Abstract
A comparison of the human genome with that of the chimpanzee is an attractive approach to attempts to understand the specificity of a certain phenotype's development. The two karyotypes differ by one chromosome fusion, nine pericentric inversions, and various additions of heterochromatin to chromosomal telomeres. Only the fusion, which gave rise to human chromosome 2, has been characterized at the sequence level. During the present study, we investigated the pericentric inversion by which chimpanzee chromosome 19 differs from human chromosome 17. Fluorescence in situ hybridization was used to identify breakpoint-spanning bacterial artificial chromosomes (BACs) and plasmid artificial chromosomes (PACs). By sequencing the junction fragments, we localized breakpoints in intergenic regions rich in repetitive elements. Our findings suggest that repeat-mediated nonhomologous recombination has facilitated inversion formation. No addition or deletion of any sequence element was detected at the breakpoints or in the surrounding sequences. Next to the break, at a distance of 10.2–39.1 kb, the following genes were found: NGFR and NXPH3 (on human chromosome 17q21.3) and GUC2D and ALOX15B (on human chromosome 17p13). The inversion affects neither the genomic structure nor the gene-activity state with regard to replication timing of these genes.
Introduction
At least four categories of genetic differences are supposed to have contributed to the separation of the hominoid lineages: biochemical changes owing to loss or functional alterations of specific proteins, differences in gene expression caused by minor sequence divergence, segmental duplications followed by evolution of new genes, and chromosomal changes. Several genes are known to be specifically altered in the human lineage. One is the human tropoelastin (ELN [MIM 130160]) gene, which lacks exon 34. This exon is retained in the ELN gene of the great apes (Szabó et al. 1999). A second gene is the human type I hair-keratin gene cluster, which contains a pseudogene that was inactivated <240,000 years ago, in contrast to the still-functional homologous gene in the chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla) (Winter et al. 2001). A third gene that is altered in humans, but not in other hominoids, is the cytidine monophosphate–sialic acid hydroxylase gene, in which the deletion of a 92-bp exon caused by an AluY replacement results in a subsequent shift in the coding sequence (Chou et al. 1998; Hayakawa et al. 2001). The truncated hydroxylase cannot convert CMP-N-acetylneuraminic acid (Neu5Ac) to CMP-N-glycolylneuraminic acid (Neu5Gc). In contrast to humans, great apes and other mammals have substantial amounts of Neu5Gc in most tissues, but not in the brain. A second uniquely human mutation that affects sialic acid biology is the missense mutation in the sialic acid–binding immunoglobulin-like lectin-like protein 1 (SIGLECL1 [MIM 606094]) gene (Angata et al. 2001). Siglecs are immunoglobulin (Ig) superfamily members that recognize sialic acids, such as Neu5Ac and Neu5Gc, differentially. The biological significance of both the human Neu5Gc deficiency and the SIGLECL1 mutation remains to be investigated. It has been suggested that they influenced pathogen susceptibility and brain development during hominoid evolution (Hayakawa et al. 2001; Varki 2001). Among other known examples of genetic differences between human and chimpanzee genomes, it is important to mention the SMN2 gene (MIM 60167), which is unique to humans (Rochette et al. 2001). In addition, several olfactory-receptor genes are suspected to have differential activity in humans and chimpanzees (Sharon et al. 1999). Recently, the rapid and complex evolution of the morpheus genes in hominoid lineages has been described by Johnson et al. (2001). Many more examples will emerge, as chimpanzee and gorilla genomic DNA sequences become available.
On the DNA-sequence level, human and chimpanzee show 98.8% overall sequence homology, a rate that is even higher if coding sequences are compared (Chen and Li 2001). These differences are spread over the whole genome, and it would be difficult, at present, to assign phenotypic differences to certain base changes.
Although considerable homology between human and great ape chromosomes has been observed by use of G-banding, researchers have found some notable structural differences, which may mark regions of evolutionarily important genetic changes. These differences include heterochromatin variability, pericentric inversions, and the telomeric fusion of two ancestral chromosomes, which resulted in the generation of human chromosome 2 (Dutrillaux et al. 1975; Yunis and Prakash 1982; Wienberg et al. 1990; IJdo et al. 1991). Since genetic disparities are assumed to be responsible for phenotypic differences between hominoid species, exact molecular characterization of the evolutionary breakpoints is important. Furthermore, these analyses will give insights into mechanisms involved in chromosome evolution. The molecular characterization of evolutionary breakpoints provides the basis for investigations of whether they coincide with human genomic regions susceptible to constitutional or somatic rearrangements and of whether, owing to intrinsic sequence features, these breakpoints have been utilized more than once during evolution.
When human and chimpanzee chromosomes have been compared, nine pericentric inversions have been observed (Yunis and Prakash 1982). Breakpoint-spanning YACs of pericentric inversions on chimpanzee chromosomes equivalent to human chromosomes 4, 9, and 12 were identified (Nickerson and Nelson 1998; Marzella et al. 2000). However, the corresponding breakpoint sequences and the genes flanking the breakpoints are still unknown. So far, only the fusion event that gave rise to human chromosome 2 has been characterized at the molecular level (IJdo et al. 1991). As a relict of this fusion event, some telomere-specific hexanucleotide repeats (5′-(TTAGGG)n-(CCCTAA)m-3′) mark the fusion on the sequence level, without an obvious alteration of the neighboring genes on either side. Here, we describe the characterization of the pericentric inversion breakpoints of the chimpanzee chromosome 19 compared with human chromosome 17 (fig. 1). We specified the sequence environment that flanks the breakpoints, identified the genes next to the breaks at each site, and addressed the contribution of this pericentric inversion to the structure, activity, and expression of at least one of the flanking genes.
Figure 1.
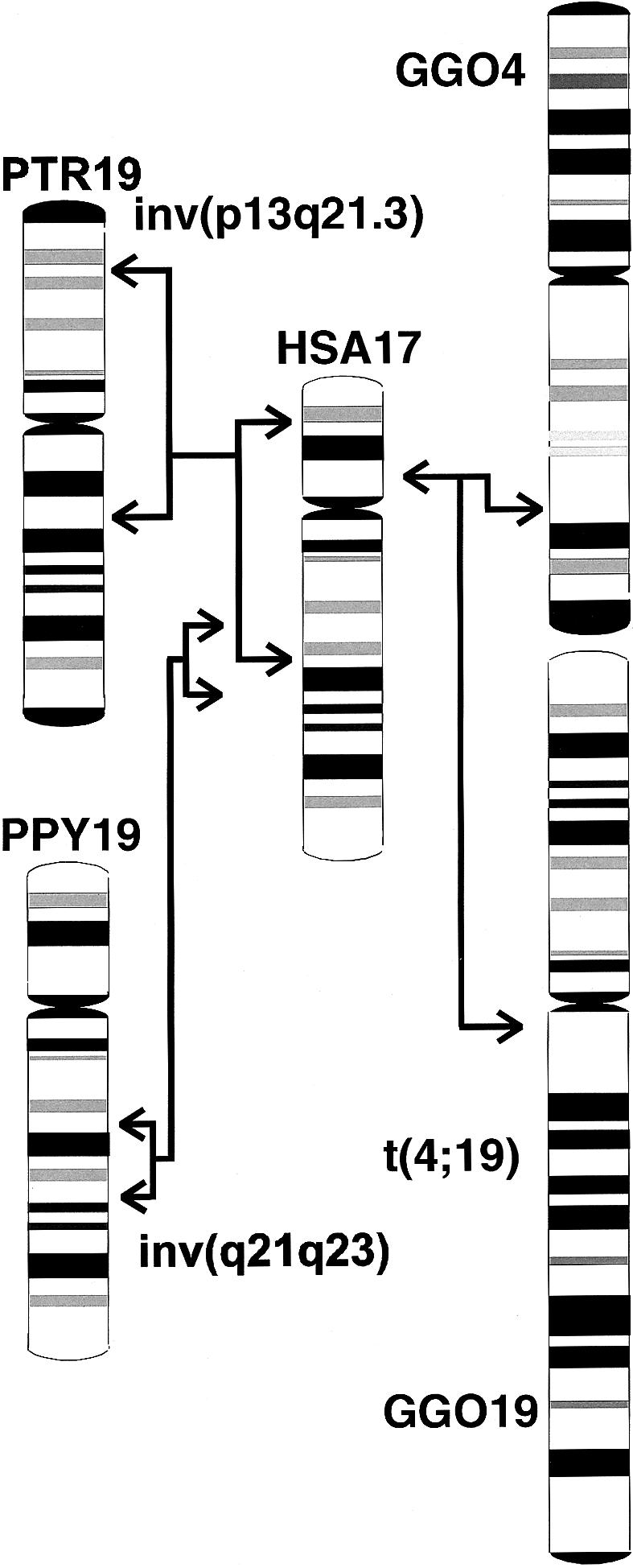
Comparison of the G-banding pattern of human chromosome 17 and the homologous chromosomes in chimpanzees, orangutans, and gorillas. Gorillas have a unique reciprocal translocation t(4;19), which arose from the ancestral chromosomes equivalent to human chromosomes 5 and 17. Arrows indicate the parts homologous to human chromosome 17 involved in the pericentric inversion of chimpanzee 19, the paracentric inversion of orangutan chromosome 19, and the position of the gorilla t(4;19) breakpoints. GGO = Gorilla gorilla; PPY = Pongo pygmaeus; PTR = Pan troglodytes; HSA = Homo sapiens.
Material and Methods
Filter Libraries and Southern Blot Hybridizations
Filters from the Roswell Park Cancer Institute (RPCI)–43 male chimpanzee BAC library and the RPCI-1,3–5 human PAC libraries were obtained from the Resource Center and Primary Database for the German Human Genome Project and the Children's Hospital Oakland–BACPAC ResourcesWeb sites. BACs and PACs were purchased from the same institutions. For Southern blot analysis, BAC and PAC DNA was digested with restriction endonucleases, electrophoresed on 0.8% agarose gels, and transferred to nylon membranes (Amersham Pharmacia). BAC and PAC filters and Southern blot membranes were hybridized in buffer containing 7% SDS, 0.5 M sodium phosphate (pH 7.2) and 1 mM EDTA, at 65°C overnight. For filter and Southern blot hybridizations, PCR products were labeled by random priming with hexamer oligonucleotides and Klenow polymerase in the presence of [32P]-dCTP (Amersham Pharmacia). PCR product p210/211, amplified with primers p210 (5′-AGGATCAAACCAAACTCAAG-3′) and p211 (5′-AAGGATGTATTTTAGGATGAA-3′), as well as BAC RP43-134L13 DNA as template DNA, was used as a probe to screen the RPCI-1,3–5 human PAC libraries and to isolate a breakpoint-spanning SmaI fragment from PAC RP1-179H24.
Cell Lines and FISH
Chimpanzee primary fibroblast cultures (PTR-CP132) and the lymphoblastoid cell line PTR-EB176 (ECACC number 89072704) were obtained from the European Collection of Cell Cultures. The chimpanzee lymphoblastoid cell line L-2008 was a generous gift from W. Schempp. The siamang gibbon (Hylobates [Symphalangus] syndactylus) lymphoblastoid cell line was established (as described by Koehler et al. [1995a]) by use of Epstein-Barr–virus transformation of peripheral lymphocytes from a gibbon housed at the Munich Zoo, and the lymphoblastoid cell line (described by Müller et al. 1998) for the white-cheeked gibbon (Hylobates [Nomascus] leucogenys) was kindly provided by the Kunming Cell Bank of the Chinese Academy of Sciences. The lymphoblastoid white-handed gibbon (Hylobates lar) cell line HY35 was the same as that described by Jauch et al. (1992) and was kindly provided by T. Ishida. BAC or PAC DNA and PCR products used as FISH probes either were labeled with biotin-16-desoxyuridine triphosphate (dUTP) (Roche Diagnostics) and detected with FITC-avidin and biotinylated antiavidin (Vector) or were labeled with digoxygenin-11-dUTP (Roche Diagnostics), detected by mouse antidigoxygenin coupled with Texas-Red and, in a second step, by antimouse antibody conjugated with Texas-Red (Dianova). Slides were counterstained with diaminophenylindole and were mounted with Vectashield antifade solution (Vector).
Long-Range PCR and Cloning of PCR Products
The reactions were performed with the Expand Long Template PCR System (Roche Diagnostics) with 1× buffer I, 350 μM dNTP, 0.25 μM each primer, 0.75 μl enzyme mix, and 100 ng template BAC DNA. The sequence and position of the primers used are listed in table 1. PCR products were cloned using the TOPO TA cloning kit (Invitrogen).
Table 1.
PCR Products Used to Identify the Inversion Breakpoint on 17q21.3
| Primer | Sequence 5′→3′ | Position on Human PAC RP5-1029K10a | Productb (size in bp) | Present on Chimpanzee BAC RP43-134L13c | FISH Signal onChimpanzeeChromosome |
| p61 | TCAAGGCGGTGGGGAAGAGA | 35594 | p61/19 | − | |
| p19 | AATCTAGACTGGAGGCCACATT | 35849 | (255) | ||
| p18 | GGGCCCTCCTGAAACTTACA | 29299 | p18/19 | − | 19p |
| p19 | AATCTAGACTGGAGGCCACATT | 35849 | (6550) | ||
| p59 | GGAGAAGAGGCCCTGCACACC | 38650 | p59/60 | − | |
| p60 | ATCCCCTTTCTTCTCTCCCTT | 39259 | (609) | ||
| p58 | GATTCATCTCTGCAGCCTCC | 40603 | p58/p38 | − | |
| p38 | CCGTGTGGAAGTGGATCTGAGT | 40738 | (135) | ||
| p37 | CACTCAGTCCTGCCCTCTTC | 34866 | p37/38 | − | 19p |
| p38 | CCGTGTGGAAGTGGATCTGAGT | 40738 | (5872) | ||
| p51 | CCTGCCTTAGGCTTAACC | 42123 | p51/52 | − | |
| p52 | AACTCTTCCCCCATCCAC | 42377 | (254) | ||
| p42 | TGCCTTCCCTGCAGAACTCCA | 43328 | p42/43 | − | |
| p43 | GAGAGGTAATTCTGTGTCC | 43550 | (222) | ||
| p45 | CGCCTTCCTGCGTAAGATAG | 43962 | p45/48 | − | |
| p48 | ACAGCAGAGCACGGAGGAA | 44214 | (252) | ||
| p62 | TCTTAATTGCTGACACTTGT | 48116 | p62/63 | − | |
| p63 | AAGGCAGTTGCCATTTGGTT | 48217 | (101) | ||
| p40 | ACCAGTAGTCGTGTCAAATA | 51178 | p40/41 | − | |
| p41 | CTGACGCAAAGGACAAAATA | 51809 | (631) | ||
| p12 | AAGTTTAGACTGCCAGGA | 58652 | p12/65 | + | |
| p65 | GTGACACTCAGACCAAGCCC | 59187 | (535) | ||
| p12 | AAGTTTAGACTGCCAGGA | 58652 | p12/13 | + | 19q |
| p13 | GCCTGTGGGGCAGAAGTGGA | 63717 | (5065) | ||
| p14 | CTCTGAGAGGAGCTGGCAGT | 66018 | p14/p15 | + | 19q |
| p15 | AGCCCCCTTTGCCCCACTCCA | 71331 | (5313) | ||
| p1 | CAAAAGGGGTGGAGGAAGGAA | 82020 | p1/17 | + | |
| p17 | GATCGCATCTCTGGAAATAGTC | 82790 | (770) | ||
| p1 | CAAAAGGGGTGGAGGAAGGAA | 82020 | p1/7 | + | 19q |
| p7 | ACAGCAGGGTGCACAATAGA | 85362 | (3342) | ||
| p3 | CCACAGGACTTGGGCAGGC | 92498 | p3/4 | nd | 19q |
| p4 | CCTGTCCACACCAGGAGCAAG | 99569 | (7071) | ||
| p5 | TTACGTAGCTATCTTCCCAA | 111880 | p5/10 | nd | 19q |
| p10 | GCTATTTGGCCAGCCTGTGTA | 114438 | (2558) | ||
| p5 | TTACGTAGCTATCTTCCCAA | 111880 | p5/6 | nd | 19q |
| p6 | GATGATTTCCCACTTTAGC | 123509 | (11629) |
Numbering according to the sequence of PAC RP5-1029K10 available for GenBank accession number AC006487.
All PCR products were amplified from DNA of PAC RP5-1029K10.
− = Absent; + = present; nd = not done.
Sequencing of Breakpoint-Spanning BAC and PAC Clones and Database Analysis
SmaI fragments of BAC RP43-141Q35 (10.5 kb) and PAC RP1-179H24 (11.5 kb) were fragmented by ultrasound nebulization. DNA ends of the resulting fragments were polished by Klenow polymerase and T4 polynucleotide kinase. Fragments were then ligated into pUC18 and were sequenced using the ABI Prism BigDye terminator kit and ABI Prism 377 and 3700 DNA sequencers. Sequence assembly and manual editing were performed with the GAP4 software (Dear and Staden 1991). The results of the assemblies were confirmed by restriction analyses of the SmaI fragments.
The sequence of the 10-kb SmaI fragment of BAC RP43-134L13 was determined using the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit on an ALF Express Sequencer (Amersham Pharmacia). After restriction with EcoRI, the 10-kb SmaI-fragment was subcloned into pUC18. Walking primers were designed, according to the end sequences of the cloned restriction fragments, to complete the sequence of all fragments. At least two clones of all restriction and PCR fragments were sequenced from both strands. Alignments and homology searches were performed using the Wisconsin Package, version 10.2 (Genetics Computer Group), and the BLAST search facilities of the National Center for Biotechnology Information. Repetitive elements were identified by Repeat Masker. Sequences around the breakpoints were compared with several sequences that are considered to be involved in chromosomal rearrangements. The sequences were topoisomerase II recognition sites topoIIv ([A/G]N[T/C]NNCNNG[T/C]NG[G/T]TN[T/C]N[T/C]) (Spitzner and Muller 1988), topoIId (GTN[T/A]A[C/T]ATTNATNN[A/G]) (Sander and Hsieh 1985), and topoIIi ([T/C][T/C]CNTA[C/G][C/T]CC[T/G][T/C][T/C]TNNC) (Kas and Laemmli 1992), as well as translin recognition sites ATGCAG and GCCC[A/T][G/C][G/C][A/T] or GCNC[A/T][G/C]CT) (Aoki et al. 1995).
Identification of Flanking Genes
The ALOX15B gene was identified adjacent to the inversion breakpoint in chimpanzee 19q on the proximal site, by use of cloning and sequencing of EcoRI and PstI restriction fragments, which overlap the breakpoint-spanning 10-kb SmaI fragment of BAC RP43-134L13. In an analogous manner, we physically mapped and sequenced the regions flanking the 11.5-kb breakpoint-spanning SmaI fragment of PAC RP1-179H24 on human 17p13 and the 10.5-kb SmaI fragment of BAC RP43-141Q35 in chimpanzee 19p. NIX analysis to search for expressed sequences in the breakpoint regions were performed through the U.K. Human Genome Mapping Project Resource Center Web site.
Expression Analysis and Real-Time PCR
Cytoplasmic RNA was isolated using the RNeasy kit (Qiagen) and lysis buffer RLN. First-strand cDNA was prepared with 2 μg total RNA, as determined by absorbance, random hexamers, and the SuperScript preamplification system (Invitrogen). The relative expression of the NXPH3 gene in lymphoblastoid and fibroblast cell lines from chimpanzee and human was quantified using real-time PCR analysis and the 2-ΔΔCT or comparative CT method (TaqMan, ABI Prism 7700 sequence detection system [PE Applied Biosystems]). We used β-actin as an internal control gene to normalize for the amount of RNA added to the real-time assays. The following primers and concentrations were used: β-actin (5′-TCA CCC ACA CTG TGC CCA TCT ACG A-3′ [forward, 300 nM] and 5′-CAG CGG AAC CGC TCA TTG CCA ATG G-3′ [reverse, 50 nM]); NXPH3 gene (5′-CAC CCT CAT TGG CCC ATT CT-3′ [forward, 300 nM] and 5′-AGC CCC GGT TCC AAG CT-3′ [reverse, 300 nM]). The PCR conditions were: denaturation at 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, using Sybr Green PCR core reagents (PE Applied Biosystems). Semiquantitative values were obtained from the cycle threshold (CT) number at which the increase in signal associated with an exponential growth of PCR product is first detected. The relative NXPH3 gene expression in a given sample was then normalized to the endogenous reference (β-actin) and calculated as ΔCT=CTNXPH3-CTβ-actin. The ΔCT value was then normalized to a calibrator, which was the ΔCT value of chimpanzee cell line PTR-L2008, for comparison of lymphoblastoid cells, or the ΔΔCT value of chimpanzee fibroblast cells CP132, for fibroblastlike cells. This normalization step is expressed by the ΔΔCT value, which results from the subtraction of the respective ΔCT sample value from the ΔCT calibrator value. The final results are presented as N-fold differences in target-gene expression, relative to endogenous β-actin and the calibrator, by use of the formula 2-ΔΔCT, according to the instruction manual of the TaqMan detection system.
Study of Replication Timing
Lymphoblastoid and fibroblast cultures of chimpanzee and human were incubated with 3×10-5 M bromodeoxyuridin (BrdU) prior to collection, to identify cells in S phase. After 2 h, cells were collected, incubated in hypotonic 0.4% KCl solution for 15 min, fixed in methanol and acetic acid (at a ratio of 3:1), and placed on slides. FISH was performed with biotin-labeled human BAC DNA. Signals were detected with FITC-avidin and biotinylated antiavidin (Vector). Cells that had incorporated BrdU were visualized by incubation with an anti-BrdU mouse antibody (Roche Diagnostics) that was diluted 1:100 and by a second antibody-detection step, using a Texas-Red–conjugated anti-mouse antibody diluted 1:100. Slides were counterstained with 4′, 6-diamino-2-phenylindole and were mounted with antifade solution. To estimate the replication timing of the sequence represented by each BAC clone during S phase, we evaluated at least 100 BrdU-labeled interphase nuclei for the presence of two double signals, one single and one double signal, or two single hybridization signals of the respective BAC clone (Simon et al. 1999).
Results
Identification of Breakpoint-Spanning BACs and PACs and Subcloning of the Breakpoints
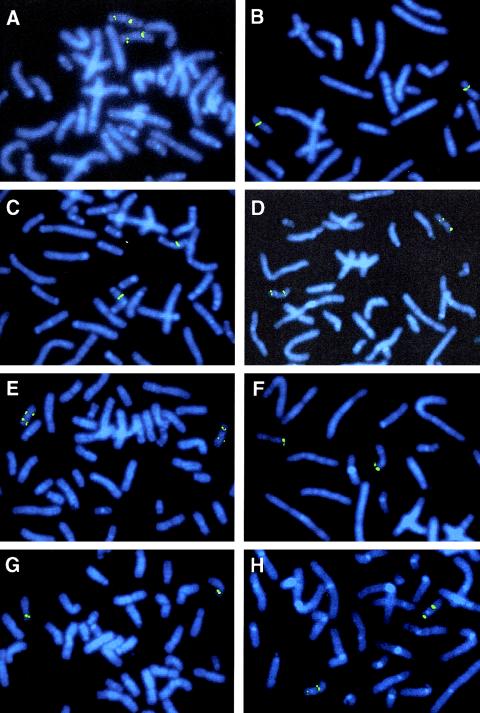
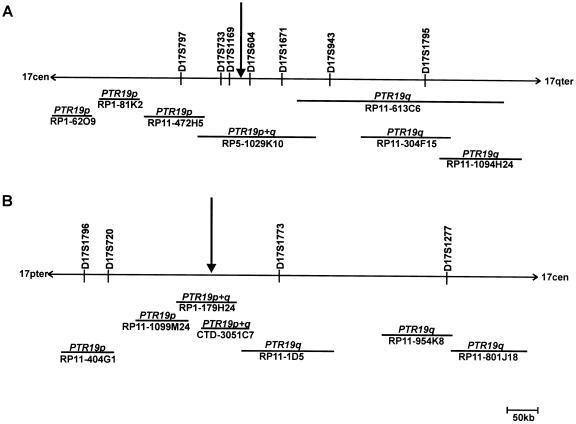
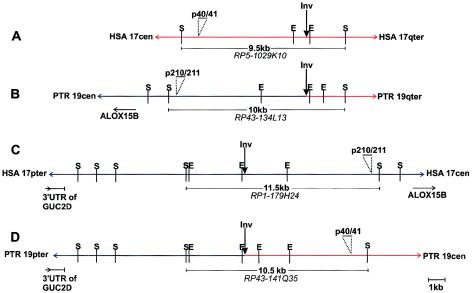
To identify BACs or PACs that span the inversion breakpoints, human clones from the 17q21.3 and the 17p13 region registered in the Ensembl database were hybridized to human and chimpanzee metaphase chromosomes. We identified PAC clone RP5-1029K10 to span the breakpoint syntenic to human 17q21.3, since a split hybridization signal is observed by FISH on chimpanzee chromosome 19, one on the p and the other on the q arm (figs. 2A and 2B). All BACs flanking clone RP5-1029K10 hybridized to human 17q21.3 and to either chimpanzee 19p or chimpanzee 19q (fig. 3A). Because the sequence of PAC RP5-1029K10 (GenBank accession number AC006487) is completely available, we were able to narrow the region harboring the inversion breakpoint by use of PCR and FISH analysis, as summarized in table 1. We amplified 2.5–11.6 kb, spanning PCR products from different regions of PAC RP5-1029K10, and hybridized these PCR products to chimpanzee and human chromosomes. PCR products that hybridized to chimpanzee 19q are located distal to the breakpoint, whereas those located on chimpanzee 19p are proximal to the breakpoint. According to the FISH results (table 1), the breakpoint was confined to the region between fragments p37/38 and p12/13 (between nucleotide positions 40738 and 58652, according to the numbering of AC006487). Using PCR fragment p1/17, which is located in the noninverted segment of PAC RP5-1029K10, as radioactively labeled probe, we screened the chimpanzee BAC library RPCI-43 and isolated BAC RP43-134L13. FISH with this BAC clone resulted in a single signal on chimpanzee 19q (fig. 2C), but two signals on human metaphase chromosomes, one at 17p13 and one at 17q21.3, substantiating that chimpanzee BAC RP43-134L13 covers the inversion breakpoint (fig. 2D). The inversion breakpoints in chimpanzee BAC RP43-134L13 and human PAC RP5-1029K10 were identified by comparative PCR with the primer pairs listed in table 1. PCR products up to primer pair p12/65 were amplified from both clones, but PCR products proximal to p12/65 could be amplified only from the human PAC RP5-1029K10, not from the chimpanzee BAC. The absence of fragments p51/52, p45/48, and p40/41 from chimpanzee BAC RP43-134L13 was confirmed by Southern blot analysis. Taken together with the FISH results, these findings strongly suggest that the breakpoint is located between fragments p12/65 and p40/41 (table 1). To isolate the breakpoint-spanning fragment from BAC RP43-134L13, Southern blot analysis was performed with probe p12/65. A 10-kb SmaI fragment was isolated from chimpanzee BAC RP43-134L13, subcloned, and sequenced (AY447035). Sequence comparison between this 10-kb SmaI fragment and human PAC RP5-1029K10 helped to identify the breakpoint on human 17q21.3 (figs. 4A and 4B) in a 0.7-kb EcoRI fragment. The breakpoint within the chimpanzee BAC RP43-134L13 could be assigned to a 2.8-kb EcoRI fragment (fig. 4B).
Figure 2.
FISH with breakpoint-spanning human PAC clones RP5-1029K10 (A and B) and RP1-179H24 (E and F) and chimpanzee BACs RP43-134L13 (C and D) and RP43-141Q35 (G and H). A, C, E, and G, Results of hybridizations on chromosome preparations of chimpanzee lymphoblastoid cell lines. B, D, F, and H, Results of FISH on human lymphocyte metaphases.
Figure 3.
Position of breakpoint-spanning human PAC clones and flanking BACs on human 17q21.3 (A) and human 17p13 (B). The FISH hybridization pattern of each BAC or PAC on chimpanzee chromosomes is given in italics above each horizontal line. The line represents the relative length of each clone. Vertical arrows point to the position of the breakpoints.
Figure 4.
Structure of the breakpoint region on human 17q21.3 (A), chimpanzee 19q (B), human 17p13 (C), and chimpanzee 19p (D). The sequences with homology to human 17p13 are shown as blue lines, whereas regions of homology to 17q21.3 are in red. SmaI and EcoRI restriction sites are indicated. PCR products p40/41 and p210/211 were used as labeled probes in Southern blot hybridizations to isolate breakpoint-spanning SmaI fragments. Their positions are indicated by horizontal bars and the respective sizes of these fragments are added. The flanking genes, which have been identified by physical mapping and sequencing of cloned fragments overlapping the breakpoint-spanning SmaI fragments, are also indicated. E = EcoRI; S = SmaI.
To find a breakpoint-spanning BAC on human 17p13, we hybridized BACs from this region that were registered in the Ensembl database to human and chimpanzee chromosomes, but a breakpoint-spanning BAC could not be identified. Using the sequence information from the 10-kb SmaI fragment, which spans the inversion breakpoint of chimpanzee BAC RP43-134L13, we generated a PCR product (p210/211) from the proximal part of the SmaI fragment with sequence homology to human 17p13 (fig. 4B). We used probe p210/211 to screen the human RPCI-1,3–5 PAC libraries and isolated PAC RP1-179H24, which spans the breakpoint, as indicated by a split hybridization signal on chimpanzee chromosome 19 (fig. 2E) and a single FISH signal at human 17p13 (fig. 2F). Partial sequencing, by BLAST analyses, of this BAC helped to identify another human BAC clone (CTD-3051C7), which also turned out to span the inversion breakpoint when analyzed by use of FISH. The map positions of these and the neighboring BACs on human 17p13, together with the corresponding hybridization pattern on chimpanzee 19, are shown schematically in figure 3B. The inversion-spanning fragment of PAC RP1-179H24 was identified by Southern blot analysis, using PCR product p210/211 as a probe. An 11.5-kb SmaI fragment (GenBank accession number AF503579) was identified and completely sequenced (fig. 4C). This sequence completely matches the sequence of BAC RP11-769H22, which has been released by GenBank (GenBank accession number AC104762) during the preparation of the manuscript. To isolate a chimpanzee BAC that spans the chimpanzee 19p inversion breakpoint, PCR product p42/43 (nucleotide position 43328–43550 on PAC RP5-1029K10 [table 1]) was used to screen the chimpanzee BAC library. By this method, we identified BAC RP43-141Q35, which turned out to span the inversion breakpoint when analyzed by FISH (figs. 2G and 2H). The breakpoint-spanning 10.5-kb SmaI fragment of BAC RP43-141Q35 was identified by use of Southern blot hybridization with PCR product p40/41 as a probe (fig. 4D). This 10.5-kb SmaI fragment was likewise subcloned and sequenced (AF503578).
Structure of the Breakpoint-Spanning Sequences
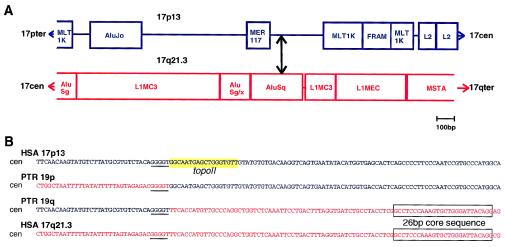
The sequence of the breakpoint-spanning fragments from the chimpanzee BACs was compared with the corresponding sequences on human 17p13 and 17q21.3. The breakpoint within PAC RP5-1029K10 on human 17q21.3 occurred within a region rich in short interspersed repetitive elements and in long terminal repeat (LTR) elements in an AluSq-element (fig. 5A and 5B). At a position 60 bp distal to the q-arm–specific breakpoint, a 26-bp Alu-core sequence is found, which is often observed close to sites of recombination (Rüdiger et al. 1995). The inversion breakpoint on human 17p13 was assigned to a 300-bp, small single-copy segment, which is also included in a repeat-rich region (fig. 5A). The actual chromosomal breaks occurred before, within, or directly after an inverted pentamer repeat, GGGGT. Addition or loss of nucleotides at the sites of rearrangement was not observed when human and chimpanzee sequences were compared. Immediately distal to the p-arm–specific breakpoint, a sequence with 83% similarity to the consensus topoisomerase IIv cleavage site was found (fig. 5B).
Figure 5.
Breakpoints on human 17p13 and 17q21.3. A, Repeat organization of the regions that harbor the inversion breakpoints on human 17p13 and human 17q21.3 as determined by the Repeat Masker program. The breakpoints were found in regions rich in repeats such as Alu elements (including a free right arm Alu-monomer [FRAM]), medium reiteration-frequency repeats (MER), parts of LINEs (L1, L2, L1MC3, and L1Mec), and LTR-element family members/mammalian retrotransposon-like superfamily (MLT1K and MSTA). The breakpoint is indicated by a double-headed arrow. B, Aligned sequences of human 17q21.3 in red letters and human 17p13 in blue letters and the respective sequences from chimpanzee 19p and 19q spanning the inversion breakpoints. The pentamer repeat GGGGT at the breakpoints is underlined. The 26-bp core sequence of the AluSq-element is marked by a box. At the human 17p breakpoint, a sequence with 83% similarity to the consensus topoisomerase IIv cleavage site is highlighted in yellow.
Identification of Genes Flanking the Inversion Breakpoints
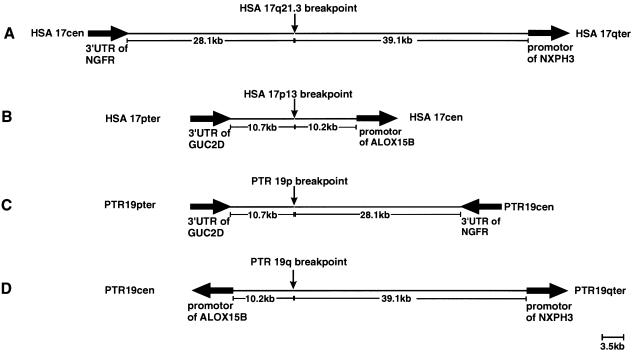
Sequence analysis and database searches of the breakpoint-covering fragments did not give any hint that a gene is directly affected by the inversion breakpoints. NIX analysis of the completely sequenced PAC RP5-1029K10 showed that the breakpoint on human 17q21.3 is 28.1 kb distal to the 3′ end of the nerve growth factor–receptor gene (NGFR [MIM 162010]) and 39.1 kb proximal to the promoter of the neurexophilin 3 gene (NXPH3 [MIM 604636]) (fig. 6A). The breakpoint on human 17p13 is 10.7 kb distal to the 3′ UTR of the retina-specific guanylyl cyclase 2D gene (GUC2D [MIM600179]) and 10.2 kb proximal to the promoter of the arachidonate 15-lipoxygenase 2 gene (ALOX15B [MIM 603697]) (fig. 6B). The position and orientation of these four breakpoint-flanking genes on chimpanzee chromosome 19 is shown in figures 6C and 6D. Because of the inversion in chimpanzee 19p, the 3′ end of the NGFR gene is located 28.1 kb proximal to the breakpoint, whereas, in chimpanzee 19q, theALOX15B and the NXPH3 genes are arranged in a head-to-head manner, as confirmed by physical mapping of the corresponding regions and by PCR analysis.
Figure 6.
Distance and orientation of the genes flanking the pericentric inversion breakpoints on human 17q21.3 (A), on human 17p13 (B), in chimpanzee 19p (C), and in chimpanzee 19q (D). Thick arrows indicate the transcription orientation of each gene.
Investigations on Replication Timing of Sequences Flanking the Breakpoints
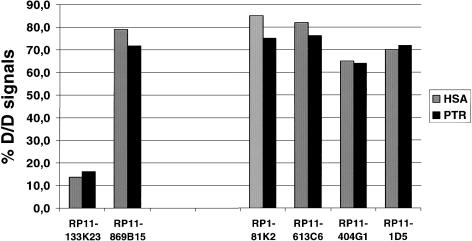
To evaluate whether the inversion affects the state of gene activity, the replication timing of flanking sequences during S-phase was determined. We assessed the number of signals of breakpoint-flanking BACs in BrdU-labeled interphase nuclei of chimpanzee and human by use of FISH. A high percentage of two double signals of a given BAC is indicative of early replication, whereas a preponderance of single dots characterizes late-replicating DNA. BAC RP11-133K23 from a late-replicating region at chromosome 7q31.2 that contains parts of the cystic fibrosis transmembrane conductance regulator gene (CFTR) and BAC RP11-869B15 from an early-replicating region at chromosome 11q13.1 spanning the myophosphorylase gene (PYGM) were used as control probes and were hybridized to slides from the same preparation as was used for the investigation of the breakpoint-flanking BACs. When fibroblasts from human and chimpanzee were compared with regard to replication timing of sequences surrounding the breakpoint regions, we did not observe any difference. A similar high percentage of S-phase cells with two doublet signals indicates early replication in human and chimpanzee (fig. 7). Nearly identical results were obtained by comparison of lymphoblastoid cell lines of chimpanzee and human (data not shown).
Figure 7.
Evaluation of replication timing of sequences flanking the pericentric inversion breakpoints with BACs and PACs from human 17q21.3 (RP1-81K2 and RP11-613C6) and from human 17p13 (RP11-404G1 and RP11-1D5) as FISH probes (see fig. 3 for position of BACs relative to the breakpoint). The percentage of two doublet signals was determined in BrdU-labeled S-phase nuclei of fibroblastlike cells from chimpanzee (chimpanzee cell line CP132) and primary human foreskin fibroblasts. FISH analysis was also performed with BAC RP11-133K23 as reference probe for a late-replicating genomic region and with BAC RP11-869B15 for an early-replicating genomic region. D/D = two doublets; HSA = Homo sapiens; PTR = Pan troglodytes.
Expression Analysis of the Genes Flanking the Inversion Breakpoints in the Chimpanzee
The expression of the genes NGFR, GUC2D, and ALOX15B in lymphoblastoid and fibroblastlike cells of chimpanzees and humans was too low for a comparison. However, the NXPH3 gene was expressed highly enough in the cell lines available to us to quantify the relative amounts by real-time PCR and the comparative CT method. The ΔCT value of chimpanzee lymphoblastoid cell line L2008 was used as a calibrator value to determine the NXPH3 expression level of a second chimpanzee cell line (PTR-EB176) and of four different human lymphoblastoid cell lines (human-L1–L4) relative to PTR-L2008. We observed 0.56–3.59-fold differences in NXPH3 expression of human cell lines compared with the expression of PTR-L2008. When we compared cell line PTR-EB176 with PTR-L2008, we noticed a 3.38-fold difference in NXPH3 expression. However, fluctuations in NXPH3 expression by a factor of 1.4–3.7 occurred, if different RNA preparations from the same cell line were evaluated. Similar ranges of differences in NXPH3 expression were observed in two human foreskin fibroblast cultures relative to chimpanzee fibroblast line CP132 (data not shown).
FISH Analysis of Breakpoint-Spanning Human PACs on Three Gibbon Species
To investigate whether the regions involved in the pericentric inversion of chimpanzee 19 have been utilized more than once during hominoid evolution, we hybridized the breakpoint-spanning PACs RP5-1029K10 and RP1-179H24 to metaphase chromosomes of H. lar, H. leucogenys, and H. syndactylus. In these gibbon species, the sequences homologous to human chromosome 17 are distributed on two chromosomes (H. leucogenys and H. syndactylus) or three chromosomes (H. lar). In contrast to findings in other great apes, numerous translocations were detected in gibbons, and these translocations led to a massive reorganization of their karyotypes (Jauch et al. 1992; Koehler et al. 1995a, 1995b). The results of these FISH analyses are summarized in table 2. With each of the human PACs used as FISH probe, we observed a single signal on gibbon chromosomes, which suggested that they do not cover an evolutionary breakpoint in one of these species.
Table 2.
FISH Pattern of Breakpoint-Spanning Human PAC Clones on Metaphase Chromosomes of Three Gibbon Species
|
FISH Signals of Human PACs Spanning Breakpoints of Pan troglodytes on Chromosomes of |
|||
| PAC | H. lar | H. leucogenys | H. syndactylus |
| RP1-179H24(human 17p13) | 8q | 19qter | 20qter |
| RP5-1029K10(human17q21.3) | 13p | 19q11-12 | 20q11-q12 |
FISH signals are from the pericentric inversion breakpoints.
Discussion
In the present study, we investigated the breakpoint regions of the pericentric inversion of chimpanzee chromosome 19, which is homologous to human chromosome 17. This inversion occurred after the divergence of humans and chimpanzees 5–6 million years ago. Since the inversion is also observed in the pygmy chimpanzee, or bonobo (Pan paniscus), the rearrangement predates at least the divergence of Pan paniscus from other Pan species ∼2 million years ago. The G-banding pattern of chimpanzee, gorilla, and orangutan (Pongo pygmaeus) chromosomes homologous to human chromosome 17 shows a distinct evolutionary history of the human 17-homologous chromosomes in each great ape species (Yunis and Prakash 1982; fig. 1). A similar scheme, as given for human 17, has been elaborated for human 4 (Marzella et al. 2000). This comparison demonstrated that the human arrangement represents the ancestral form and that the homologous chromosomes of other hominoid species are derived forms resulting from single species-specific inversion events. Comparative-mapping studies in hominoids suggest that human and orangutan karyotypes are most similar to the ancestral primate karyotype (Müller and Wienberg 2001).
It has been suggested that pericentric inversions have been important for the establishment of reproductive isolation and speciation of hominoids. King (1993) reviewed the possible speciation mechanism by multiple pericentric inversions. The single pericentric inversion is thought to have no deleterious effect on fertility in the heterozygous state, owing to overriding mechanisms such as changes in the position of chiasma formation. If there are no meiotic effects in F1 hybrids, the fixation of pericentric inversions by stochastic processes is facilitated. Successively further pericentric inversions, each with no or only minimal depressive effects on fertility, are repaired. However, further interbreeding is interrupted by postmeiotic effects when multiple inversions meet in the heterozygous state in an individual of the chromosomally derived and the parental populations. Recombinational effects of multiple pericentric inversions might enforce sterility barriers in F2 generations and backcross matings.
According to this scenario, it will be difficult to assign genetic effects to single pericentric inversions. Nevertheless, other contributions of pericentric inversions to evolution may result from changes of the sequence environment or gene structure at the breakpoint sites. Therefore, the molecular characterization of the rearrangements by which human and chimpanzee differ from each other is necessary. Our analysis of the pericentric inversion breakpoints of chimpanzee 19 showed that one breakpoint is located between the NGFR and the NXPH3 genes on human 17q21.3 and that the other occurred between the GUC2D and the ALOX15B gene on human 17p13. Concerning their genomic structure, neither of these genes is affected by the breakpoints (fig. 6). As demonstrated with BACs from the flanking regions, the inversion apparently has no effects on the replication timing of these genes during S phase (fig. 7), yet it remains possible that the inversion influences the transcription of the genes flanking the breakpoints. Unfortunately, the expression of the GUC2D, ALOX15B, and NGFR genes in lymphoblastoid and fibroblastlike cells of chimpanzee and human was too low to be assessed quantitatively. Only the NXPH3 gene was expressed strongly enough in the cell lines available to us to compare the expression level by real-time PCR. Significant differences between NXPH3 expression in chimpanzee cell lines and that in human cell lines were not observed. The relevance of the comparative-expression studies of NXPH3 in lymphoblastoid and fibroblastlike cell lines, however, is limited. Neurexophilins function as neuropeptidelike glycoproteins via α-neurexins, which are neuronal membrane proteins with a domain structure similar to that of cell surface receptors (Missler and Südhof 1998). More extended comparative expression studies, especially including neuronal cells or brain tissue, as performed by Enard et al. (2002), are needed to assess possible expression differences of the breakpoint-flanking genes in humans and chimpanzees. Conspicuously, two genes, NXPH3 and NGFR, which encode for proteins with neurospecific functions, are located in direct proximity to the breakpoints.
To our knowledge, this is the first inversion by which chimpanzees differ from humans that has been cloned and characterized on the molecular level. The breakpoints were identified in regions rich in members of the repetitive-gene family (fig. 5A). The breakpoint on human 17p occurred in a small single-copy segment flanked by a medium reiteration-frequency repeat (MER117) and a MLT1K element, a member of the mammalian retrotransposon-like superfamily. At this breakpoint, a sequence with strong similarity to the consensus topoisomerase IIv cleavage site was found, which suggests the involvement of topoisomerase IIv in the creation of this break (fig. 5B). Topoisomerase II generates double-strand breaks and, by this means, catalyzes the unknotting (or decatenation) of DNA. It has been proposed that staggered breaks created by topoisomerase II are transiently stabilized by the binding of translin to single-stranded DNA ends (Kanoe et al. 1999). We could not identify translin-binding sites at either inversion breakpoint. However, the breakpoint on human 17q21.3 occurred in an Alu element, which may mediate translin binding (Jeffs et al. 1998). The breakpoint within the Alu element is located proximal to the highly conserved 26-bp core sequence suspected to enhance recombination (fig. 5B; Rüdiger et al. 1995). Since the break in the p arm occurred in a short single-copy sequence, Alu-mediated nonhomologous recombination between the q- and p-arm sequences, with the creation of a large inversion loop, may have facilitated the inversion formation. Precisely at the sites of breakage, the pentamer sequence GGGGT was found, which has been shown to facilitate DNA cleavage by endonucleases (Lyon et al. 1996).
Sequence analysis of the breakpoint-spanning fragments in chimpanzee and human did not reveal any loss or addition of nucleotides at the sites of rearrangement. Therefore, it is unlikely that filling of overhanging ends, rearrangement, and religation underlie the inversion. These mechanisms have been proposed to be involved in a number of leukemia-associated inversions and translocations of human chromosomes (Zhang et al. 1995; Super et al. 1997; Romana et al. 1999; Todd et al. 2001). Our findings imply that the molecular basis of the evolutionary inversion of chimpanzee 19 was nonhomologous recombination, which was not associated with complex additional rearrangements, such as deletions or duplications at the breakpoint regions. This is in contrast to the findings of Stankiewicz et al. (2001), who investigated the evolutionary translocation t(4;19) in the gorilla. Stankiewicz et al. suggested that a duplication of ∼250 kb preceded the translocation. This duplication and insertion event might have rendered the genomic region unstable, promoting further rearrangements like the nonhomologous exchange between gorilla chromosomes 4 and 19. The duplication in gorilla chromosome 19p involved a fragment homologous to the chromosomal region surrounding the proximal Charcot-Marie-Tooth disease type 1A (CMT1A) repeat on human 17p12 and flanking the proximal Smith-Magenis syndrome (SMS) repeat. The CMT1A repeat and the SMS repeat are low-copy repeats that cause frequent constitutional genomic rearrangements, such as duplications and deletions, that lead to the above-mentioned disorders (Chen et al. 1997; Lupski 1998). The assignment of the t(4;19) breakpoint and the associated duplication observed in gorillas to a region on human 17p with low-copy repeats suggests that these genomic regions are not only susceptible to syndrome-associated constitutional rearrangements in humans but that they also played a role in hominoid karyotype evolution. In the sequence environment of the pericentric inversion breakpoints of chimpanzee 19, low-copy repeats or duplicated regions are not observed. The breakpoint regions are not involved in other evolutionary rearrangements (table 2), and we are not aware of common tumor-associated breakpoints in the corresponding genomic regions. The fact that the inversion took place in regions enriched with Alu and LTR elements suggests that repetitive elements themselves are recombinogenic and that they contributed to evolutionary rearrangements. Quantitative analyses of specific Alu subfamilies in the genomes of humans and the great apes demonstrated that the rate of Alu insertion increased during hominid evolution (Zietkiewicz et al. 1994; Schmid 1998).Therefore, Alu-related events most probably played a significant role in primate diversification.
Acknowledgments
We thank Helene Spöri and Antje Kollak for excellent technical assistance.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/ (for alignments and homology searches)
- Children's Hospital Oakland—BACPAC Resources, http://www.chori.org/bacpac (for chimpanzee BAC and human PAC libraries)
- Ensembl, http://www.ensembl.org/ (for human BACs)
- European Collection of Cell Cultures, http://www.ecacc.org.uk (for chimpanzee cell lines)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SIGLECL1 [MIM 606094], NGFR [MIM 162010], NXPH3 [MIM 606436], ALOX15B [MIM 603697], GUC2D [MIM 600179], SMN2 [MIM 601627}, and ELN [MIM 130160])
- Repeat Masker, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
- Resource Center and Primary Database for the German Human Genome Project, http://www.rzpd.de/ (for chimpanzee BAC and human PAC libraries)
- UK Human Genome Mapping Project Resource Center, http://www.hgmp.mrc.ac.uk/ (for NIX analysis)
References
- Angata T, Varki NM, Varki A (2001) A second uniquely human mutation affecting sialic acid biology. J Biol Chem 276:40282–40287 [DOI] [PubMed] [Google Scholar]
- Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M (1995) A novel gene, translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet 10:167–174 [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH (2001) Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 68:444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A (1998) A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci USA 95:11751–11756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear S, Staden R (1991) A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res 19:3907–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrillaux B, Rethore MO, Lejeune J (1975) Comparison of the karyotype of the orangutan (Pongo pygmaeus) to those of man, chimpanzee, and gorilla. Ann Genet 18:153–161 [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, Doxiadis GM, Bontrop RE, Pääbo S (2002) Intra- and interspecific variation in primate gene expression patterns. Science 296:340–343 [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N (2001) Alu-mediated inactivation of the human CMP-N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci USA 98:11399–11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA (1991) Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA 88:9051–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T (1992) Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci USA 89:8611–8615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs AR, Benjes SM, Smith TL, Sowerby SJ, Morris CM (1998) The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukaemia. Hum Mol Genet 7:767–776 [DOI] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE (2001) Positive selection of a gene family during the emergence of humans and African apes. Nature 413:514–519 [DOI] [PubMed] [Google Scholar]
- Kanoe H, Nakayama T, Hosaka T, Murakami H, Yamamoto H, Nakashima Y, Tsuboyama T, Nakamura T, Ron D, Sasaki MS, Toguchida J (1999) Characteristics of genomic breakpoints in TLS-CHOP translocations in liposarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene 18:721–729 [DOI] [PubMed] [Google Scholar]
- Kas E, Laemmli UK (1992) In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J 11:705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M (1993) Species evolution: the role of chromosome change. Cambridge University Press, Cambridge [Google Scholar]
- Koehler U, Arnold N, Wienberg J, Tofanelli S, Stanyon R (1995a) Genomic reorganization and disrupted chromosomal synteny in the siamang (Hylobates syndactylus) revealed by fluorescence in situ hybridization. Am J Phys Anthropol 97:37–47 [DOI] [PubMed] [Google Scholar]
- Koehler U, Bigoni F, Wienberg J, Stanyon R (1995b) Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30:287–292 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–22 [DOI] [PubMed] [Google Scholar]
- Lyon CJ, Miranda GA, Piao JS, Aguilera RJ (1996) Characterization of an endonuclease activity which preferentially cleaves the G-rich immunoglobulin switch repeat sequences. Mol Immunol 33:157–169 [DOI] [PubMed] [Google Scholar]
- Marzella R, Viggiano L, Miolla V, Storlazzi CT, Ricco A, Gentile E, Roberto R, Surace C, Fratello A, Mancini M, Archidiacono N, Rocchi M (2000) Molecular cytogenetic resources for chromosome 4 and comparative analysis of phylogenetic chromosome IV in great apes. Genomics 63:307–313 [DOI] [PubMed] [Google Scholar]
- Missler M, Südhof TC (1998) Neurexophilins form a conserved family of neuropeptide-like glycoproteins. J Neurosci 18:3630–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wienberg J (2001) Bar-coding primate chromosomes: molecular cytogenetic screening for the ancestral hominoid karyotype. Hum Genet 109:85–94 [DOI] [PubMed] [Google Scholar]
- Müller S, O'Brien PC, Ferguson-Smith MA, Wienberg J (1998) Cross-species colour segmenting: a novel tool in human karyotype analysis. Cytometry 33:445–452 [PubMed] [Google Scholar]
- Nickerson E, Nelson DL (1998) Molecular definition of pericentric inversion breakpoints occurring during the evolution of humans and chimpanzees. Genomics 50:368–372 [DOI] [PubMed] [Google Scholar]
- Rochette CF, Gilbert N, Simard LR (2001) SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet 108:255–266 [DOI] [PubMed] [Google Scholar]
- Romana S, Poirel H, Della Valle V, Mauchauffe M, Busson-Le Coniat M, Berger R, Bernard OA (1999) Molecular analysis of chromosomal breakpoints in three examples of chromosomal translocation involving the TEL gene. Leukemia 13:1754–1759 [DOI] [PubMed] [Google Scholar]
- Rüdiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well-conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Hsieh TS (1985) Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res 13:1057–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H (1999) Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401:929–932 [DOI] [PubMed] [Google Scholar]
- Schmid CW (1998) Does SINE evolution preclude Alu function? Nucleic Acids Res 26:4541–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D (1999) Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics 61:24–36 [DOI] [PubMed] [Google Scholar]
- Spitzner JR, Muller MT (1988) A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res 16:5533–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Park SS, Inoue K, Lupski JR (2001) The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res 11:1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Super HG, Strissel PL, Sobulo OM, Burian D, Reshmi SC, Roe B, Zeleznik-Le NJ, Diaz MO, Rowley JD (1997) Identification of complex genomic breakpoint junctions in the t(9;11) MLL-AF9 fusion gene in acute leukemia. Genes Chromosomes Cancer 20:185–195 [PubMed] [Google Scholar]
- Szabó Z, Levi-Minzi SA, Christiano AM, Struminger C, Stoneking M, Batzer MA, Boyd CD (1999) Sequential loss of two neighboring exons of the tropoelastin gene during primate evolution. J Mol Evol 49:664–671 [DOI] [PubMed] [Google Scholar]
- Todd R, Bia B, Johnson E, Jones C, Cotter F (2001) Molecular characterization of a myelodysplasia-associated chromosome 7 inversion. Br J Haematol 113:143–152 [DOI] [PubMed] [Google Scholar]
- Varki A (2001) Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol 33:54–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Stanyon R, Cremer T (1990) Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization. Genomics 8:347–350 [DOI] [PubMed] [Google Scholar]
- Winter H, Langbein L, Krawczak M, Cooper DN, Jave-Suarez LF, Rogers MA, Praetzel S, Heidt PJ, Schweizer J (2001) Human type I hair keratin pseudogene phihHaA has functional orthologs in the chimpanzee and gorilla: evidence for recent inactivation of the human gene after the Pan-Homo divergence. Hum Genet 108:37–42 [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O (1982) The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530 [DOI] [PubMed] [Google Scholar]
- Zhang JG, Goldman JM, Cross NC (1995) Characterization of genomic BCR-ABL breakpoints in chronic myeloid leukaemia by PCR. Br J Haematol 90:138–146 [DOI] [PubMed] [Google Scholar]
- Zietkiewicz E, Richer C, Makalowski W, Jurka J, Labuda D (1994) A young Alu subfamily amplified independently in human and African great apes lineages. Nucleic Acids Res 22:5608–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]