.32 Here, we showed that Dmp1 receives signals from cyclin D1 overexpression and binds to the Dmp1/Ets site on the Ink4a promoter, which is different from the Ets consensus characterized in the previous study. Thus the Ink4a gene is transactivated by different oncogenes through distinct Ets consensus sequences.
The Dmp1-binding site is within approximately 200 bp from the transcription initiation site of the murine Ink4a gene. Although Dmp1 directly binds to both mouse and human INK4a promoters, the affinity was lower than that in the Arf promoter. Of note, reduced affinity binding of Dmp1 has been reported on other promoters such as Areg,22 indicating that the extent of Dmp1 involvement in gene regulation is dependent on the sequences of the promoter, where CCCG(G/T)ATG(T/C) (Dmp1/Ets core is underlined) shows high affinity (eg, the Arf14, CD1318 promoters) and XXCG(G/T)ATGX (Dmp1/Ets core is underlined; X could be any nucleotide) has low affinity in Dmp1 binding (eg, the Ink4a, Areg promoters).
Previous studies from our group showed that Dmp1 regulates the Arf-p53 pathway by activating Arf14 and interacting with p53 to neutralize its antagonism by Mdm2.33 Although Dmp1 has been isolated as a cyclin D2–binding transcription factor, very little is known about its role in cyclin D–mediated signal transduction and tumorigenesis. Because cyclin D1 does not have a DNA-binding domain, it needs to interact with transcription factors to regulate gene expression. Dmp1 is the critical binding partner of Ink4a/Arf transcription. Both Arf and Ink4a promoters were transactivated by cyclin D1, which was confirmed by mRNA analyses of pre-malignant mammary glands from cyclin D1 and D1T286A transgenic mice. In the process of increased p19Arf and p16Ink4a expression and induction of cell-cycle arrest or apoptosis, wild-type cyclin D1 and its constitutively active mutant D1T286A had very similar effect, suggesting that high expression of wild-type cyclin D1 is sufficient to induce Arf and Ink4a responses in normal cells to quench potentially oncogenic signals. In the activation of both Ink4a/Arf promoters, cyclin D1–Dmp1 interaction was necessary for this process due to lack of DNA binding of cyclin D1. Consistently, the cyclin D1 mutant D1Δ142–253, deficient in interacting with Dmp1, did not activate these two promoters, and cyclin D1 did not bind to these promoters in Dmp1-deficient cells. Thus, we conclude that Dmp1 plays critical roles in Arf and Ink4a gene activation in response to cyclin D1 overexpression. The fact that the MMTV-cyclin D1 and D1T286A mice eventually developed mammary tumors is possibly due to loss of Dmp1 or overexpression of Ink4a/Arf repressors (such as Bmi1, Tbx2/3, Twist, Pokemon) during tumor development. Further studies are needed to clarify the roles of these repressors in cyclin D1–driven mammary tumorigenesis.
We found that endogenous cyclin D1 bound to endogenous DMP1 and E2F1 in MCF7 breast cancer cells (DMP1 is shown in Supplemental Figure S6; E2F1 data not shown). C/EBPβ is known to bind to cyclin D1,4 and our data showed that cyclin D1 did not activate the Arf promoter with deleted C/EBP-consensus sequence (data not shown). Thus, we cannot exclude the possibility that other transcriptional factors are also involved in cyclin D1–mediated activation of the Arf promoter. Recent studies indicated that cyclin D1 interacts with transcription factors including NF-Y, STAT, CREB2, ELK1, ZNF423, and CUX1,11 and we also identified their consensus sequences in the Arf/Ink4a promoters (data not shown). Because our data showed the Dmp1-dependent Arf/Ink4a activation, future studies will be needed to determine whether Dmp1 collaborates with C/EBPs or other transcription factors in regulating Arf/Ink4a expression.
Our results showed the levels of p21Cip1 induction did not change significantly between Dmp1+/+ and Dmp1−/− MEFs, consistent with a previous study that Arf-induced cell-cycle arrest independent of p21Cip1.34 ARF directly binds to DP1 and inhibits DP1-E2F1 interaction.35 The activity of E2F1 can also be inhibited by INK4a.14 Therefore, it would be needed to study the roles of E2F1 and its target genes in cyclin D1 overexpression-induced cell-cycle arrest.
We showed that both cyclin D1– and D1T286A-driven mammary carcinogenesis was accelerated in Dmp1+/− mice. Importantly, mammary tumors from Dmp1+/− mice exhibited significantly increased Ki-67 in comparison to those from Dmp1+/+ mice because Ki-67 is one of the target genes for E2Fs,36 suggesting that the activity of E2Fs stimulated by cyclin D1 is suppressed by Dmp1. Our study showed that the Dmp1 locus was deleted in 4 of 19 (21%) mammary tumors from MMTV-cyclin D1 and MMTV-D1T286A mice, in which neither Ink4a/Arf nor p53 deletion was observed. Frequent deletion of one Dmp1 allele was also observed in MMTV-neu tumors,17 suggesting the critical role of Dmp1-loss in cyclin D1– and neu-induced mammary tumorigenesis. The current study showed that the p16Ink4a was only significantly down-regulated in Dmp1−/− mammary glands, but not in Dmp1+/− tissues, in comparison to Dmp1+/+ samples, whereas p19Arf was significantly down-regulated in both Dmp1+/− and Dmp1−/− mammary glands. Similar findings were observed in mammary tumors from MMTV-neu mice as well as in the lungs from three different Dmp1 backgrounds.16,17 The difference of p16Ink4a expression in Dmp1+/− background from that in Dmp1−/− background may explain more accelerated tumor development in Dmp1−/− mice than that in Dmp1+/− mice although the difference was not statistically significant.13,16
Given the link between Ras signaling and Dmp1, it is noteworthy that neither overexpression of cyclin D1 nor hemizygous loss of Ink4a and Arf accelerated tumorigenesis in MMTV-ErbB2 mice.37 The above study is consistent with a previous report that HER2/neu-driven mammary carcinogenesis was not observed in cyclin D1–null mice,38 suggesting that cyclin D1 is a critical target of HER2/neu/ErbB2 in promoting mammary tumorigenesis. Our published study showed that both Ink4a and Arf inductions in response to HER2/neu overexpression were markedly attenuated (>80%) in mammary tissue from Dmp1−/− mice.17 Our current study also showed the reduced p16Ink4a/p19Arf mRNA levels in pre-malignant mammary tissues from MMTV-cyclin D1 and D1T286A mice in both Dmp1+/− and Dmp1−/− backgrounds compared to that in Dmp1 wild-type mice (Figure 4, C and D). The above data explain the difference between no acceleration of cyclin D1–induced mammary tumor developed in Ink4a/Arf+/− mice37 and the significantly accelerated carcinogenesis in Dmp1+/− mice (Figure 5A). Meanwhile, the data also suggest that other targets of Dmp1 may be involved in cyclin D1–induced mammary carcinogenesis.
Our study also showed the significantly increased metastasis of keratin-positive cyclin D1–induced mammary tumors in Dmp1+/− mice in comparison to Dmp1+/+ mice. Most cyclin D1 tumors were adenosquamous ductal carcinomas that metastasized to liver, ovary, uterus, and intestines, although we did not see any brain/bone metastasis that is commonly found in human breast cancer patients. Nevertheless, our results imply an important role of Dmp1 in preventing the mammary tumor metastasis induced by cyclin D1 and provide a potential mechanism of breast cancer metastasis. Currently, the frequency of human breast cancer with high cyclin D1 overexpression with or without hDMP1 deletion, their prognostic values, and correlation with clinical stages of patients are unknown. Thus, it will be necessary to analyze human breast cancer samples for the correlation between cyclin D1/Dmp1 alterations and patient survival data to determine the clinical values of our discovery.
Acknowledgments
We thank Dr. Robert Weinberg for HMEC cells, Drs. Charles Sherr and Martin Roussel for providing DNA constructs, Dr. Emmett V. Schmitt for MMTV-D1 mice, Dr. J. Alan Diehl for MMTV-TA mice, Dr. Eiji Hara for human p16INK4a promoter constructs, Dr. Gustavo Leone for E2f1/2/3 triple knockout MEFs, Megan J. Whelen for editing, and Dejan Maglic for helpful discussions.
Footnotes
Supported by ACSRSG-07-207-01-MGO, 5R01CA106314, and Director’s Challenge Award #20595 from Wake Forest University Health Sciences (K.I.); R01CA118329 (G.K.); ACS116403-RSG-09-082-01-MGO and 5R01CA106314 (G.S.); institutional grant IRSC-GTS: 37540 (S.Z.); and the Susan G. Komen Foundation postdoctoral fellowship KG080179 (P.T.). The Cell and Virus Vector Core Laboratory of the Comprehensive Cancer Center at Wake Forest University Health Sciences provided access to cell culture materials.
Contributor Information
Guangchao Sui, Email: gsui@wakehealth.edu.
Kazushi Inoue, Email: kinoue2@triad.rr.com.
Supplemental Data
Dmp1 binds and activates the p16 promoter. A: The murine p16Ink4a promoter (pro). The nucleotide sequence of −316 bp 5′ to the transcriptional start site (+1) is shown. Putative binding site for DMP1/Ets/Elk-1 is underlined. B: Electrophoretic mobility shift assays were performed with a recombinant DMP1 protein. Lane 1: 32P-labeled probe containing binding site for DMP1 on Ink4a or Arf promoter, lane 2: probe with 5 μL of DMP1; lane 3: probe with DMP1 and 50 fold of nonspecific competitor; and lane 4: probe with DMP1 and 50-fold of unlabeled probe. C: NIH 3T3 cells were cotransfected with increased amount of Dmp1 and the luciferase reporter constructs encoding wild-type p16Ink4a (−400) promoter or its deletion mutant or point mutant (A to T) deficient in binding to Dmp1. The numbers show the fold activation of the luciferase reporters normalized by internal control. D: Electrophoretic mobility shift assays were performed with radiolabeled probes containing Dmp1-binding consensus on human INK4a promoter using recombinant DMP1 made in insect Sf9 cells. Lanes 1 and 7: probe with 5 μL of DMP1; lanes 2 and 8: probe with DMP1 and 50-fold of nonspecific competitor; lanes 3 and 9: probe with DMP1 and 50-fold of unlabeled probe; lanes 4 and 10: probe with DMP1 and nonimmune rabbit serum; and lanes 5, 6, 11, and 12: probe with DMP1 and different antibodies against DMP1. E: NIH 3T3 cells were cotransfected with the luciferase reporter construct encoding −3000 p16INK4a promoter and increased amount of Dmp1 using endocrine alkaline phosphatase as an internal control.
Specific deletion of Dmp1 and expression of Arf/Ink4a mRNA in Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mammary tumors. A: Real-time PCR analysis of the Dmp1 copy numbers showing hemizygous Dmp1 deletion in 20% of Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mammary tumors. HZ, hemizygous. B and C: Expression of the Arf (B) and Ink4a (C) mRNA in Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mammary tumors analyzed by real-time PCR. β-Actin was used as an internal control.
Establishment of Dmp1+/−; and Dmp1−/−; MMTV-cyclin D1 and MMTV-D1T286A compound mice. A male MMTV-cyclin D1 or MMTV-D1T286A mouse was crossed with Dmp1+/− females to obtain Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice. Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A compound transgenic mice then were further crossed with Dmp1+/− mice to obtain >15 mice with each genetic background. Littermate wild-type mice were used as controls.
Expression of cyclin D1 and Dmp1 proteins in mammary glands from Dmp1+/+; and Dmp1+/−; MMTV-cyclin D1 and MMTV-D1T286A mice. Mammary glands were isolated from 8- to 10-month-old females from each group of cohorts, and were stained for cyclin D1 (A) and Dmp1 (B). Scale bars: 100 μm.
Expression of ERα in mammary tumors from Dmp1+/+; and Dmp1+/−; MMTV-cyclin D1 and MMTV-D1T286A mice. Representative images of immunostaining for ERα (red) in mammary tumors from Dmp1+/+;MMTV-cyclin D1, Dmp1+/+;MMTV-D1T286A, Dmp1+/−;MMTV-cyclin D1, and Dmp1+/−;MMTV-D1T286A mice. Nuclei were counterstained with hematoxylin (blue). Scale bars: 50 μm.
Endogenous hDMP1-Cyclin D1 binding assay in MCF7 breast cancer cells. Endogenous cyclin D1-hDMP1 binding assay was conducted using 1 mg of total MCF7 cell lysates. Approximately 10% of hDMP1 was immunoprecipitated with the cyclin D1 antibody and 1% to 2% of cyclin D1 was immunoprecipitated with the Dmp1 antibodies.
References
- 1.Sutherland R.L., Musgrove E.A. Cyclins and breast cancer. J Mammary Gland Biol. 2004;9:95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- 2.Arnold A., Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 3.Fu M.F., Wang C.G., Li Z.P., Sakamaki T., Pestell R.G. Minireview: cyclin D1: normal and abnormal functions. Endocrinol. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 4.Lamb J., Ramaswamy S., Ford H.L., Contreras B., Martinez R.V., Kittrell F.S., Zahnow C.A., Patterson N., Golub T.R., Ewen M.E. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 5.Musgrove E.A., Caldon C.E., Barraclough J., Stone A., Sutherland R.L. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 6.Zwijsen R.M., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R.J. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 7.Knudsen K.E., Cavenee W.K., Arden K.C. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59:2297–2301. [PubMed] [Google Scholar]
- 8.Horstmann S., Ferrari S., Klempnauer K.H. Regulation of B-Myb activity by cyclin D1. Oncogene. 2000;19:298–306. doi: 10.1038/sj.onc.1203302. [DOI] [PubMed] [Google Scholar]
- 9.Fu M.F., Wang C.G., Rao M., Wu X.F., Bouras T., Zhang X.P., Li Z.P., Jiao X.M., Yang J.G., Li A.P., Perkins N.D., Thimmapaya B., Kung A.L., Munoz A., Giordano A., Lisanti M.P., Pestell R.G. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280:29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 10.Rubio M.F., Fernandez P.N., Alvarado C.V., Panelo L.C., Grecco M.R., Colo G.P., Martínez-Noel G.A., Micenmacher S.M., Costas M.A. Cyclin D1 is a NF-κB corepressor. Biochim Biophys Acta. 2012;1823:1119–1131. doi: 10.1016/j.bbamcr.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Bienvenu F., Jirawatnotai S., Elias J.E., Meyer C.A., Mizeracka K., Marson A., Frampton G.M., Cole M.F., Odom D.T., Odajima J., Geng Y., Zagozdzon A., Jecrois M., Young R.A., Liu X.S., Cepko C.L., Gygi S.P., Sicinski P. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue K., Wen R., Rehg J.E., Adachi M., Cleveland J.L., Roussel M.F., Sherr C.J. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue K., Zindy F., Randle D.H., Rehg J.E., Sherr C.J. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Gene Dev. 2001;15:2934–2939. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue K., Roussel M.F., Sherr C.J. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci U S A. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maglic D., Zhu S., Fry E.A., Taneja P., Kai F., Kendig R.D., Sugiyama T., Miller L.D., Willingham M.C., Inoue K. Prognostic value of the hDMP1-ARF-Hdm2-p53 pathway in breast cancer. Oncogene. 2012 doi: 10.1038/onc.2012.423. [Epub ahead of press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallakin A., Sugiyama T., Taneja P., Matise L.A., Frazier D.P., Choudhary M., Hawkins G.A., D’Agostino R.B., Willingham M.C., Inoue K. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007;12:381–394. doi: 10.1016/j.ccr.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taneja P., Maglic D., Kai F., Sugiyama T., Kendig R.D., Frazier D.P., Willingham M.C., Inoue K. Critical roles of DMP1 in human epidermal growth factor receptor 2/neu-Arf-p53 signaling and breast cancer development. Cancer Res. 2010;70:9084–9094. doi: 10.1158/0008-5472.CAN-10-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue K., Sherr C.J., Shapiro L.H. Regulation of the CD13/aminopeptidase N gene by DMP1, a transcription factor antagonized by D-type cyclins. J Biol Chem. 1998;273:29188–29194. doi: 10.1074/jbc.273.44.29188. [DOI] [PubMed] [Google Scholar]
- 19.Sreeramaneni R., Chaudhry A., McMahon M., Sherr C.J., Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diehl J.A., Zindy F., Sherr C.J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 21.Inoue K., Sherr C.J. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallakin A., Sugiyama T., Kai F., Taneja P., Kendig R.D., Frazier D.P., Maglic D., Matise L.A., Willingham M.C., Inoue K. The Arf-inducing transcription factor Dmp1 encodes a transcriptional activator of amphiregulin, thrombospondin-1, JunB and Egr1. Int J Cancer. 2010;126:1403–1416. doi: 10.1002/ijc.24938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taneja P., Mallakin A., Matise L.A., Frazier D.P., Choudhary M., Inoue K. Repression of Dmp1 and Arf transcription by anthracyclins: critical roles of the NF-kappa B subunit p65. Oncogene. 2007;26:7457–7466. doi: 10.1038/sj.onc.1210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallakin A., Taneja P., Matise L.A., Willingham M.C., Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its regulation by E2Fs. Oncogene. 2006;25:7703–7713. doi: 10.1038/sj.onc.1209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirai H., Sherr C.J. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16:6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T.C., Cardiff R.D., Zukerberg L., Lees E., Arnold A., Schmidt E.V. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 27.Lin D.I., Lessie M.D., Gladden A.B., Bassing C.H., Wagner K.U., Diehl J.A. Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene. 2008;27:1231–1242. doi: 10.1038/sj.onc.1210738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherr C.J. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 29.Han E.K., Ng S.C., Arber N., Begemann M., Weinstein I.B. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis. 1999;4:213–219. doi: 10.1023/a:1009618824145. [DOI] [PubMed] [Google Scholar]
- 30.Lee R.J., Albanese C., Fu M.F., D’Amico M., Lin B., Watanabe G., Haines G.K., Siegel P.M., Hung M.C., Yarden Y., Horowitz J.M., Muller W.J., Pestell R.G. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20:672–683. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue K., Mallakin A., Frazier D.P. Dmp1 and tumor suppression. Oncogene. 2007;26:4329–4335. doi: 10.1038/sj.onc.1210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohtani N., Zebedee Z., Huot T.J., Stinson J.A., Sugimoto M., Ohashi Y., Sharrocks A.D., Peters G., Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 33.Frazier D.P., Kendig R.D., Kai F., Maglic D., Sugiyama T., Morgan R.L., Fry E.A., Lagedrost S.J., Sui G., Inoue K. Dmp1 physically interacts with p53 and positively regulates p53’s stability, nuclear localization, and function. Cancer Res. 2012;72:1740–1750. doi: 10.1158/0008-5472.CAN-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modestou M., Puig-Antich V., Korgaonkar C., Eapen A., Quelle D.E. The alternative reading frame tumor suppressor inhibits growth through p21-dependent and p21-independent pathways. Cancer Res. 2001;61:3145–3150. [PubMed] [Google Scholar]
- 35.Datta A., Sen J., Hagen J., Korgaonkar C.K., Caffrey M., Quelle D.E., Hughes D.E., Ackerson T.J., Costa R.H., Raychaudhuri P. ARF directly binds DP1: interaction with DP1 coincides with the G(1) arrest function of ARF. Mol Cell Biol. 2005;25:8024–8036. doi: 10.1128/MCB.25.18.8024-8036.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zambon A.C. Characterization of the human Ki67 promoter for high-throughput functional genomic screens of G1-S phase cell-cycle regulation (abstract) FASEB J. 2008;22:636.3. [Google Scholar]
- 37.Yang C.W., Ionescu-Tiba V., Burns K., Gadd M., Zukerberg L., Louis D.N., Sgroi D., Schmidt E.V. The role of the cyclin D1-dependent kinases in ErbB2-mediated breast cancer. Am J Pathol. 2004;164:1031–1038. doi: 10.1016/S0002-9440(10)63190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Q., Geng Y., Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dmp1 binds and activates the p16 promoter. A: The murine p16Ink4a promoter (pro). The nucleotide sequence of −316 bp 5′ to the transcriptional start site (+1) is shown. Putative binding site for DMP1/Ets/Elk-1 is underlined. B: Electrophoretic mobility shift assays were performed with a recombinant DMP1 protein. Lane 1: 32P-labeled probe containing binding site for DMP1 on Ink4a or Arf promoter, lane 2: probe with 5 μL of DMP1; lane 3: probe with DMP1 and 50 fold of nonspecific competitor; and lane 4: probe with DMP1 and 50-fold of unlabeled probe. C: NIH 3T3 cells were cotransfected with increased amount of Dmp1 and the luciferase reporter constructs encoding wild-type p16Ink4a (−400) promoter or its deletion mutant or point mutant (A to T) deficient in binding to Dmp1. The numbers show the fold activation of the luciferase reporters normalized by internal control. D: Electrophoretic mobility shift assays were performed with radiolabeled probes containing Dmp1-binding consensus on human INK4a promoter using recombinant DMP1 made in insect Sf9 cells. Lanes 1 and 7: probe with 5 μL of DMP1; lanes 2 and 8: probe with DMP1 and 50-fold of nonspecific competitor; lanes 3 and 9: probe with DMP1 and 50-fold of unlabeled probe; lanes 4 and 10: probe with DMP1 and nonimmune rabbit serum; and lanes 5, 6, 11, and 12: probe with DMP1 and different antibodies against DMP1. E: NIH 3T3 cells were cotransfected with the luciferase reporter construct encoding −3000 p16INK4a promoter and increased amount of Dmp1 using endocrine alkaline phosphatase as an internal control.
Specific deletion of Dmp1 and expression of Arf/Ink4a mRNA in Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mammary tumors. A: Real-time PCR analysis of the Dmp1 copy numbers showing hemizygous Dmp1 deletion in 20% of Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mammary tumors. HZ, hemizygous. B and C: Expression of the Arf (B) and Ink4a (C) mRNA in Dmp1+/+;MMTV-cyclin D1 and Dmp1+/+;MMTV-D1T286A mammary tumors analyzed by real-time PCR. β-Actin was used as an internal control.
Establishment of Dmp1+/−; and Dmp1−/−; MMTV-cyclin D1 and MMTV-D1T286A compound mice. A male MMTV-cyclin D1 or MMTV-D1T286A mouse was crossed with Dmp1+/− females to obtain Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A mice. Dmp1+/−;MMTV-cyclin D1 and Dmp1+/−;MMTV-D1T286A compound transgenic mice then were further crossed with Dmp1+/− mice to obtain >15 mice with each genetic background. Littermate wild-type mice were used as controls.
Expression of cyclin D1 and Dmp1 proteins in mammary glands from Dmp1+/+; and Dmp1+/−; MMTV-cyclin D1 and MMTV-D1T286A mice. Mammary glands were isolated from 8- to 10-month-old females from each group of cohorts, and were stained for cyclin D1 (A) and Dmp1 (B). Scale bars: 100 μm.
Expression of ERα in mammary tumors from Dmp1+/+; and Dmp1+/−; MMTV-cyclin D1 and MMTV-D1T286A mice. Representative images of immunostaining for ERα (red) in mammary tumors from Dmp1+/+;MMTV-cyclin D1, Dmp1+/+;MMTV-D1T286A, Dmp1+/−;MMTV-cyclin D1, and Dmp1+/−;MMTV-D1T286A mice. Nuclei were counterstained with hematoxylin (blue). Scale bars: 50 μm.
Endogenous hDMP1-Cyclin D1 binding assay in MCF7 breast cancer cells. Endogenous cyclin D1-hDMP1 binding assay was conducted using 1 mg of total MCF7 cell lysates. Approximately 10% of hDMP1 was immunoprecipitated with the cyclin D1 antibody and 1% to 2% of cyclin D1 was immunoprecipitated with the Dmp1 antibodies.