Abstract
Membrane-associated serine protease matriptase is widely expressed by epithelial/carcinoma cells in which its proteolytic activity is tightly controlled by the Kunitz-type protease inhibitor, hepatocyte growth factor activator inhibitor (HAI-1). We demonstrate that, although matriptase is not expressed in lymphoid hyperplasia, roughly half of the non-Hodgkin B-cell lymphomas analyzed express significant amounts of matriptase. Furthermore, a significant proportion of these tumors express matriptase in the absence of HAI-1. Aggressive Burkitt lymphoma was more likely than indolent follicular lymphoma to express matriptase alone (86% versus 36%). In the absence of significant HAI-1 expression, the lymphoma cells activate and shed active matriptase when the cells are stimulated with mildly acidic buffer or the hypoxia-mimicking agent, CoCl2. The shed active matriptase can initiate pericellular proteolytic cascades by activating urokinase-type plasminogen activator on the cell surface of monocytes, and it can activate prohepatocyte growth factor. In addition, matriptase knockdown suppressed proliferation and colony-forming ability of neoplastic B cells in culture and growth as tumor xenografts in mice. Furthermore, exogenous expression of HAI-1 significantly suppressed proliferation of neoplastic B cells. These studies suggest that dysregulated pericellular proteolysis as a result of unregulated matriptase expression with limited HAI-1 may contribute to the pathological characteristics of several human B-cell lymphomas through modulation of the tumor microenvironment and enhanced tumor growth.
Pericellular proteolysis plays crucial roles in the modulation of the tumor microenvironment through activation of cytokines and growth factors, remodeling of the extracellular matrix (ECM), and release of sequestered growth factors and cytokines from the ECM.1 Matriptase, a type II transmembrane serine protease, has recently been recognized as an important pericellular protease that may affect tumor microenvironments through the initiation of a protease cascade and the activation of growth factors.2–4 Matriptase and its cognate inhibitor, hepatocyte growth factor (HGF) activator inhibitor (HAI)-1, are broadly co-expressed in epithelial tissues,5,6 where critical interactions between the protease and the inhibitor are required for the maintenance of the integrity of the epithelium, epidermal differentiation and barrier functions, and the development of the placenta.7–9 Both matriptase and HAI-1 are commonly dysregulated in human tumors of epithelial origin and may contribute to the development and progression of carcinomas.10 Mild overexpression of matriptase in the absence of a parallel increase in HAI-1 expression in mouse skin upsets a tightly regulated balance between these proteins in favor of increased proteolysis, which results in the spontaneous development of squamous cell carcinomas and enhances sensitivity to chemical carcinogens.11 HAI-1, a Kunitz-type serine protease inhibitor, modulates matriptase proteolytic activity through the formation of extremely stable complexes with active matriptase, thereby avoiding undesired proteolysis and the potential toxicity of active matriptase.12,13 The inhibition of unregulated matriptase activation by HAI-1 appears to be important for the biosynthesis, intracellular trafficking, and even zymogen activation of matriptase.14,15
Although most matriptase studies have focused on epithelial and carcinoma cells, in which HAI-1 plays a pivotal role in intracellular trafficking, zymogen activation, and inhibition of matriptase, growing evidence has shown that altered matriptase expression or regulation may be important in hematological cells and neoplasms. Matriptase expression has been detected in THP-1 human monocytic cells, and the protease was reported to be responsible for accelerating plasmin generation via activation of urokinase plasminogen activator (uPA).16 Matriptase has also been reported to be expressed by, and involved in, the activation of peritoneal macrophages through the activation of macrophage-stimulating protein-1 and the recepteur d'origine nantais.17 Matriptase was also detected in two Burkitt lymphoma (BL) cells (Daudi and ST 486) in our previous study18 and, more recently, in human leukemia.19 In contrast to the situation in epithelial/carcinoma cells, these hematological cells express no or low levels of HAI-1. Regulation and even function of matriptase in hematological cells and tumors may, therefore, be different from that in epithelial and carcinoma cells. In the current study, we set out to investigate the role and regulation of matriptase in human B-cell lymphomas. Our data show that matriptase is expressed in a variety of non-Hodgkin B-cell lymphomas in the absence of, or with limited, HAI-1 expression, with important implications for tumor behavior.
Materials and Methods
Chemicals and Reagents
Gelatin and cobalt chloride (CoCl2) were purchased from Sigma-Aldrich (St. Louis, MO); N-tert-butoxycarbonyl-Gln-Ala-Arg-7-amido-4-methylcoumarin was obtained from Enzo Life Sciences (Farmingdale, NY); pyro-Glu-Gly-Arg-p-nitroaniline hydrochloride (p-NA) for uPA was from Aniara (Hyphen BioMed, West Chester, OH); and human pro-uPA was purchased from American Diagnostica Inc. (Stamford, CT). Silver stain plus was obtained from Bio-Rad (Hercules, CA).
Cell Lines and shRNA Knockdown
Hematological and breast cancer cells were obtained from the Tissue Culture Shared Resource in the Lombardi Comprehensive Cancer Center, Georgetown University (Washington, DC). The hematological cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. The T-47D human breast carcinoma cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS. All cells were maintained at 37°C in a humidified atmosphere with 5% CO2. The human BL cells Raji and diffuse large B-cell lymphoma (DLBCL) cells SU-DHL-4 were used to establish stable matriptase knockdown pools. The cells were transduced with lentiviral vectors expressing either a nontargeting control shRNA sequence (NT) or shRNAs targeting the human matriptase gene (ST14) (Open Biosystems, Lafayette, CO). The shRNA targeting sequences were as follows: NT, 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′; MTP 1, 5′-CCGGCGTGTCCAGAAGGTCTTCAATCTCGAGATTGAAGACCTTCTGGACACGTTTTTG-3′; and MTP 2, 5′-CCGGCAATGACTTCACCTTCGACTACTCGAGTAGTCGAAGGTGAAGTCATTGTTTTTG-3′. Transduced cells were selected with 1 μg/mL puromycin, and stable pools of resistant cells were established. Knockdown of matriptase expression was confirmed by using Western blot analysis. The doxycycline-inducible HAI-1–expressing cell line was generated using a Lenti-X Tet-On 3G inducible expression system from Clontech Laboratories, Inc. (Mountain View, CA). Briefly, the open reading frame of the HAI-1 gene (SPINT-1) was generated by PCR amplification and ligated into the pLVX-TER-IRES expression vector, along with the puromycin-resistant marker. This lentiviral expression vector was stably transduced into a preselected neomycin-resistant BL Raji line that stably expressed Tet-On 3G–transactivated regulator proteins. Transduced cells were selected in the presence of 1 μg/mL puromycin, and stable pools of resistant cells were established. Cell pools derived from this dual selection process were then selected on the basis of induction of HAI-1 expression in the presence of 2 μg/mL tetracycline analogue, doxycycline. Cell extracts of stable clones were analyzed by using Western blot analysis with M19 monoclonal antibody (mAb).
IHC Data
The study was reviewed and approved as being of minimal to no risk by the Institutional Review Board at the University of Maryland, School of Medicine (Baltimore), and the University of Texas MD Anderson Cancer Center (Houston). Lymphoma tissue and nonneoplastic lymph node specimens were obtained from the University of Maryland Greenebaum Cancer Center Pathology Biorepository and Research Core and the MD Anderson Cancer Center Tumor Bank. Immunohistochemistry (IHC) was performed following a standard protocol provided by the reagent manufacturer (Dako, Carpinteria, CA), as described in our previous studies.20,21 The staining results were blinded and independently scored by two experienced hematopathologists (X.F.Z. and K.H.Y.), according to published scaled scoring criteria.22 Both membranous and focal cytoplasmic staining patterns were considered positive. A score was assigned to each specimen according to both a scaled criterion (the proportion of cells positive for staining) and staining intensity. The scaled criteria were as follows: 0, <25% neoplastic cells stained; 1+, 26% to 50% neoplastic cells stained; 2+, 50% to 75% neoplastic cells stained; and 3+, >75% neoplastic cells stained. The lymphoma staining intensities were divided into negative, weak, and strong staining patterns. Because the staining intensity score might vary from assay to assay, our scaled criteria overrule the staining intensity if only a few lymphoma cells showed strong staining. Occasional interobserver inconsistencies were reconciled by rereview by the two hematopathologists together to reach agreement. In combination, the lymphomas were roughly divided into negative (0 and/or negative), low (1+ and/or weak), and high (2+, 3+, and strong) expresser groups. Weak staining and 1+ positivity were considered a low level of expression, whereas strong staining and 2+ or greater were considered a high level or strong expression.
Cleavage of Protein Substrates
Samples of single-chain pro-HGF or pro-uPA were incubated with purified active matriptase in 100 mmol/L Tris-HCl, pH 8.5, at 37°C for the indicated times. Incubation was stopped by adding SDS sample buffer containing dithiothreitol to the reactions and boiling the resultant mixture. The cleavage products were separated by SDS-PAGE and analyzed by using Western blot analysis for HGF or by silver staining for uPA.
Matriptase Activity Assay
For gelatin zymography, 1 mg/mL gelatin was copolymerized in 7.5% SDS polyacrylamide gels. The details of the gelatin zymography assay can be found in our previous study.23 For fluorogenic peptide substrate assays, 125 nmol/L of N-tert-butoxycarbonyl-Gln-Ala-Arg-amido-4-methylcoumarin was mixed with the matriptase samples in a reaction buffer containing 100 mmol/L Tris-HCl, pH 8.5, and 100 μg/mL bovine serum albumin in a total volume of 200 μL. The increase in fluorescence resulting from hydrolysis of the peptide substrates was recorded using a DTX 880 multimode detector (Beckman Coulter, Fullerton, CA), with an excitation wavelength of 360 nm and a detection emission of 480 nm.
uPA Activation on the Cell Surface of U937 Cells
The U937 monocytes were washed to remove any uPA present on the cell surface, as described previously.16,24 Briefly, the cells were washed twice in RPMI 1640 medium supplemented with 25 mmol/L HEPES, pH 7.4, and then treated with 50 mmol/L glycine-hydrochloride buffer, pH 3.0, containing 0.1 mmol/L sodium chloride for 5 minutes, followed by neutralization with 0.5 mmol/L HEPES buffer, pH 7.5, containing 0.1 mmol/L sodium chloride. The cells were then washed once with RPMI 1640 medium supplemented with 25 mmol/L HEPES, pH 7.4, and resuspended at a density of 1 × 107 cells/mL. The cells were incubated with 1.4 nmol/L pro-uPA for 20 minutes at 37°C, after which the unbound pro-uPA was washed away with RPMI 1640 medium supplemented with 25 mmol/L HEPES, pH 7.4. Cells were incubated at a final density of 1 × 106 cells/mL in 0.05 mmol/L Tris-HCl, pH 7.4, 0.1 mmol/L NaCl, 0.01% Tween-80 in the presence of the chromogenic substrate, pyro-Glu-Gly-Arg-p-NA, and active matriptase at 37°C. The amidolytic activity of uPA was measured against the chromogenic substrate, and the rate at which p-NA was released was measured photometrically at 405 nm on a DTX 880 multimode detector.
In Vitro Cell Proliferation Assay
An equal number of cells were plated in complete growth media, and the cells were maintained at 37°C in 5% CO2 for 4 days. Aliquots were taken from the wells every 24 hours, mixed with trypan blue (Invitrogen, Grand Island, NY) at a 1:1 ratio to identify viable cells, which were then counted using a hemocytometer and a microscope.
Clonogenic Assay
The clonogenic assay was conducted using a modified version of the protocol provided by R&D Systems, Inc., Minneapolis, MN. Briefly, 5 × 103 cells were plated in quadruplicate in 0.5 mL of plating medium consisting of 1.2% methylcellulose, 10% FBS, and 200 mmol/L l-glutamine in Iscove's modified Dulbecco's medium (Invitrogen). The plates were maintained at 37°C in 5% CO2 for 10 days, when they were stained with 1 mg/mL iodonitrotetrazolium. Colonies consisting of >50 cells were automatically counted and analyzed using a colony counter.
Xenograft Tumors
Animal experiments were conducted under a protocol approved by the Georgetown University Animal Care and Use Committee. Before injection into animals, the cells were tested and confirmed to be negative for mycoplasma using a mycoplasma detection kit (Lonza, Basel, Switzerland). Eight-week-old female severe combined immunodeficient (SCID) mice (Taconic Farms, Inc., Hudson, NY) were divided into three groups (NT, 1, and 2; n = 10) and injected into the flank s.c. with 107 cells in 0.1 mL of medium. Tumor volume was determined by measuring the length, width, and height of tumors and calculating their product. The mice were euthanized as a cohort 24 days after tumor inoculation, and the tumors were removed and weighed. The resected tumors were fixed in 10% neutral-buffered formalin, embedded in paraffin, and divided into sections for histological analysis. The IHC staining for matriptase was performed as previously described, modified to include the use of M.O.M blocking reagent (Vector Labs, Burlingame, CA) to suppress background staining of endogenous mouse immunoglobulin. For histopathological examination, sections were stained with H&E. The levels of proliferation and apoptosis were assessed separately by staining sections with a mouse antibody against the proliferation marker, Ki-67 (Leica Biosystems, Buffalo Grove, IL), and for apoptosis using the ApopTag Plus system (Millipore, Billerica, MA).
Statistical Analysis
The data were analyzed using the Student's t-test for two samples assuming equal variances. P < 0.05 was considered significant.
Results
Human Hematological Malignancies Express Matriptase in the Absence of Significant HAI-1 Expression
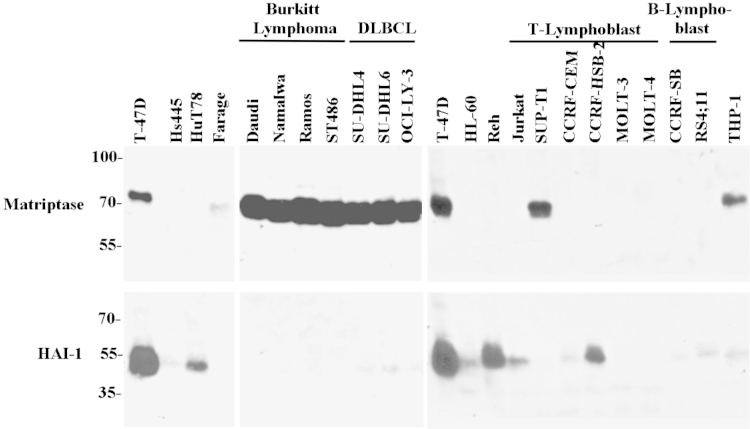
To understand the pathophysiological role of matriptase and HAI-1 in the context of human hematological cells and their cancers, we first analyzed their protein expression levels by using Western blot analysis in a panel of 21 lymphoma and leukemia cell lines (Figure 1). The T47-D breast cancer cells were used as a representative control for the ratio of matriptase/HAI-1 typically seen in epithelial cells (Figure 1). Previous data have demonstrated almost complete concordance of expression of matriptase and HAI-1 in a variety of human epithelial and carcinoma cell lines.17,25 Surprisingly, we observed that the expression of matriptase and HAI-1 was almost mutually exclusive in the 21 hematopoietic cell lines tested (Figure 1). Matriptase was found to be expressed at high levels in four BL lines, all three DLBCL lines, one (SUP-T1) of six T-cell leukemia lines, and THP-1 monocytes. Among these nine cell lines, HAI-1 was either undetectable or only detected or at low levels. The highest expression of HAI-1 was found in acute lymphoblastic leukemia-derived REH cells; high expression of HAI-1 was also found in the T-cell leukemia line, CCRF-HSB-2, and the T-cell lymphoma line, HuT-78. None of the four HAI-1–expressing cells expressed matriptase. These data demonstrate that some B-cell lymphoma lines, including BL and DLBCL, express matriptase in the context of low HAI-1 expression, in stark contrast to the large HAI-1/matriptase ratio commonly found in epithelial and carcinoma cells.
Figure 1.
Expression of matriptase and HAI-1 in human lymphoma cells. Cell lysates prepared from 21 hematological cancer cells and T-47D breast cancer cells, as indicated, were analyzed by immunoblot for matriptase using mAb M24 and HAI-1 using mAb M19.
Matriptase Expression Is Detected in a Variety of B-Cell Lymphomas, But Not in Reactive B Cells
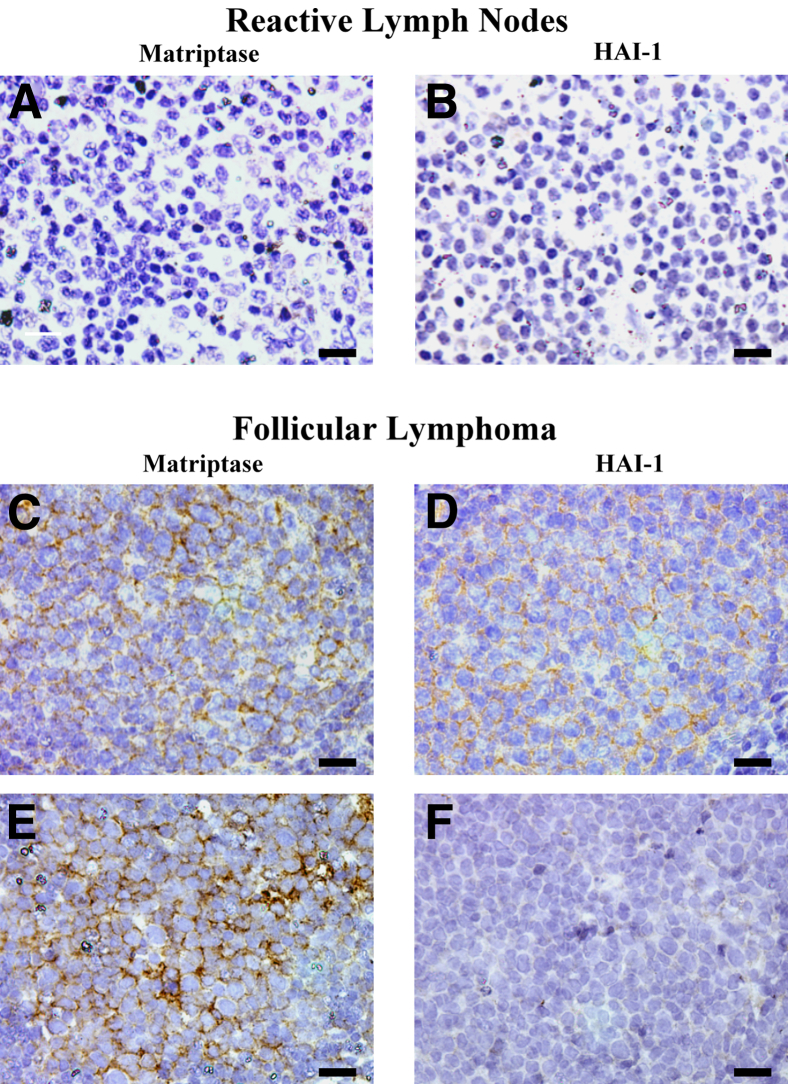
We next determined the expression of matriptase and HAI-1 in non-Hodgkin lymphomas (NHLs) by IHC staining of formalin-fixed, paraffin-embedded human tissue specimens. A total of 65 NHLs were examined, which included 25 follicular lymphomas (FLs), 27 DLBCLs, and 13 BLs. In addition, three reactive lymph nodes were examined as nontumor controls. Neither matriptase nor HAI-1 was detected in the three reactive lymph nodes (Table 1). Of the 25 FLs, 11 (44%) were positive for matriptase and 14 (56%) were positive for HAI-1. Of the 11 FLs that scored positive for matriptase, 7 (64%) were positive for HAI-1; 9 (64%) of the 14 FLs that were scored positive for HAI-1 were positive for matriptase. Correlation analysis revealed that the matriptase protein expression level was not correlated with HAI-1 protein expression levels (Pearson correlation coefficient r = 0.0141, P = 0.9467). Representative examples of matriptase and HAI-1 staining of the reactive lymph nodes and FLs are shown in Figure 2.
Table 1.
Summary of Matriptase and HAI-1 IHC Staining
| Variable | Case group (no.) | MTP+ | HAI-1+ | (MTP+, HAI-1−)/MTP+ | (MTP+, HAI-1+)/HAI-1+ |
|---|---|---|---|---|---|
| Reactive lymph nodes | 3 | 0/3 (0) | 0/3 (0) | 0/0 (0) | 0/0 (0) |
| FLs | 25 | 11/25 (44) | 14/25 (56) | 4/11 (36.4) | 9/14 (64.3) |
| DLBCLs | 27 | 12/27 (44.4) | 19/27 (70.4) | 3/12 (25.0) | 9/19 (47.4) |
| BLs | 13 | 7/13 (53.8) | 5/13 (38.5) | 6/7 (85.7) | 1/5 (20) |
Data are given as number/total number (percentage) unless otherwise indicated.
Figure 2.
Expression of matriptase and HGF activator inhibitor (HAI)-1 in human reactive lymph nodes and follicular lymphoma. Tissue sections from three paraffin-embedded cases of reactive lymph nodes and 25 cases of FL were analyzed by IHC for matriptase using mAb M24 and HAI-1 using mAb M19. Representative staining for matriptase (A, C, and E) and HAI-1 (B, D, and F). Scale bar = 20 μm.
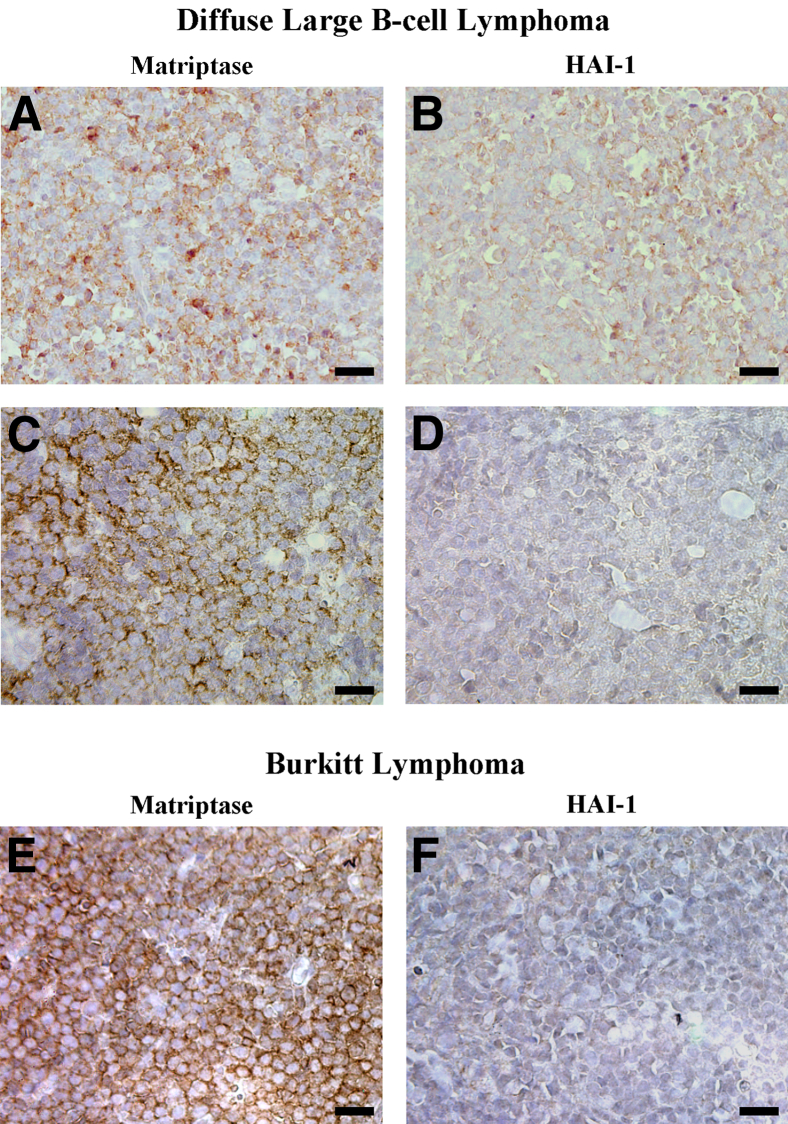
Of the 27 DLBCLs, 12 (44%) were positive for matriptase and 19 (70%) were positive for HAI-1. Of the 12 DLBCLs that scored positive for matriptase, 9 (75%) were positive for HAI-1; 9 (47%) of the 19 DLBCLs that were scored positive for HAI-1 were positive for matriptase. Correlation analysis revealed that matriptase protein expression in DLBCL was not correlated with HAI-1 protein expression (Pearson correlation coefficient r = 0.08479, P = 0.6741). Representative examples of matriptase and HAI-1 staining of DLBCL are shown in Figure 3.
Figure 3.
Expression of matriptase and HAI-1 in human DLBCLs and BLs. Tissue sections from 27 paraffin-embedded DLBCL cases and 13 BL cases were analyzed by IHC for matriptase using mAb M24 and for HAI-1 using mAb M19. Representative staining for matriptase (A, C, and E) and HAI-1 (B, D, and F). Scale bar = 20 μm.
Of the 13 BLs, 7 (54%) were positive for matriptase and 5 (39%) were positive for HAI-1. Of the 7 BLs that scored positive for matriptase, 1 (14%) was positive for HAI-1; 1 (20%) of the 5 BLs that were scored positive for HAI-1 was positive for matriptase. Correlation analysis revealed that matriptase protein expression in BLs was not correlated with HAI-1 protein expression (Pearson correlation coefficient r = −0.33071, P = 0.2697). Representative examples of matriptase and HAI-1 staining of BLs are shown in Figure 3.
In summary, our analysis demonstrates that roughly half of the NHLs examined express matriptase (30 of 65) and HAI-1 (38 of 65), in contrast to the reactive lymph nodes that exhibited no staining for either protein. No statistically significant relationship was found between the expression of matriptase and HAI-1 within the three groups of NHLs. Matriptase-positive BLs were, however, less likely to express HAI-1 compared with matriptase-positive FLs (Fisher's exact test, P = 0.0573) or matriptase-positive DLBCLs (Fisher's exact test, P = 0.019).
Expression of Matriptase in the Absence of HAI-1 by Lymphoma Cells Allows the Shedding of Free Active Matriptase
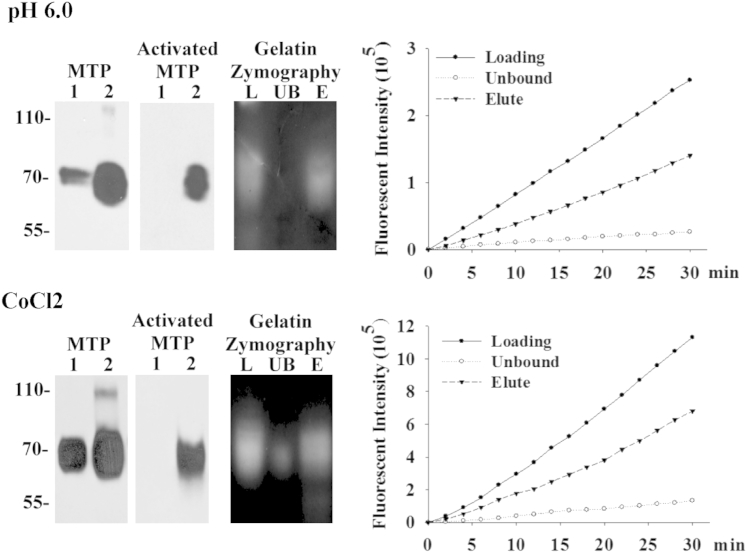
The lack of significant HAI-1expression in some lymphomas suggests that these tumor cells may regulate matriptase in a profoundly different way than that used by epithelial and carcinoma cells. Some lymphomas express matriptase in the absence of significant HAI-1 expression, raising the possibility that these tumors may produce significant amounts of free active matriptase. Furthermore, this active enzyme may have a significant half-life once liberated from the cells, in stark contrast to the case in epithelial cell systems and tumors, where we have shown that activated matriptase has a short half-life, being almost immediately inactivated by binding to HAI-1.26 To test this possibility, we induced matriptase activation in Raji BL cells by transiently exposing the cells to a phosphate buffer, pH 6.0, for 20 minutes (Figure 4), a method that we have previously used to stimulate epithelial and carcinoma cells to robustly activate matriptase.26,27 Activation of matriptase was also induced by incubation of the BL cells with 200 μmol/L CoCl2 overnight (Figure 4). The proteins shed into the cell-conditioned buffer (for pH 6.0) and media (for CoCl2) were collected and analyzed for the presence of active matriptase using multiple assays. Immunoblot analyses of the shed proteins using a matriptase mAb revealed that more matriptase was shed from the cells exposed to the mildly acidic buffer or CoCl2 containing media compared with those cells exposed to basal media (Figure 4). When an activated matriptase-specific mAb was used, a significant amount of active matriptase was present in the conditioned buffer and media of the acid- or CoCl2-treated cells, respectively, but not in the controls (Figure 4). The presence of a matriptase-like proteolytic activity in the conditioned buffer and media was demonstrated by gelatin zymography (Figure 4) and by measuring amidolytic activity with a synthetic fluorogenic substrate commonly used for matriptase (Figure 4). By using the activated matriptase mAb, active matriptase was found to be responsible for the proteolytic activity. The strong gelatinocytic activity detected at a position consistent with the expected size of active matriptase (Figure 4) was immunodepleted using the active matriptase-specific antibody (Figure 4) and then recovered using mAb (Figure 4). Similarly, the strong amidolytic activity (Figure 4) was almost completely depleted by the activated matriptase mAb (Figure 4). The proteolytic activity was then eluted and recovered from the mAb (Figure 4). These data demonstrate that the lack of significant levels of HAI-1 in these cells results in the release of free active matriptase into the extracellular milieu. Matriptase proteolytic activity was only detected in the conditioned buffer and media and not in lysates of cells that conditioned the buffer and media. This suggests that active matriptase is rapidly shed to the extracellular milieu by the cells and that this shedding would allow the protease to act close to the cancer cells and in the proximity of the stromal cells.
Figure 4.
Lymphoma cells produce free, active matriptase. Raji BL cells were induced to activate matriptase (lanes 2) by exposing cells to 150 mmol/L phosphate buffer, pH 6.0, for 20 minutes or 200 μmol/L CoCl2 in basal medium overnight, or basal medium as a nonactivation control (lanes 1). The shed proteins were analyzed by immunoblot for total matriptase using mAb M24 (MTP) and for activated matriptase using mAb M69 (activated MTP). The shed proteins collected from matriptase activation were immunoprecipitated using the activated matriptase mAb M69. The total shed protein (lanes L), unbound (lanes UB), and the proteins eluted from the M69 antibody (lanes E) were analyzed for matriptase gelatinolytic activity by gelatin zymography and by measuring amidolytic activity against a synthetic fluorescent substrate.
Matriptase Activates uPA and HGF
The production and shedding of free active matriptase by B-cell lymphoma cells might contribute to the malignancy of the cells by mediating the activation of other proteases and growth factors. Among the protease substrates of matriptase, uPA has received the most attention.2,3,16 In solution, active matriptase can cleave single-chain pro-uPA into two-chain uPA in a dose-dependent manner after an 8-hour incubation. By using a uPA chromogenic substrate, pyro-Glu-Gly-Arg-p-NA, the activation of pro-uPA was assessed by increasing amounts of active matriptase (0 to 0.2 nmol/L). At the top concentration of 0.2 nmol/L, active matriptase does not cleave the chromogen substrate. Although uPA activation determined by the proteolytic activity was observed to occur in a dose-dependent manner, the activation of uPA by active matriptase appeared to be modest (Figure 5B). Activation of pro-uPA is known to proceed more rapidly when pro-uPA is bound to its membrane receptor, urokinase-type plasminogen activator receptor (uPAR). Incubating both active matriptase and pro-uPA with human monocytic U937 cells, which can retain pro-uPA via uPAR and do not express matriptase, resulted in robust active uPA generation (Figure 5C). The level of uPA activity resulting from incubation of the pro-uPA with 0.2 nmol/L active matriptase in the context of U937 cells was approximately five times higher than the level of activity produced by incubating the same amount of pro-uPA in cell-free solution, containing 0.2 nmol/L active matriptase during the same time period (approximately 80 minutes) (Figure 5C).
Figure 5.
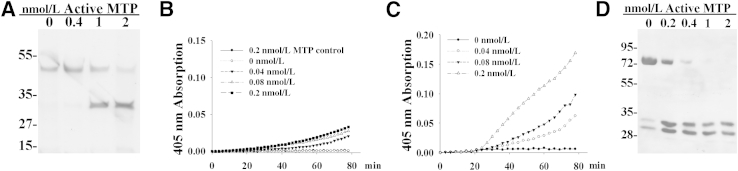
Activation of pro-uPA and HGF by active matriptase. A: Pro-uPA was incubated with increasing amounts of purified active matriptase (0 to 2 nmol/L) at 37°C for 8 hours. The activation was analyzed by SDS-PAGE, and the protein bands were visualized by silver stain. B and C: Pro-uPA and its chromogenic substrate were incubated with increasing amounts of active matriptase (0 to 0.2 nmol/L) in solution (B) or with U937 human monocytic cells (C) at 37°C. The generation of active uPA was monitored during the course of 80 minutes by measuring the liberation of the chromophore from the substrate by absorbance at 405 nm. D: Pro-HGF was incubated with increasing amounts of purified active matriptase (0 to 2 nmol/L) at 37°C for 30 minutes. The activation was analyzed by immunoblot for HGF cleavage using an antibody directed against the β subunit of HGF.
Matriptase has been shown to be able to activate several growth factors, including HGF, platelet-derived growth factors C and D, and macrophage-stimulating factor-1,2,4,17 among which pro-HGF is particularly relevant in the context of DLBCLs.28 Incubation of pro-HGF with subnanomolar concentrations of active matriptase for 30 minutes rapidly converted the single-chain pro-HGF into its two-chain form in a dose-dependent manner, as assessed by the appearance of the HGF α-chain at the cost of pro-HGF using immunoblot analysis (Figure 5D). These data suggest that the free active matriptase shed by lymphoma cells can modulate the tumor microenvironment by initiating the proteolytic cascade and activating growth factors.
Suppression of Matriptase Expression or Overexpression of HAI-1 Blunts the Proliferation of B-Cell Lymphoma
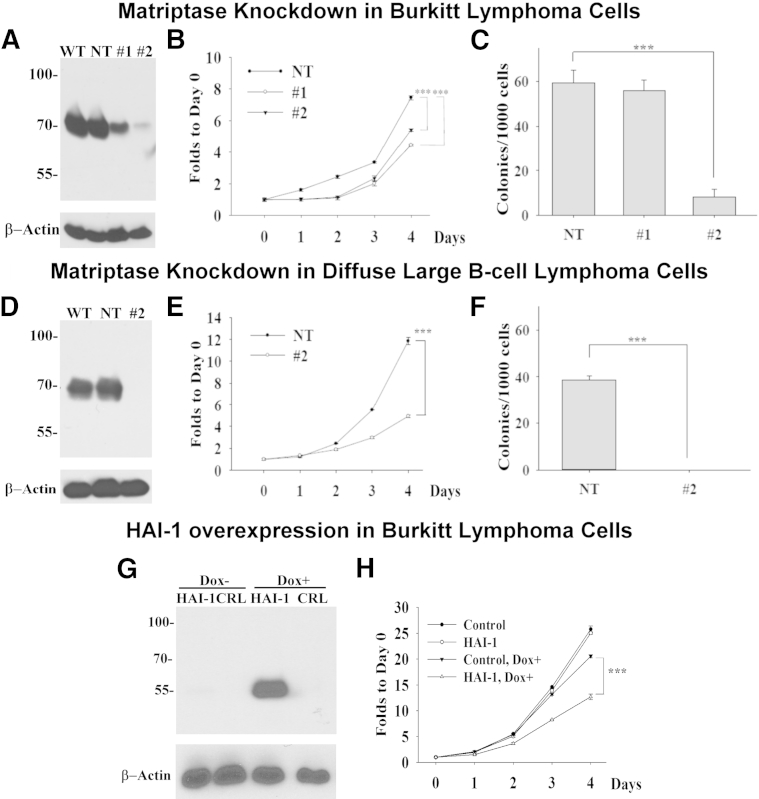
To evaluate the contribution of matriptase to the malignant behavior of B-cell lymphomas, we set out to determine the effect of suppressing matriptase expression, or HAI-1 overexpression, on the behavior of Raji BL cells, which express matriptase in the absence of HAI-1, such as other BL cells. Two shRNA constructs targeting different regions of the matriptase mRNA were used to generate stable matriptase-knockdown (MTPKD) pools: MTPKD 1 with >52% reduction in expression and MTPKD 2 with >88% reduction in matriptase expression at the protein level, when compared with that of the NT population or the parental cells (wild type) (Figure 6A). The reduced expression of matriptase affects the proliferation of the cells, because both matriptase knockdown cultures grew more slowly than the nontarget control by 27% for MTPKD 1 and 40% for MTPKD 2 (Figure 6B). The colony-forming ability of Raji cells was reduced by >80% in MTPKD 2, but only by a negligible amount in MTPKD 1 (Figure 6C). This difference may be attributed to the moderate levels of matriptase suppression in MTPKD 1. The reduction in proliferation and clonogenicity associated with strong suppression of matriptase expression was also observed using the DLBCL cell line, SU-DHL-4. When matriptase expression was almost completely suppressed by shRNA-mediated knockdown (Figure 6D), the DLBCL cells proliferated 50% slower than the control cells (Figure 6E) and lost their ability to form colonies (Figure 6F). These data suggest that the role of matriptase in tumor growth is not limited to BLs.
Figure 6.
Modulation of matriptase and HA-1 expression alters in vitro growth of lymphoma cells. A and D: Cell lysates prepared from the wild-type (WT), NT, and matriptase knockdown populations of Raji BL cells (1 and 2) and SU-DHL-4 DLBCL cells (2) were analyzed by immunoblot for matriptase using matriptase mAb M24 and for β-actin as a loading control. B and E: The proliferation rates of the Raji and SU-DHL-4 NT and the matriptase knockdown pools (1 and 2) were analyzed. The data are presented as means ± SD of triplicate determinations. Statistical significance was determined by Student's t-test. C and F: A total of 5 × 103 cells of the NT and the matriptase knockdown pools (1 and 2) were grown in methylcellulose for 10 days, and the resultant number of colonies was counted. The means ± SD of triplicate determinations are presented, and statistical significance was assessed by Student's t-test. G: Cell lysates prepared from the doxycycline-inducible HAI-1 line (HAI-1) and the empty vector line (CRL) were prepared from cells grown in the absence (Dox−) or presence (Dox+) of doxycycline and analyzed by immunoblot for HAI-1 using mAb M19 and for actin as a loading control. H: The proliferation rates of the doxycycline-inducible HAI-1 line (HAI-1) and the empty vector line (CRL) were determined in the absence (Control and HAI-1) or presence (Control, Dox+ and HAI-1, and Dox+) of doxycycline. The data are presented as means ± SD of triplicate determinations. Statistical significance was determined by Student's t-test. ∗∗∗P < 0.001 (B, C, E, F, and H).
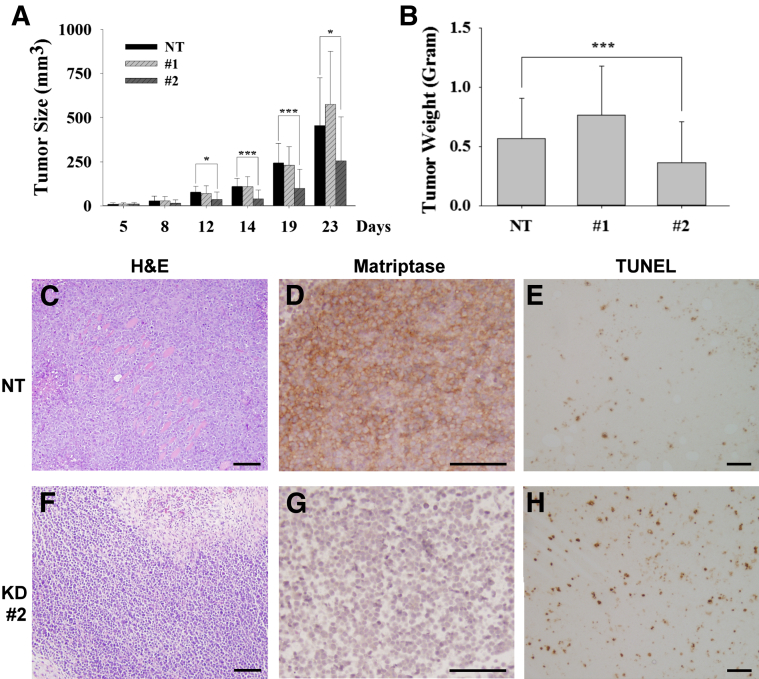
Consistent with the suppression of the proliferation of the cancer cells by the reduced matriptase expression, increased HAI-1 expression in Raji cells, mediated through the use of a doxycycline-induced expression system, also suppresses the proliferation of the cells (Figure 6, G and H). Significant levels of HAI-1 were induced by doxycycline treatment in the Raji clone transduced with a lentiviral vector, containing the HAI-1 cDNA under the control of a tetracycline-responsive promoter, and a viral vector expressing the tetracycline transactivator protein (Figure 6G). Negligible HAI-1 expression was seen in this clone in the absence of doxycycline. No HAI-1 expression was detected in the control clone containing only the vector expressing the tetracycline transactivator, regardless of the presence of doxycycline. Transduction with the HAI-1–inducible construct does not appear to alter cell proliferation in the absence of doxycycline when compared with the Raji clone containing tetracycline transactivator alone (Figure 6H). Although the treatment with doxycycline does slightly alter the growth of the control clone, the induced HAI-1 expression further suppressed the proliferation of the Raji cells in a statistically significant manner. Collectively, our study suggests that the unopposed proteolysis resulting from the overexpression of matriptase in the absence of significant HAI-1 expression confers an increased growth potential on lymphoma cells. The impact of matriptase suppression on tumor growth was further investigated by xenografting the cells into SCID mice. The two MTPKD populations (1 and 2) and the NT-control (107 cells/0.5 mL) were injected s.c. into SCID mice. The tumors were measured and the volume was calculated on the indicated dates (Figure 7A), and the tumors were harvested and weighed when the animals were euthanized (Figure 7B). The data are presented as the means ± SE of nine tumors in the NT, 10 tumors in MTPKD 1, and seven tumors in MTPKD 2. The sizes and weights of MTPKD 2 tumors were significantly smaller than those of MTPKD 1 or nontarget control tumors (Figure 7, A and B, respectively). The overall morphological characteristics of the NT control and MTPKD 2 Raji tumors were similar based on the H&E staining (Figure 7, C and F, respectively). Both tumors showed areas of viable cancer cells, particularly around the periphery, with areas of necrosis, particularly in the center of the tumors. The differential expression of matriptase in the NT control and the MTPKD 2 tumors was confirmed by IHC, with much higher levels in the NT control compared with the MTPKD 2 tumors (Figure 7, D and G). To explore the reason(s) behind smaller MTPKD 2 tumor size compared with the NT control, sections of each tumor type were stained for a marker of proliferation (Ki-67) and for apoptosis using the TUNEL assay. The Ki-67 staining was variable in different portions of the tumors, but when considering the tumors as a whole, did not appear to be significantly different between the NT control and MTPKD 2 tumors (data not shown). In contrast, TUNEL staining appeared to demonstrate that there was a consistently higher level of apoptosis in the MTPKD 2 tumors compared with the NT control tumors (Figure 7, E and H), suggesting that the smaller size of the MTPKD 2 tumors might be the result of increased apoptosis.
Figure 7.
Suppression of matriptase expression reduces the growth of B-cell lymphoma tumor xenografts. Three Raji pools of the NT and the two matriptase-knockdown populations (1 and 2) of Raji cells (107 cells in 0.1 mL) were injected s.c. into SCID mice. The growth of tumors was monitored by measuring their dimensions using calipers and calculating their volume on the indicated dates (A) and assessed by the weight when the animals were euthanized (B). The data are presented as means ± SEM of nine tumors for the NT control, 10 tumors in matriptase-knockdown 1 cells, and seven tumors in the knockdown 2 cells. Statistical significance was determined by Student's t-test. ∗P < 0.05, ∗∗∗P < 0.001. Tumor sections were analyzed by H&E staining for histological assessment (C and F), by IHC for matriptase (D and G) and Ki-67 (data not shown), and by TUNEL assay for apoptosis (E and H).
Discussion
In this study, we have examined the role of matriptase-mediated pericellular proteolysis in NHLs, and revealed interesting contrasts between carcinoma and B-cell lymphoma with respect to how matriptase activity is dysregulated. First, matriptase is expressed in a cancer-specific manner in NHLs, because the enzyme was detected at significant levels in approximately 50% of the specimens examined, whereas matriptase is not expressed in reactive B cells. This is in contrast to the widespread expression of matriptase in both carcinoma and normal epithelial cells. Second, HAI-1 does not play nearly as important a role in matriptase biological characteristics in B-cell lymphomas as it does in epithelial tumors, where, in addition to rapidly binding to and inactivating active matriptase, HAI-1 also facilitates the expression and zymogen activation of the protease.15,29 Expression correlation between matriptase and HAI-1 in NHL is not significant. Furthermore, in many cases of matriptase-positive FLs and DLBCLs, the IHC staining of HAI-1 was always relatively weak and in some cases did not appear to be localized at the cell surface.
Matriptase is synthesized as a zymogen that has limited intrinsic enzymatic activity, and undergoes autoactivation to acquire its full proteolytic activity.30 Epithelial and carcinoma cells activate matriptase in response to mild acidity and CoCl2, a hypoxia-mimicking agent. A consequence of this unique feature of matriptase zymogen activation is that matriptase is likely active in solid tumors, in which hypoxia is common and results in an acidic milieu. Although both hypoxia and its resultant extracellular acidosis are not thought to be as important to the progression of B-cell lymphoma as in carcinoma, this unique feature of matriptase activation allowed us to demonstrate that neoplastic B cells are able to generate and shed active matriptase with prolonged life span, because of the lack, or limited expression, of HAI-1 or other matriptase inhibitors in NHL cells. This is in a stark contrast to carcinoma cells, in which most active matriptase is rapidly inhibited by high levels of HAI-1 and is likely, therefore, to only activate and process those downstream substrates that are close to matriptase in the secretory pathway and on the surface of the cells. The lack of large amounts of HAI-1 means that activity of the matriptase will only be inhibited when it encounters other canonical matriptase inhibitors in the extracellular environment, such as antithrombin, α-1 antiprotease, and α-2 antiplasmin.18
The presence of this potent active protease in the tumor microenvironment of NHLs raises the distinct possibility that the disease pathological characteristics are influenced by processes such as matriptase-initiated proteolytic cascades, altered ECM remodeling, and modulation of the function of growth factors, cytokines, and chemokines. The area over which the active matriptase may exert an effect is also probably greater than in epithelial systems, with the result that the stromal components of the tumor are also exposed to the influence of free active matriptase. The shed active matriptase in the extracellular milieu can, therefore, initiate pericellular proteolysis by activating uPA in those cells that express uPAR. Because uPAR is abundantly expressed by stromal cells, including macrophages, fibroblasts, and endothelial cells, but not by lymphoma cells,31 the matriptase-accelerated activation of the uPA/plasmin system in NHL provides an important mechanism for cancer cells to regulate pericellular proteolysis in stromal cells.
Among the potential downstream events resulting from the presence of active matriptase, the activation of pro-HGF has the potential to have a profound impact on B-cell lymphomas. HGF, through engagement with its membrane receptor, c-MET, promotes adhesion of lymphoma cells to ECM molecules through the activation of integrins, an important step for the invasion and dissemination of lymphoma cells.32 In human B-cell lymphomas, HGF is mainly produced by macrophages in the stromal components of the tumor, and in the context of DLBCLs (eg, approximately one third of tumors express c-MET). HGF is secreted as a single-chain, inactive precursor, and is mainly deposited in the ECM or on nearby cell surfaces.33 The extracellular activation of HGF is the principle switch regulating HGF activity, and the activation of HGF may, therefore, occur in the close vicinity of its target cells. Several unrelated serine proteases have been reported to be involved in the activation of HGF/scatter factor (SF), including the blood coagulation factor XIIa, HGF activator (HGFA), uPA, tissue plasminogen activator, and matriptase.34–36 Although HGFA was reported to be the most potent activator and is expressed by DLBCL and multiple myeloma in vitro and in vivo, HGFA must itself be activated by another protease to activate HGF; thus, it apparently depends on the presence of another protease, such as thrombin, at least in the in vitro setting.28,37 Among the serine protease activators of pro-HGF, matriptase is unique in the following ways: i) its potency is similar to that of HGFA38; ii) it is co-expressed and colocalized with the c-MET receptor at the basolateral plasma membrane in the target epithelial, carcinoma,39 and some neoplastic B cells; and iii) it has the ability to autoactivate, with the result that active matriptase can be generated without dependence on other proteases.30
In addition to serving as an important mediator that facilitates the cross talk between neoplastic B cells and their microenvironment, matriptase may also affect the cancer cells (Figures 6 and 7). Induced expression of the matriptase inhibitor, HAI-1, reduced BL cell proliferation, and reduced matriptase expression resulting from shRNA knockdown produced a decrease in the in vitro and in vivo proliferation and colony-forming ability of BL and DLBCL cells. The degree by which matriptase expression is suppressed seems to be important to the nature of the altered behaviors of the cancer cells. Previously, the loss of skin barrier function and neonatal death in matriptase knockout mice can be rescued by the re-expression of as little as 1% of wild-type matriptase levels.40 This suggests that low levels of matriptase are sufficient to maintain its physiological functions. An increase in the level of matriptase activation, combined with the prolonged survival of active matriptase, provides a plausible mechanism for cells with moderate suppression of matriptase expression to compensate for the reduced matriptase expression, allowing the maintenance of the parental phenotype.
Given the role of matriptase in the modulation of the tumor microenvironment and in the promotion of tumor growth, the expression of matriptase may confer a malignant advantage to matriptase-positive NHL and may, therefore, have clinical implications for disease progression and outcome. This hypothesis is supported by the fact that matriptase was 1 of 76 genes identified in a 6912-gene array that can distinguish activated B-cell (ABC)-like from germinal center B-cell–like DLBCL.8 In this study, high matriptase expression was detected in the DLBCL ABC-like OCI-LY10 cells and not in FLs. It will be interesting in the future studies to correlate matriptase expression with the ABC subtype of DLBCL and determine whether matriptase-expressing NHLs have worse overall survival than the matriptase-negative counterparts.
Footnotes
Supported by National Cancer Institute (NCI) grant R01 CA 123223 (M.J. and C.-Y.L.), Maryland Cigarette Reconstitute Fund (C.-Y.L.), and the assistance of the following shared resources at the Lombardi Comprehensive Cancer Center, which are supported in part by NIH/NCI grant P30-CA051008: Tissue Culture Shared Resource, Microscopy and Imaging Shared Resource, and Histology and Tissue Shared Resource.
Disclosures: C.-Y.L. is an inventor on US patents 6,077,938 and 6,677,377, and M.J. and C.-Y.L. are inventors on US patent 7,355,015 (all related to matriptase).
References
- 1.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91:439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 2.Lee S.L., Dickson R.B., Lin C.Y. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi T., Harris J.L., Huang W., Yan K.W., Coughlin S.R., Craik C.S. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 4.Ustach C.V., Huang W., Conley-LaComb M.K., Lin C.Y., Che M., Abrams J., Kim H.R. A novel signaling axis of matriptase/PDGF-D/ss-PDGFR in human prostate cancer. Cancer Res. 2010;70:9631–9640. doi: 10.1158/0008-5472.CAN-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberst M.D., Singh B., Ossandon M., Dickson R.B., Johnson M.D., Lin C.Y. Characterization of matriptase expression in normal human tissues. J Histochem Cytochem. 2003;51:1017–1025. doi: 10.1177/002215540305100805. [DOI] [PubMed] [Google Scholar]
- 6.Kataoka H., Suganuma T., Shimomura T., Itoh H., Kitamura N., Nabeshima K., Koono M. Distribution of hepatocyte growth factor activator inhibitor type 1 (HAI-1) in human tissues: cellular surface localization of HAI-1 in simple columnar epithelium and its modulated expression in injured and regenerative tissues. J Histochem Cytochem. 1999;47:673–682. doi: 10.1177/002215549904700509. [DOI] [PubMed] [Google Scholar]
- 7.Szabo R., Kosa P., List K., Bugge T.H. Loss of matriptase suppression underlies spint1 mutation-associated ichthyosis and postnatal lethality. Am J Pathol. 2009;174:2015–2022. doi: 10.2353/ajpath.2009.090053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo R., Molinolo A., List K., Bugge T.H. Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene. 2007;26:1546–1556. doi: 10.1038/sj.onc.1209966. [DOI] [PubMed] [Google Scholar]
- 9.Carney T.J., von der Hardt S., Sonntag C., Amsterdam A., Topczewski J., Hopkins N., Hammerschmidt M. Inactivation of serine protease Matriptase1a by its inhibitor Hai1 is required for epithelial integrity of the zebrafish epidermis. Development. 2007;134:3461–3471. doi: 10.1242/dev.004556. [DOI] [PubMed] [Google Scholar]
- 10.Bugge T.H., List K., Szabo R. Matriptase-dependent cell surface proteolysis in epithelial development and pathogenesis. Front Biosci. 2007;12:5060–5070. doi: 10.2741/2448. [DOI] [PubMed] [Google Scholar]
- 11.List K., Szabo R., Molinolo A., Sriuranpong V., Redeye V., Murdock T., Burke B., Nielsen B.S., Gutkind J.S., Bugge T.H. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005;19:1934–1950. doi: 10.1101/gad.1300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C.Y., Anders J., Johnson M., Dickson R.B. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J Biol Chem. 1999;274:18237–18242. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- 13.Benaud C., Dickson R.B., Lin C.Y. Regulation of the activity of matriptase on epithelial cell surfaces by a blood-derived factor. Eur J Biochem. 2001;268:1439–1447. doi: 10.1046/j.1432-1327.2001.02016.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee M.-S., Kiyomiya K., Benaud C., Dickson R.B., Lin C.Y. Simultaneous activation and HAI-1-mediated inhibition of matriptase induced at activation foci in immortal human mammary epithelial cells. Am J Physiol Cell Physiol. 2005;288:C932–C941. doi: 10.1152/ajpcell.00497.2004. [DOI] [PubMed] [Google Scholar]
- 15.Oberst M.D., Chen L.Y., Kiyomiya K.I., Williams C.A., Lee M.S., Johnson M.D., Dickson R.B., Lin C.Y. Hepatocyte growth factor activator inhibitor 1 (HAI-1) regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol. 2005;289:C462–C470. doi: 10.1152/ajpcell.00076.2005. [DOI] [PubMed] [Google Scholar]
- 16.Kilpatrick L.M., Harris R.L., Owen K.A., Bass R., Ghorayeb C., Bar-Or A., Ellis V. Initiation of plasminogen activation on the surface of monocytes expressing the type II transmembrane serine protease matriptase. Blood. 2006;108:2616–2623. doi: 10.1182/blood-2006-02-001073. [DOI] [PubMed] [Google Scholar]
- 17.Bhatt A.S., Welm A., Farady C.J., Vasquez M., Wilson K., Craik C.S. Coordinate expression and functional profiling identify an extracellular proteolytic signaling pathway. Proc Natl Acad Sci U S A. 2007;104:5771–5776. doi: 10.1073/pnas.0606514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng I.C., Chou F.P., Su S.F., Oberst M., Madayiputhiya N., Lee M.S., Wang J.K., Sloane D.E., Johnson M., Lin C.Y. Purification from human milk of matriptase complexes with secreted serpins: mechanism for inhibition of matriptase other than HAI-1. Am J Physiol Cell Physiol. 2008;295:C423–C431. doi: 10.1152/ajpcell.00164.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L., Liu M., Dong N., Jiang Y., Lin C.Y., Huang M., Wu D., Wu Q. Matriptase is highly upregulated in chronic lymphocytic leukemia and promotes cancer cell invasion. Leukemia. 2013;27:1191–1194. doi: 10.1038/leu.2012.289. [DOI] [PubMed] [Google Scholar]
- 20.Wang J.K., Lee M.S., Tseng I.C., Chou F.P., Chen Y.W., Fulton A., Lee H.S., Chen C.J., Johnson M.D., Lin C.Y. Polarized epithelial cells secrete matriptase as a consequence of zymogen activation and HAI-1-mediated inhibition. Am J Physiol Cell Physiol. 2009;297:C459–C470. doi: 10.1152/ajpcell.00201.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.J., Wu B.Y., Tsao P.I., Chen C.Y., Wu M.H., Chan Y.L., Lee H.S., Johnson M.D., Eckert R.L., Chen Y.W., Chou F., Wang J.K., Lin C.Y. Increased matriptase zymogen activation in inflammatory skin disorders. Am J Physiol Cell Physiol. 2011;300:C406–C415. doi: 10.1152/ajpcell.00403.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai B., Zhao X.F., Mazan-Mamczarz K., Hagner P., Corl S., Bahassie M., Lu S., Stambrook P.J., Shapiro P., Gartenhaus R.B. Functional and molecular interactions between ERK and CHK2 in diffuse large B-cell lymphoma. Nat Commun. 2011;2:402. doi: 10.1038/ncomms1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C.Y., Wang J.K., Torri J., Dou L., Sang Q.A., Dickson R.B. Characterization of a novel, membrane-bound, 80-kDa matrix-degrading protease from human breast cancer cells: monoclonal antibody production, isolation, and localization. J Biol Chem. 1997;272:9147–9152. [PubMed] [Google Scholar]
- 24.Duval-Jobe C., Parmely M.J. Regulation of plasminogen activation by human U937 promonocytic cells. J Biol Chem. 1994;269:21353–21357. [PubMed] [Google Scholar]
- 25.Oberst M., Anders J., Xie B., Singh B., Ossandon M., Johnson M., Dickson R.B., Lin C.Y. Matriptase and HAI-1 are expressed by normal and malignant epithelial cells in vitro and in vivo. Am J Pathol. 2001;158:1301–1311. doi: 10.1016/S0002-9440(10)64081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tseng I.C., Xu H., Chou F.P., Li G., Vazzano A.P., Kao J.P., Johnson M.D., Lin C.Y. Matriptase activation, an early cellular response to acidosis. J Biol Chem. 2010;285:3261–3270. doi: 10.1074/jbc.M109.055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M.S., Tseng I.C., Wang Y., Kiyomiya K., Johnson M.D., Dickson R.B., Lin C.Y. Autoactivation of matriptase in vitro: requirement for biomembrane and LDL receptor domain. Am J Physiol Cell Physiol. 2007;293:C95–C105. doi: 10.1152/ajpcell.00611.2006. [DOI] [PubMed] [Google Scholar]
- 28.Tjin E.P., Groen R.W., Vogelzang I., Derksen P.W., Klok M.D., Meijer H.P., van E.S., Pals S.T., Spaargaren M. Functional analysis of HGF/MET signaling and aberrant HGF-activator expression in diffuse large B-cell lymphoma. Blood. 2006;107:760–768. doi: 10.1182/blood-2005-05-1929. [DOI] [PubMed] [Google Scholar]
- 29.Oberst M.D., Johnson M.D., Dickson R.B., Lin C.Y., Singh B., Stewart M., Williams A., al Nafussi A., Smyth J.F., Gabra H., Sellar G.C. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res. 2002;8:1101–1107. [PubMed] [Google Scholar]
- 30.Oberst M.D., Williams C.A., Dickson R.B., Johnson M.D., Lin C.Y. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol Chem. 2003;278:26773–26779. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- 31.Petersen L.C. Kinetics of reciprocal pro-urokinase/plasminogen activation–stimulation by a template formed by the urokinase receptor bound to poly(D-lysine) Eur J Biochem. 1997;245:316–323. doi: 10.1111/j.1432-1033.1997.00316.x. [DOI] [PubMed] [Google Scholar]
- 32.Weimar I.S., de Jong D., Muller E.J., Nakamura T., van Gorp J.M., de Gast G.C., Gerritsen W.R. Hepatocyte growth factor/scatter factor promotes adhesion of lymphoma cells to extracellular matrix molecules via alpha 4 beta 1 and alpha 5 beta 1 integrins. Blood. 1997;89:990–1000. [PubMed] [Google Scholar]
- 33.Schuppan D., Schmid M., Somasundaram R., Ackermann R., Ruehl M., Nakamura T., Riecken E.O. Collagens in the liver extracellular matrix bind hepatocyte growth factor. Gastroenterology. 1998;114:139–152. doi: 10.1016/s0016-5085(98)70642-0. [DOI] [PubMed] [Google Scholar]
- 34.Miyazawa K., Shimomura T., Kitamura A., Kondo J., Morimoto Y., Kitamura N. Molecular cloning and sequence analysis of the cDNA for a human serine protease responsible for activation of hepatocyte growth factor: structural similarity of the protease precursor to blood coagulation factor XII. J Biol Chem. 1993;268:10024–10028. [PubMed] [Google Scholar]
- 35.Shimomura T., Miyazawa K., Komiyama Y., Hiraoka H., Naka D., Morimoto Y., Kitamura N. Activation of hepatocyte growth factor by two homologous proteases, blood-coagulation factor XIIa and hepatocyte growth factor activator. Eur J Biochem. 1995;229:257–261. doi: 10.1111/j.1432-1033.1995.tb20463.x. [DOI] [PubMed] [Google Scholar]
- 36.Mars W.M., Zarnegar R., Michalopoulos G.K. Activation of hepatocyte growth factor by the plasminogen activators uPA and tPA. Am J Pathol. 1993;143:949–958. [PMC free article] [PubMed] [Google Scholar]
- 37.Tjin E.P., Derksen P.W., Kataoka H., Spaargaren M., Pals S.T. Multiple myeloma cells catalyze hepatocyte growth factor (HGF) activation by secreting the serine protease HGF-activator. Blood. 2004;104:2172–2175. doi: 10.1182/blood-2003-12-4386. [DOI] [PubMed] [Google Scholar]
- 38.Kirchhofer D., Peek M., Li W., Stamos J., Eigenbrot C., Kadkhodayan S., Elliott J.M., Corpuz R.T., Lazarus R.A., Moran P. Tissue expression, protease specificity, and Kunitz domain functions of hepatocyte growth factor activator inhibitor-1B (HAI-1B), a new splice variant of HAI-1. J Biol Chem. 2003;278:36341–36349. doi: 10.1074/jbc.M304643200. [DOI] [PubMed] [Google Scholar]
- 39.Satomi S., Yamasaki Y., Tsuzuki S., Hitomi Y., Iwanaga T., Fushiki T. A role for membrane-type serine protease (MT-SP1) in intestinal epithelial turnover. Biochem Biophys Res Commun. 2001;287:995–1002. doi: 10.1006/bbrc.2001.5686. [DOI] [PubMed] [Google Scholar]
- 40.List K., Currie B., Scharschmidt T.C., Szabo R., Shireman J., Molinolo A., Cravatt B.F., Segre J., Bugge T.H. Autosomal ichthyosis with hypotrichosis syndrome displays low matriptase proteolytic activity and is phenocopied in ST14 hypomorphic mice. J Biol Chem. 2007;282:36714–36723. doi: 10.1074/jbc.M705521200. [DOI] [PubMed] [Google Scholar]