Significance
Retinoic acid inducible gene I-like RNA helicases (RLHs) act as virus sensors in mammalian antiviral immunity. RLHs also play an essential role in antiviral RNAi in the nematode Caenorhabditis elegans through a currently unknown mechanism. Here, we show that the retinoic acid inducible gene I domains involved in virus sensing functionally replace the corresponding domain of Dicer-related RNA helicase 1 (DRH-1), one of the worm RLHs, suggesting that DRH-1 contributes to antiviral RNAi by acting as a virus sensor. Consistent with this observation, virus-derived primary siRNAs were significantly reduced in drh-1 mutants. We further show that DRH-3, another worm RLH that shares domain structure with DRH-1, is required for the biogenesis of virus-derived secondary, but not primary, siRNAs, suggesting that DRH-1 and DRH-3 contribute to antiviral RNAi through distinct mechanisms.
Abstract
RNAi-mediated antiviral immunity in Caenorhabditis elegans requires Dicer-related helicase 1 (DRH-1), which encodes the helicase and C-terminal domains homologous to the mammalian retinoic acid inducible gene I (RIG-I)-like helicase (RLH) family of cytosolic immune receptors. Here we show that the antiviral function of DRH-1 requires the RIG-I homologous domains as well as its worm-specific N-terminal domain. We also demonstrate that the helicase and C-terminal domains encoded by either worm DRH-2 or human RIG-I can functionally replace the corresponding domains of DRH-1 to mediate antiviral RNAi in C. elegans. Notably, substitutions in a three-residue motif of the C-terminal regulatory domain of RIG-I that physically interacts with viral double-stranded RNA abolish the antiviral activity of C-terminal regulatory domains of both RIG-I and DRH-1 in C. elegans. Genetic analysis revealed an essential role for both DRH-1 and DRH-3 in C. elegans antiviral RNAi targeting a natural viral pathogen. However, Northern blot and small RNA deep sequencing analyses indicate that DRH-1 acts to enhance production of viral primary siRNAs, whereas DRH-3 regulates antiviral RNAi by participating in the biogenesis of secondary siRNAs after Dicer-dependent production of primary siRNAs. We propose that DRH-1 facilitates the acquisition of viral double-stranded RNA by the worm dicing complex for the subsequent processing into primary siRNAs. The strong parallel for the antiviral function of RLHs in worms and mammals suggests that detection of viral double-stranded RNA may activate completely unrelated effector mechanisms or, alternatively, that the mammalian RLHs have a conserved activity to stimulate production of viral siRNAs for antiviral immunity by an RNAi effector mechanism.
Innate immunity refers to ancient host defense systems that act immediately after pathogen attack and are under the control of germline-encoded immune receptors. Pathogen sensing by several classes of immune receptors such as retinoic acid inducible gene I (RIG-I)-like cytosolic sensors in mammals leads to the transcriptional activation of effector genes in the nucleus (1). In contrast, detection of viral double-stranded RNA (dsRNA) by Dicer endonucleases is associated with the production of small interfering RNAs (siRNAs), which subsequently direct sequence-specific antiviral immunity through RNA interference (RNAi) (2–5). Antiviral RNAi is a major antiviral defense mechanism in fungi, plants, insects, and nematodes, so that suppression of the defense by a virus-encoded suppressor of RNAi is essential to establish infection (6–8). Although the RNAi machinery is highly conserved in mammals, conclusive evidence for a natural antiviral role of RNAi in mammals is not available (9).
The nematode Caenorhabditis elegans has recently emerged as a small-animal model for innate immunity studies (10, 11). Genetic studies indicate that antiviral RNAi in C. elegans requires the genes essential for RNAi induced by exogenous dsRNA (12–17). These include Dicer-1 and dsRNA-binding protein RDE-4(RNA interference-deficient 4), involved in the biogenesis of primary siRNAs predominantly 23 nt in length, and Argonaute protein RDE-1 (RNA interference-deficient 1) and RNA-dependent RNA polymerase RRF-1, required for the production of secondary siRNAs (18–22). Worm secondary siRNAs are 22-nt single-stranded RNAs with G as the 5′-terminal nt and antisense to the target mRNA, and are often referred as 22G RNAs (23–25). Intriguingly, antiviral RNAi in C. elegans is also regulated by Dicer-related RNA helicase 1 (DRH-1), which is highly homologous to mammalian RIG-I and is largely dispensable for exogenous RNAi (15). Whereas antiviral RNAi induced by a Flock house virus (FHV)-based RNA replicon is dependent on drh-1, a reduced replication of the FHV replicon is detected in mutant nematodes defective in drh-2, which does not encode the N-terminal domains (NTDs) found in drh-1 and drh-3 (15, 18).
The homologous DExD/H-box helicase domain of mammalian RIG-I is flanked by the N-terminal caspase recruitment domains (CARDs) and a C-terminal regulatory domain (CTD). RIG-I binding of the viral 5′-triphosphate dsRNA through the helicase domain and CTD activates the downstream signaling events, leading to the induction of IFN-dependent antiviral immunity (26–28). The mammalian genomes also encode two additional RIG-I–like RNA helicases (RLHs), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). Both MDA5 and LGP2 contain CTD, but the N-terminal CARDs are found only in MDA5. MDA5 senses viruses that are not detected by RIG-I via a distinct strategy, whereas LGP2 seems to function as a regulator of RIG-I and MDA5 activities through a currently unknown mechanism (29, 30, 31–34).
In this study, we show that drh-1 and drh-3 of C. elegans are essential for antiviral RNAi but exhibit distinct antiviral activities. Using a transgenic approach developed to assay for the antiviral function of drh-1 in whole animals, we examined the antiviral activity of the predicted domains in DRH-1 and determined whether C. elegans antiviral RNAi could be mediated by any of the conserved domains of human RIG-I protein. Our results indicate that C. elegans antiviral RNAi requires an essential activity of DRH-1 to detect viral dsRNA in a manner analogous to virus dsRNA sensing by RIG-I in mammals.
Results
DRH-1 Is Required for Nematode Defense Against Infection by a Natural Virus Pathogen.
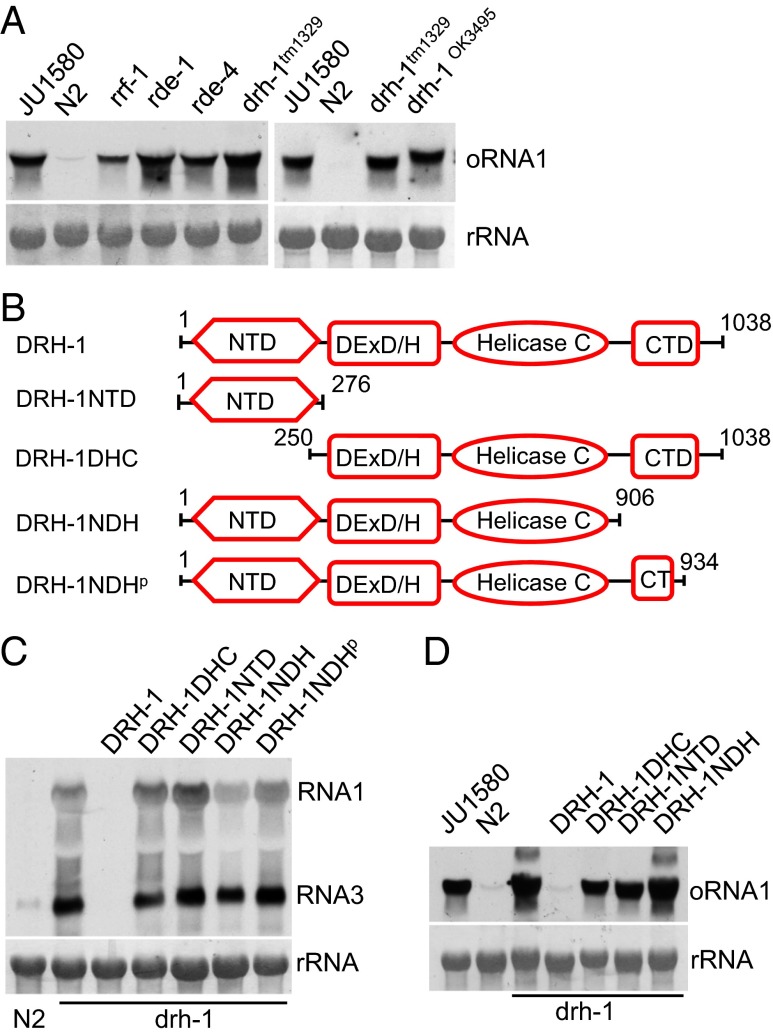
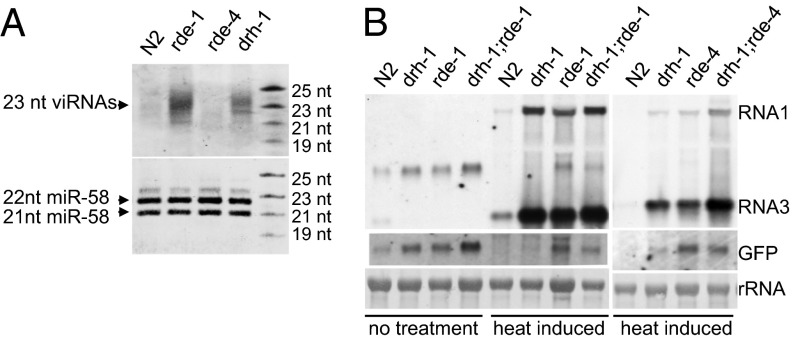
A antiviral role of DRH-1 was first identified through the characterization of antiviral RNAi induced by replication of FHV, which does not naturally infect C. elegans (15). A recent report described identification of a natural C. elegans virus, Orsay virus, which is closely related to FHV and has been shown to accumulate to significantly enhanced levels in RNAi-defective worm mutants such as rde-1 and rde-4 (16). We found that Orsay virus infection was also markedly enhanced in worm mutants carrying either of the two drh-1 loss-of-function alleles (Fig. 1A). This finding indicates that DRH-1 is also required for natural antiviral defense in C. elegans.
Fig. 1.
Both the NTD and CTD of DRH-1 are indispensable for antiviral RNAi. (A) Accumulation of Orsay virus RNA1 detected by Northern blot in wild-type worms and genetic mutants, as indicated. Methylene blue-stained ribosomal RNA (rRNA) serves as equal loading control. JU1580, an isolate of C. elegans that is defective in antiviral RNAi; N2, the Bristol isolate of wild type C. elegans. (B) Schematic structure of DRH-1 and its derivatives used in the DRH-1 function rescue assay. CTD, C-terminal regulatory domain originally identified in RIG-I; helicase C, the RNA helicase superfamily C-terminal domain; NTD, N-terminal domain. DExD/H, the DEAD-box superfamily domain. (C) Northern blot detection of FR1gfp transcripts in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1 or its derivatives, as indicated, 48 h after heat induction. Methylene blue-stained ribosomal RNA serves as equal loading control. (D) Orsay virus RNA1 detected by Northern blot in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1 or its derivatives, as indicated, 72 h after virus inoculation.
To verify the antiviral RNAi function of DRH-1, we carried out a transgene complementation assay in drh-1 mutant animals, as illustrated in Fig. S1A. In this assay, plasmid Psur-5::DRH-1 that directed DRH-1 expression under the constitutively active sur-5 promoter was injected into drh-1 mutant animals that carried the chromosomally integrated transgene FR1gfp controlled by a heat-inducible promoter (15). Heat treatment of drh-1;FR1gfp worms is expected to induce replication of the FHV-based RNA replicon and expression of the replicon-encoded green fluorescence protein (GFP) from a subgenomic mRNA synthesized by the FHV replicase. Because FR1gfp does not encode the RNAi suppressor protein B2 (15), green fluorescence would be undetectable if antiviral RNAi were restored in drh-1 mutant animals by ectopic expression of DRH-1. We found that green fluorescence indeed became undetectable in drh-1;FR1gfp worms after gonad microinjection with Psur-5::DRH-1 (compare animals carrying Psur-5::DRH-1, marked by the red fluorescence expressed from the coinjected mCherry reporter plasmid, with animals showing no red fluorescence in heads; Fig. S1B). This finding illustrated that ectopic expression of DRH-1 restored antiviral RNAi in the drh-1 mutants.
To further confirm this result, drh-1;FR1gfp worms carrying a stably integrated Psur-5::DRH-1 transgene were generated and used for the heat induction of FR1gfp or for Orsay virus infection. Northern blotting analysis showed that FR1gfp replication and Orsay virus infection were both suppressed in drh-1 mutants containing the Psur-5::DRH-1 transgene (Fig. 1 C and D). These results together show that DRH-1 plays an essential role in the worm antiviral RNAi induced by either FHV replication or Orsay virus infection.
Both the NTD and CTD of DRH-1 Are Indispensable for Antiviral RNAi.
The DRH-1 function rescue assay established earlier made it possible for us to map the domain requirement in the antiviral function of DRH-1. In addition to the DEAD-box RNA helicase domain, DRH-1 contains a worm-specific NTD and a conserved C-terminal regulatory domain originally identified in RIG-I (35). We generated four domain deletion mutants of DRH-1 (Fig. 1B), named DRH-1NTD, DRH-1DHC, DRH-1NDH, and DRH-1NDHp, and examined their antiviral activity using the DRH-1 function rescue assay. We found that ectopic expression of none of these four DRH-1 mutants restored antiviral RNAi in drh-1;FR1gfp worms (Fig. S1B), indicating that both of the terminal domains of DRH-1 are essential for its antiviral function. As a further confirmation, we generated chromosomal integrants carrying nuclear transgenes corresponding to each of these DRH-1 deletion mutants and assayed for both FR1gfp replication and Orsay virus infection in the selected animal lines. Northern blot analysis (Fig. 1 C and D) showed that neither FR1gfp replication nor Orsay virus infection was suppressed in integrants carrying any of the DRH-1 domain deletion mutants in contrast to the control integrants carrying the wild-type DRH-1 transgene. Western blotting suggested stable expression of both DRH-1NTD and DRH-1NDH examined in transgenic worms by C-terminal tagging with an HA epitope (Fig. S1 C and D). These results together indicate that in addition to the central helicase domains, both NTD and CTD of DRH-1 are indispensable for antiviral RNAi.
The Predicted Domains of DRH-2 Were Functional in a Fusion Protein with the NTD of DRH-1.
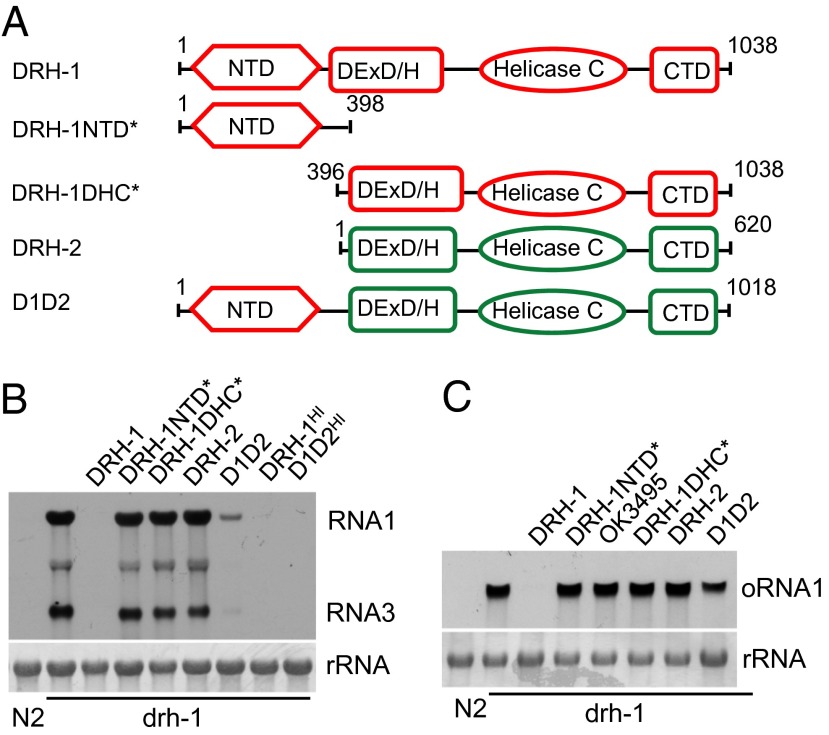
The predicted DRH-2 protein is highly homologous to DRH-1 (Fig. S2) but lacks the worm-specific NTD plus one of the three motifs in the DEAD-box helicase domain. We determined whether use of the predicted protein domains of DRH-2 to replace the corresponding domains of DRH-1 produced a chimeric protein capable of mediating antiviral RNAi in drh-1 mutant background. We found that ectopic expression of such a chimeric protein, D1D2 (Fig. 2A), was associated with loss of GFP expression in drh-1;FR1gfp worms (Fig. S3A), indicating rescue of DRH-1 function in antiviral RNAi by D1D2 driven by the constitutive sur-5 promoter. We next generated stable animal lines expressing D1D2 driven by either sur-5 or the heat-inducible promoter in drh-1;FR1gfp background. Northern blot analysis showed that expression of D1D2 driven by the sur-5 promoter suppressed the replication of both FR1gfp and Orsay virus in the drh-1 mutant background (Fig. 2 B and C). Notably, expression of D1D2 using the heat-inducible promoter conferred stronger antiviral RNAi targeting FR1gfp replicon (Fig. 2B). However, transgenic expression of DRH-2, the region of DRH-1 equivalent to DRH-2 (DRH-1DHC*), or the N-terminal region of DRH-1 absent in DRH-2 (DRH-1NTD*), was insufficient to rescue DRH-1 function (Fig. 2 A–C and Fig. S3A). Epitope-tagging and Western blotting assay suggested stable expression of DRH-1NTD*, DRH-1DHC,* and DRH-1 in transgenic worms (Fig. S3B). Thus, these findings indicate that when expressed as a fusion protein with the NTD of DRH-1, the predicted helicase and CTD domains of DRH-2 mediate antiviral RNAi as effectively as the corresponding domains of DRH-1.
Fig. 2.
The putative domains encoded by DRH-2 mediate antiviral RNAi when fused with the NTD of DRH-1. (A) Schematic structure of DRH-1, DRH-2, and their domain variants. See Fig. 1B and main text for more details. (B) Accumulation of FR1gfp transcripts detected by Northern blot in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1, putative DRH-2, or their domain variants, as indicated. HI denotes DRH-1 function rescue using a heat-inducible promoter for candidate gene expression. (C) Accumulation of Orsay virus RNA1 detected by Northern blot in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1, putative DRH-2, or their domain variants, as indicated. OK3495, a DRH-1 variant encoded by the ok3495 allele.
The Helicase and CTD Domains of RIG-I Were Functional in a Fusion Protein with the NTD of DRH-1.
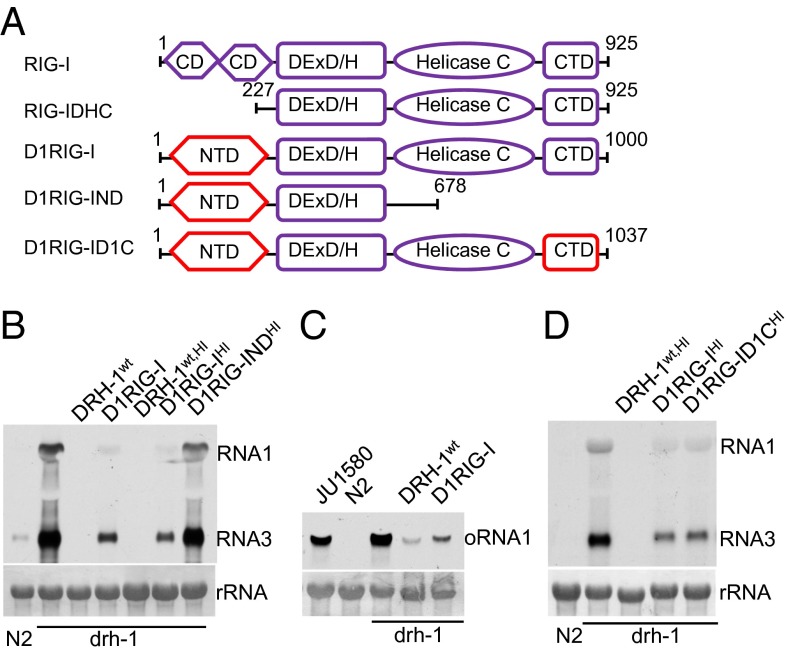
An essential role for both the NTD and CTD of DRH-1 in the antiviral defense is analogous to the domain requirement of mammalian RIG-I (36). DRH-1 shares significant sequence homology in the RNA helicase and CTD domains with RIG-I (Fig. S4), although by comparison, DRH-1 and RIG-I are more distantly related than DRH-1 and DRH-2. We found that transgenic expression of human RIG-I or a truncation mutant removing the N-terminal CARD domains (RIG-IDHC) in drh-1;FR1gfp worms did not suppress replication of FR1gfp (Fig. 3A and Fig. S5A). Because the worm-specific NTD of DRH-1 was indispensable in antiviral RNAi, we next determined the antiviral activity of a DRH-1/RIG-I chimeric protein termed D1RIG-I, in which the N-terminal CARD domains of human RIG-I were replaced with the NTD of DRH-1 (Fig. 3A). As shown in Fig. S5A, ectopic expression of D1RIG-I, but not D1RIG-IND, effectively restored antiviral RNAi in drh-1;FR1gfp worms, indicating that the helicase domain and CTD of RIG-I confer biological function similar to what the corresponding DRH-1 domains do. Similarly, antiviral RNAi was also restored by another DRH-1/RIG-I chimeric protein, D1RIG-ID1C, in which only the central helicase domain of DRH-1 was replaced by the corresponding domain of RIG-I (Fig. 3A and Fig. S5A). This finding showed that antiviral RNAi was mediated by the helicase domain of RIG-I in the context of the NTD and CTD of DRH-1.
Fig. 3.
The RNA helicase and CTD domains of RIG-I functionally replace the corresponding domains of DRH-1 in antiviral RNAi. (A) Schematic structure of RIG-I and its domain variants. (B) Accumulation of FR1gfp transcripts detected by Northern blot in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1 or RIG-I domain variants, as indicated. HI denotes DRH-1 function rescue using a heat-inducible promoter for candidate gene expression. (C) Accumulation of Orsay virus RNA1 detected by Northern blot in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1 and D1RIG-I. (D) Accumulation of FR1gfp transcripts detected by Northern blot in drh-1 mutants carrying the integrated transgenes corresponding to wild-type DRH-1, D1RIG-I, and D1RIG-ID1C.
As described earlier, we further generated stable animal lines carrying each of these DRH-1/RIG-I chimeric constructs and assayed for both FR1gfp replication and Orsay virus infection in the selected animal lines. Consistent with earlier observations, replication of both FR1gfp and Orsay virus was significantly inhibited in stable transgenic animals constitutively expressing D1RIG-I, and heat-inducible expression of D1RIG-ID1C restored antiviral RNAi to an extent comparable with that restored by D1RIG-I (Fig. 3 B–D). As observed earlier for D1D2, the heat-inducible promoter directed a more efficient rescue on DRH-1 function in antiviral FR1gfp silencing than the constitutive promoter (Fig. 3B). These results together indicate that either of the RNA helicase and CTD domains of RIG-I is competent to functionally replace the homologous domain of DRH-1 in antiviral RNAi in C. elegans.
Previous structural studies showed that a KWK motif in the CTD of RIG-I mediates physical interaction between RIG-I and dsRNA, although it is unknown whether the interaction is functionally important (26, 27, 37). Because the KWK motif is conserved in DRH-1 CTD (Fig. S5B, Upper), we next determined whether this motif is critical for antiviral RNAi in C. elegans. To this end, we introduced alanine substitutions into the KWK motif in the CTD domains of both DRH-1 and D1RIG-I and subjected the resulting mutant constructs, DRH-1AAA and D1RIG-IAAA, to DRH-1 function rescue assay. As shown in Fig. S5B (Lower) and Fig. S5C, expression of either mutant was unable to restore DRH-1 function in antiviral RNAi against FR1gfp. These findings strongly suggest that antiviral RNAi mediated by DRH-1 involves an essential activity of DRH-1 to detect viral dsRNA in a manner analogous to virus dsRNA sensing by RIG-I in mammals.
DRH-3 Regulates Antiviral RNAi by a Mechanism Distinct from DRH-1.
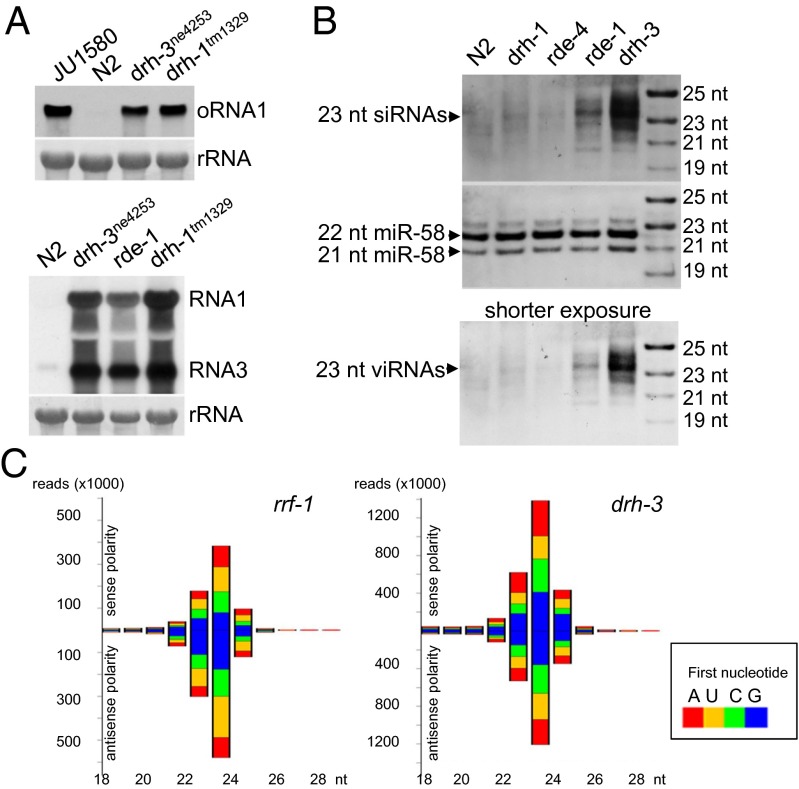
DRH-3 shares a similar domain structure with DRH-1 but contains more divergent sequences compared with DRH-1 and DRH-2 (Fig. S6). Recent studies have shown that DRH-3 is required for germline development and RNAi by participating in the biogenesis of 22G RNAs (25, 38, 39), suggesting a possible role for DRH-3 in antiviral RNAi. We investigated FHV replication and Orsay virus infection in worms containing the ne4253 allele of drh-3. A single amino acid change occurred in the helicase C domain of DRH-3 encoded by the ne4253 allele, and as a result, worms containing this allele are defective in dsRNA-induced RNAi in the somatic tissues (25). We found that drh-3 mutant worms were highly susceptible to Orsay virus infection (Fig. 4A, Upper). Northern blot analysis detected a similarly high-level replication of Orsay virus in drh-3, drh-1, and JU1580 mutant worms. Moreover, we found that FR1gfp also replicated to higher levels in drh-3 mutant worms compared with wild-type N2 worms (Fig. 4A, Lower). These findings indicate that DRH-3 indeed plays a role in worm antiviral RNAi.
Fig. 4.
DRH-3 regulates antiviral RNAi by a mechanism distinct from DRH-1. (A, Upper) Accumulation of Orsay virus RNA1 detected by Northern blot in JU1580, wild-type N2 worms, and genetic mutants defective in drh-3 or drh-1, as indicated. (Lower) Accumulation of FR1gfp transcripts detected by Northern blot in wild-type N2 worms and genetic mutants defective in drh-3, rde-1, or drh-1, as indicated. (B) Accumulation of FR1gfp-derived siRNAs detected by Northern blot in different genetic backgrounds, as indicated. (Lower) Shorter-exposure image of the same blot. (C) Primary and secondary FR1gfp-derived siRNA populations in rrf-1 and drh-3 mutants. The reads are grouped based on polarity and according to length and the identity of the first nucleotide.
To define a role for DRH-3 in antiviral RNAi, we compared the accumulation of FR1gfp-derived viral siRNAs in drh-1 and drh-3 mutants, using control mutants known to be defective in the biogenesis of primary siRNAs (rde-4) or secondary siRNAs (rde-1 and rrf-1) (21, 40, 41). Consistent with previous deep sequencing results (40, 42), several species of viral siRNAs were detected in the rde-1 mutants, with a major band detected at the position corresponding to 23 nt (Fig. 4B). Viral siRNAs also accumulated to readily detectable levels in both drh-3 and rrf-1 mutants, with a dominant 23-nt band similar to that in rde-1 mutants (Fig. 4B and Fig. S7), indicating robust production of viral primary siRNAs in drh-3, rde-1, and rrf-1 mutants. In comparison, viral siRNAs including the 23-nt species reproducibly accumulated to much lower levels in both drh-1 and rde-4 mutants than in rde-1 and drh-3 mutants, even though the FR1gfp replicated to comparable levels in rde-1, drh-1, and drh-3 mutants (Fig. 4 A and B and Fig. S7).
Our findings suggested a major defect of drh-1 mutant animals in the biogenesis of primary viral siRNAs as in rde-4 mutant animals. In contrast, loss of antiviral RNAi in drh-3 mutant animals was not associated with defective biogenesis of viral primary siRNAs, suggesting that DRH-3 may regulate the biogenesis of viral secondary siRNAs, as demonstrated for its role in exogenous RNAi (25). To test this idea, we compared the viral siRNA profiles in drh-3 and rrf-1 mutant animals through deep sequencing, using a cloning protocol capturing both primary and secondary siRNAs. As found in studies on exogenous RNAi (21, 40, 41), rrf-1 mutant animals produced a typical population of primary siRNAs, as the sequenced viral siRNAs were predominantly 23 nt in length, contained similar reads number for positive and negative strands, and were not enriched for 22-nt siRNAs with 5′-terminal guanine nt (Fig. 4C, Left). We found that the profile of viral siRNAs sequenced from drh-3 mutant animals (Fig. 4C, Right) was highly similar to that from rrf-1 mutant animals and lacked a population of 22G RNAs. These observations together suggest a role for DRH-3 in the biogenesis of viral secondary siRNAs.
DRH-1 Is Dispensable for RNAi Targeting Cellular Transcripts.
In contrast to antiviral RNAi, DRH-1 is largely dispensable in RNAi induced artificially to target cellular transcripts of endogenous genes such as skn-1, dpy-13, and unc-22 (15) or transgene (Fig. S8). Our recent study has detected RNAi of cellular mRNAs targeted by virus-derived siRNAs in C. elegans (43). Because low levels of FR1gfp-derived viral siRNAs remained detectable in drh-1 mutants (Fig. 4B), we next determined whether these viral siRNAs were active in RNAi targeting cellular transcripts in the absence of DRH-1. To test this hypothesis, we used a transgenic worm strain carrying a Psur-5::GFP transgene and a previously described replicon transgene, FR1fp, which does not express GFP because of deletion of the 5′-half of the GFP coding sequence but triggers potent silencing of the homologous cellular GFP transcripts (43) (Fig. S9A). As expected, FR1fp replication triggered potent gfp silencing, manifested as a reduction in the intensity of green fluorescence in the wild-type N2 worms but not in RNAi-defective mutants corresponding to rde-1 or rde-4 (Fig. S9B). Consistently, FR1fp-derived siRNAs accumulated at lower levels in drh-1 mutants than in rde-1 mutants (Fig. 5A). We also detected gfp silencing in drh-1 mutants, albeit to a lesser extent (Fig. S9B). Northern blot analysis showed that the gfp transcripts were markedly reduced in wild-type N2 worms and drh-1 mutants, but not in drh-1 double mutants with rde-1 or rde-4 (Fig. 5B). In contrast, both RNAs 1 and 3 from replicon FR1fp accumulated to high levels in all of the examined drh-1, rde-1, and rde-4 single and double mutants (Fig. 5B). These findings showed that drh-1 was essential for silencing a replicating viral replicon, but not a homologous cellular mRNA targeted by the same pool of siRNAs in the same animals. In an independent set of experiments, we compared the susceptibility of the replicating FR1fp and cellular GFP mRNA to feeding RNAi and found that cellular transcripts remained highly susceptible, unlike the viral replicon (Fig. S10). Therefore, we propose that unlike drh-3, which is required for both exogenous RNAi and antiviral RNAi, participation of an additional drh-1 pathway is necessary to inhibit virus infection.
Fig. 5.
DRH-1 is not required for the silencing of cellular transcripts mediated by viral siRNAs. (A) Accumulation of FR1fp-derived siRNAs detected by Northern blot in wild-type N2 worms and genetic mutants defective in rde-1, rde-4, and drh-1, as indicated. (B) Northern blot detection of GFP silencing triggered by FR1fp replication in different genetic backgrounds, as indicated, 48 h after heat induction. The GFP transcripts were detected using oligo probes that would not hybridize to the FP region of FR1fp.
Discussion
It is intriguing that, as an essential component of the nematode antiviral RNAi, DRH-1 encodes the helicase domain and CTD homologous to mammalian RLHs, as RLHs initiate the IFN-dependent antiviral immunity that is absent in C. elegans (15). In this study, we developed an assay to dissect the domain requirement of DRH-1 in antiviral RNAi against both the FHV replicon and Orsay virus by transgenic expression of the wild-type and mutant forms of DRH-1 in animals carrying a loss-of-function allele of drh-1. Our results show that the antiviral activity of DRH-1 requires its helicase domain and CTD, as well as the worm-specific NTD. We also demonstrate that the homologous helicase and CTD domains encoded by either the worm DRH-2 or human RIG-I can functionally replace the corresponding domains of DRH-1 to mediate antiviral RNAi in C. elegans. Strikingly, three amino acid substitutions in the KWK motif predicted to prevent the physical interaction of RIG-I with viral dsRNA abolished the antiviral activity of the CTDs of both RIG-I and DRH-1 in C. elegans. These findings strongly suggest that antiviral RNAi in C. elegans requires an activity of DRH-1 to detect viral dsRNA known to be essential for the virus sensing by RIG-I.
Available data illustrate an identical genetic requirement for antiviral RNAi triggered by either the replication of FHV or the infection of Orsay virus (12–16). This is probably because induction of nematode antiviral RNAi requires the recognition and processing (into siRNAs) of dsRNA produced during RNA virus replication, and Orsay virus is closely related to FHV (16). Consistently, we found in this study that DRH-1 identified in a feeding RNAi screen on the basis of the FHV replicon-induced antiviral RNAi is also necessary for the nematode defense against Orsay virus infection. Moreover, we show that antiviral RNAi induced by both the FHV replicon and Orsay virus requires DRH-3. We note that DRH-3 is known to participate in exogenous and endogenous RNAi of C. elegans (25, 38) in contrast to DRH-1, which is largely dispensable for RNAi targeting cellular transcripts, whether or not the silencing siRNAs are processed from a replicating viral RNA or an exogenous long dsRNA. Using an improved protocol for Northern detection of small RNAs (43), we found that the 23-nt primary siRNAs targeting the FHV replicon reproducibly accumulated to higher levels in drh-3 mutants than in drh-1 mutants, although both mutants supported similar replication levels of the replicon. Deep sequencing further indicated that the abundant viral siRNAs produced in drh-3 mutants did not include a population of secondary siRNAs. These observations together indicate that DRH-3 regulates RNAi targeting both cellular transcripts and viruses by participating in the biogenesis of secondary siRNAs after Dicer-dependent production of primary siRNA biogenesis, as proposed previously for its role in endogenous RNAi (25). In contrast, DRH-1 may act to specifically enhance production of viral primary siRNAs by facilitating the recruitment of the heterodimer DCR-1/RDE-4 to the viral dsRNA bound by DRH-1 for subsequent processing into primary siRNAs by DCR-1. Our model is consistent with a previous study that detected physical interaction of RDE-4 with DRH-1, but not with DRH-3 (18), and would also explain why the function of DRH-1 is essential only for RNAi against virus infection.
Viral RNA replication occurs in discrete subcellular compartments such as the spherules on the outer membrane of mitochondria shown for FHV (44). Virus dsRNA replicative intermediates are also found in complexes with viral replicase and host cofactor proteins. These physical barriers may prevent the DCR-1/RDE-4 complex from gaining access to viral dsRNA. DRH-1-mediated detection of the unique viral dsRNA for the production of primary siRNAs may be particularly important to ensure effective RNAi against the replicating viral RNAs in C. elegans, which encodes only one Dicer, unlike multi-Dicer plant and insect hosts that process distinct siRNA and miRNA precursors with dedicated Dicer proteins (4). However, production of primary siRNAs in the absence of DRH-1 may still be sufficient for RNAi to target the nonreplicating cellular transcripts.
Rapid progress has been made in understanding the IFN-regulated effector mechanism of the mammalian antiviral immunity initiated by RIG-I, MDA5, and LGP2 in the cytosol (45). Our study provides an interesting parallel for a three-member family of RLHs to regulate antiviral defense in C. elegans, using RNAi as the effector mechanism. Analogous to the role of the N-terminal CARDs of RIG-I in the downstream immune signaling (45), the interaction of DRH-1 with RDE-4 may involve the worm-specific NTD of DRH-1, which is critical for the use of RNAi as the effector mechanism. Therefore, detection of viral dsRNA by related RLHs in nematodes and mammals activates unrelated effector mechanisms, illustrating functional diversification of RLHs during evolution via acquisition of specific N-terminal signaling domains. An alternative model is that the mammalian RLHs have a conserved activity to recruit DCR-1 for the production of viral siRNAs to activate antiviral immunity by an RNAi effector mechanism.
Materials and Methods
Genetics.
The Bristol isolate of C. elegans, N2, was used as the reference strain in this study. Other N2-derived mutants used in this study include rde-1(ne300), rde-4(ne337), drh-1 (tm1329 and ok3495), and drh-3 (ne4253). The genotypes of rde-1 and rde-4 worms were confirmed using skn-1 feeding RNAi combined with genomic DNA sequencing. The genotype for drh-1 allele tm1329 was identified using PCR, as described previously (15). The genotypes of drh-3 mutants were confirmed through genomic DNA sequencing. All worm strains were maintained on NGM (nematode growth medium) plates seeded with OP50 bacteria unless otherwise indicated. All transgenes were delivered into various genetic backgrounds through standard genetic crosses.
Plasmid Constructs and Transgenic Worms.
All plasmids constructed for constitutive expression of the target genes were based on the LR50 vector (46); all of the constructs featuring inducible expression used the pPD49.83 vector (kind gift from A. Fire, Stanford University, Stanford, California).
Infectious Filtrate Preparation and Orsay Virus Inoculation.
Orsay virus was maintained using the JU1580 isolate at room temperature following a protocol described previously (16). To prepare Orsay virus inoculum, infected JU1580 worms were washed off from slightly starved 10-cm plates using M9 buffer, 5 mL per plate. The virus-containing liquid was then filtered through a 0.22-µm filter unit (Millipore) and used to resuspend pelleted OP50 Escherichia coli for NGM plate seeding.
Protein and RNA Gel Blot Analysis.
Total proteins were extracted from worms of mixed stages and were resolved on 8% (wt/vol) SDS/PAGE gel. After being transferred to Hybond-P PVDF membrane (GE Healthcare Life Science), the target proteins were detected with anti-HA as primary antibody and goat anti-rabbit secondary antibody (Cell Signaling Technology). As equal loading control, the same set of samples of equal amount was subjected to protein gel blot analysis using anti–β-actin antibody.
Total RNA extraction, small RNA enrichment, detection of viral and cellular transcripts, and small RNAs were carried out following protocols described previously (43, 46). DNA oligos with sequence ATTGCCGTACTGAACGATCTCA were labeled and used for the detection of miR-58.
Small RNA Sequencing.
RNA extracts containing enriched small RNAs were used for the construction of small RNA libraries. To capture virus-derived secondary siRNAs, all input RNA samples were treated with tobacco acid pyrophosphatase (Epicentre) before being used for small RNA library construction. All small RNA libraries were generated using TruSeq RNA Sample Preparation Kit (Illumina), following the manufacture’s instructions, and sequenced using the Illumina HiSeq2000. After the adaptor was removed, the small RNA sequences were analyzed with in-house pipelines, as described previously (47).
Imaging Microscopy.
GFP and mCherry fluorescence images were recorded using a Nikon p7000 digital camera mounted on a Nikon SMZ1500 microscope.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center for some of the worm strains used in this study; Dr. Erik Jorgensen for the Pmyo-2::mCherry construct; Dr. Zhijian Chen for the human RIG-I cDNA clone; and Dr. Félix, Dr. Miska, and Dr. Wang for the Orsay virus and the C. elegans isolate JU1580. We also thank Dr. Yanghong Han and Jinfeng Lu for their help with the small RNA library construction and sequencing. This work was supported by the Research Competitiveness Subprogram, Louisiana Board of Regents, and the National Institutes of Health (1R03AI092159-01A1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307453110/-/DCSupplemental.
References
- 1.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 2.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler J-L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7(6):590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 3.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9(12):1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 4.Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 5.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313(5783):68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 6.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130(3):413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Mierlo JT, et al. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 2012;8(8):e1002872. doi: 10.1371/journal.ppat.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA. 1999;96(24):14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen BR. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol. 2006;7(6):563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 10.Ding S-W, Lu R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr Opin Virol. 2011;1(6):533–544. doi: 10.1016/j.coviro.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10(1):47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436(7053):1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436(7053):1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 14.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102(51):18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5(2):e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Félix M-A, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9(1):e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rechavi O, Minevich G, Hobert O (2011) Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147(6):1248–1256. [DOI] [PMC free article] [PubMed]
- 18.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109(7):861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 19.Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16(2):207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- 20.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99(2):123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 21.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107(4):465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 23.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315(5809):241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 24.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315(5809):244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 25.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36(2):231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang F, et al. (2011) Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479(7373):423–427. [DOI] [PMC free article] [PubMed]
- 27.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147(2):409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Q-X, Chen ZJ. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO Rep. 2012;13(1):7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 30.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 31.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175(8):5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 32.Pippig DA, et al. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37(6):2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Satoh T, et al. (2010) LGP2 is a positive regulator of RIG-I and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA 107(4):1512–1517. [DOI] [PMC free article] [PubMed]
- 34. Wu B, et al. (2012) Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152(1–2):276–289. [DOI] [PubMed]
- 35.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 36.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29(2):178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Lu C, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18(8):1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124(2):343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 39. Claycomb JM, et al. (2009) The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139(1):123–134. [DOI] [PMC free article] [PubMed]
- 40. Wu Q, et al. (2010) Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci USA 107(4):1606–1611. [DOI] [PMC free article] [PubMed]
- 41.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127(4):747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 42.Parameswaran P, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6(2):e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, Li W-X, Lu R. Silencing of host genes directed by virus-derived short interfering RNAs in Caenorhabditis elegans. J Virol. 2012;86(21):11645–11653. doi: 10.1128/JVI.01501-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8(1):77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng CS, Kato H, Fujita T. Recognition of viruses in the cytoplasm by RLRs and other helicases—how conformational changes, mitochondrial dynamics and ubiquitination control innate immune responses. Int Immunol. 2012;24(12):739–749. doi: 10.1093/intimm/dxs099. [DOI] [PubMed] [Google Scholar]
- 46.Guo X, Lu R. Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J Virol. 2013;87(10):5414–5423. doi: 10.1128/JVI.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X-B, et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell. 2011;23(4):1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.