Significance
Germ cells, meaning the mature sperm and egg and their developmental precursors, carry the DNA that is passed from one generation to the next. Although the sperm and egg are highly specialized, differentiated cells, they meet at fertilization to produce a totipotent zygote, a cell that can generate any other cell type. We report the finding that a set of developmentally important genes is kept in a “poised” state in the germ cells: although these genes are never expressed in the germ cells themselves, they maintain a chromatin state usually associated with the potential for rapid gene activation. We propose that maintenance of this poised state in the germ cells contributes to the generation of totipotency in the fertilized zygote.
Keywords: spermatogenesis, epigenetics, chromatin, totipotency
Abstract
In multicellular organisms, germ cells carry the hereditary material from one generation to the next. Developing germ cells are unipotent gamete precursors, and mature gametes are highly differentiated, specialized cells. However, upon gamete union at fertilization, their genomes drive a totipotent program, giving rise to a complete embryo as well as extraembryonic tissues. The biochemical basis for the ability to transition from differentiated cell to totipotent zygote is unknown. Here we report that a set of developmentally critical genes is maintained in an epigenetically poised (bivalent) state from embryonic stages through the end of meiosis. We performed ChIP-seq and RNA-seq analysis on flow-sorted male and female germ cells during embryogenesis at three time points surrounding sexual differentiation and female meiotic initiation, and then extended our analysis to meiotic and postmeiotic male germ cells. We identified a set of genes that is highly enriched for regulators of differentiation and retains a poised state (high H3K4me3, high H3K27me3, and lack of expression) across sexes and across developmental stages, including in haploid postmeiotic cells. The existence of such a state in embryonic stem cells has been well described. We now demonstrate that a subset of genes is maintained in a poised state in the germ line from the initiation of sexual differentiation during fetal development and into postmeiotic stages. We propose that the epigenetically poised condition of these developmental genes is a fundamental property of the mammalian germ-line nucleus, allowing differentiated gametes to unleash a totipotent program following fertilization.
Mammalian germ cells are unipotent cells that give rise to a single, highly differentiated, sexually dimorphic cell type, the gamete. During development, germ cells undergo a series of specialized cellular processes, including migration, sexual differentiation, and meiosis, and mature male and female gametes have few cellular features in common (1, 2). Nevertheless, germ cells of both sexes and all life stages are unified by the shared ability to contribute genetic material to the next generation: unlike any other cell type, terminally differentiated germ cells can give rise to a totipotent zygote. The biochemical features conferring this ability to transition to totipotency are unknown. In many species, a specialized cytoplasm, often referred to as the “germ plasm,” provides biochemical continuity and helps define the germ line (3, 4), but mammals lack a well-defined germ plasm; mammalian germ cells are instead specified from multipotent tissue during embryogenesis. It is therefore unclear whether the potential to contribute to a new individual can be connected to any consistent molecular features in the mammalian germ line (5, 6).
One possibility is that maintenance of a transcriptionally flexible state in the germ-cell nucleus facilitates reprogramming to totipotency following fertilization (7). In vitro studies have provided insight into how such a state might be maintained. For example, in embryonic stem cells (ESCs), the poised (bivalent) promoter state is associated with the potential to give rise to multiple tissues. In ESCs, poised chromatin is defined by the presence of both an activating modification (H3K4me3) and a repressive modification (H3K27me3), and is usually found near promoters of transcriptionally silent developmental regulatory genes (8). Once an ESC begins to differentiate, poised promoters resolve toward either an active or repressed state (8, 9). Intriguingly, the presence of poised genes in relatively undifferentiated mammalian fetal germ cells has recently been reported (10, 11). If a subset of these genes is kept in a poised state beyond early fetal time points and throughout the germ-cell life cycle, this state could provide a biochemical foundation for the ability to transition from differentiated gamete to totipotent zygote.
Mammalian germ cells pass through several critical transitions during which poised chromatin configurations might be reset. In mice, migrating germ cells enter the fetal gonad by embryonic day 11.5 (E11.5). Immunohistological and genome-wide studies of DNA methylation and demethylation (12, 13), and immunohistological studies of chromatin state (14), have identified the time of gonad entry as a period of epigenetic reprogramming, and germ cells initiate transcription of a specific set of regulatory genes, including Dazl (deleted in azoospermia-like) and Ddx4 (DEAD box polypeptide 4, also called mouse vasa homolog) at this time (15, 16). By E13.5, germ-cell transcriptional profiles differ significantly between sexes, and both male and female germ cells have lost the ability to form pluripotent cell lines in vitro (17–19). Female germ cells initiate meiotic prophase by E14.5, whereas male germ cells enter a cell cycle arrest at E15.5 and begin meiosis several days after birth (20). Meiosis itself represents a period of dramatic transition, as germ cells undergo reductional cell division to become haploid gamete precursors, a process that involves chromosome repackaging and recombination between homologous chromosomes. Finally, differentiation into mature gametes requires extensive nuclear and cytoplasmic remodeling in both sexes.
We now provide high-resolution, genome-wide chromatin data, including both sexes and multiple developmental time points, to show that mammalian germ cells maintain a poised chromatin state at developmentally important transcriptional regulators from at least E12.5 through postmeiotic stages. We performed ChIP-seq and RNA-seq experiments in male and female germ cells at E12.5, E13.5, and E14.5, as well as in male meiotic spermatocytes and postmeiotic spermatids. We report a set of genes that is maintained in a poised state in both male and female fetal germ cells, as well as in meiotic and postmeiotic germ cells in the male. This set of germ-cell–poised genes is associated with differentiation of each of the three embryonic germ layers, trophectoderm, and primitive endoderm. We propose that the epigenetically poised state of this set of genes confers upon germ cells the ability to generate a complete conceptus, including both embryonic and extraembryonic structures, following fertilization.
Results
Chromatin Data Support Known Germ Cell Biology.
To better understand the dynamics of transcriptional regulation in the fetal germ cells, we performed ChIP-seq for the H3K4me3 mark (associated with active promoters) and the H3K27me3 mark (associated with facultatively repressed promoters), as well as RNA-seq, on germ cells isolated by flow cytometry from male and female embryos at E12.5, E13.5, and E14.5 (Fig. S1 and Table S1). For ChIP-seq experiments, we used a protocol adapted for small numbers of cells, similar to protocols that have recently been used successfully in similar cell populations (11, 21–23). We first tested our protocol on equivalent numbers of mouse ESCs (mESCs), with highly reproducible results that correspond to previously published mESC data (Fig. S2) (9). Germ-cell ChIP-seq experiments were performed on two or three biological replicates for each condition, with strong positive correlation between replicates (P < 10−7, Student t test) (Fig. 1A, Fig. S2, and Table S2). For each germ-cell condition, the most significantly enriched Gene Ontology (GO) categories were biosynthesis and housekeeping genes for H3K4me3 and developmental and transcriptional regulators for H3K27me3, consistent with their association with active and repressed promoters, respectively (Table S3). As expected, highly expressed genes had high H3K4me3 and low H3K27me3 signal (Fig. S3).
Fig. 1.
ChIP-seq and RNA-seq data reflect the known biology of mouse fetal germ cells. (A) Sample gene tracks showing biological replicates of female E13.5 H3K4me3 and H3K27me3 data. (B) Correlated changes between H3K4me3 ChIP signal and gene expression for three core pluripotency regulators (Oct4, Sox2, and Nanog) and three genes associated with meiotic initiation (Stra8, Rec8, and Sycp3).
Between E12.5 and E13.5, germ cells of both sexes lose the ability to give rise to pluripotent cell lines in culture and begin to down-regulate pluripotency-associated genes. Both male and female germ cells begin to express components of the meiotic program, and at E14.5 female germ cells enter the first stages of meiotic prophase. Male cells do not initiate meiotic prophase, but instead arrest at G1 until after birth. We asked whether our data were consistent with these known aspects of germ cell biology. As expected, expression of the pluripotency regulators Oct4 (Pou5f1), Sox2, and Nanog decreased over time, with a corresponding decrease in H3K4me3 signal (Fig. 1B). Conversely, expression and H3K4me3 signal increased over time at the promoters of the meiosis-associated genes Rec8, Stra8, and Sycp3, with more dramatic increases in female compared with male germ cells (Fig. 1B). In general, there were statistically significant increases in H3K4me3 signal at promoters of genes included in the “meiosis” GO category (GO:0007126) between E12.5 and E14.5 in both sexes [P = 0.0065 (female) and P = 0.00017 (male), Mann–Whitney U test], with more dramatic increases in females (Fig. S4 and Table S4). H3K27me3 signal at this class of promoters did not change significantly. In contrast, H3K27me3 signal associated with genes included in the GO category “cellular response to retinoic acid” (GO:0071300) exhibited significant differences between time points [P = 0.016 (female) and P = 0.026 (male), Mann–Whitney U test], but the direction of change varied by gene: ChIP signal increased at some promoters and decreased at others (Fig. S4 and Table S4). Retinoic acid is integral to meiotic initiation in the germ cells (24, 25), and also plays a critical role in regulation of pluripotency and differentiation (26, 27); it is likely to have multiple regulatory roles in this cell population.
Identification of Poised Promoters in Fetal Germ Cells.
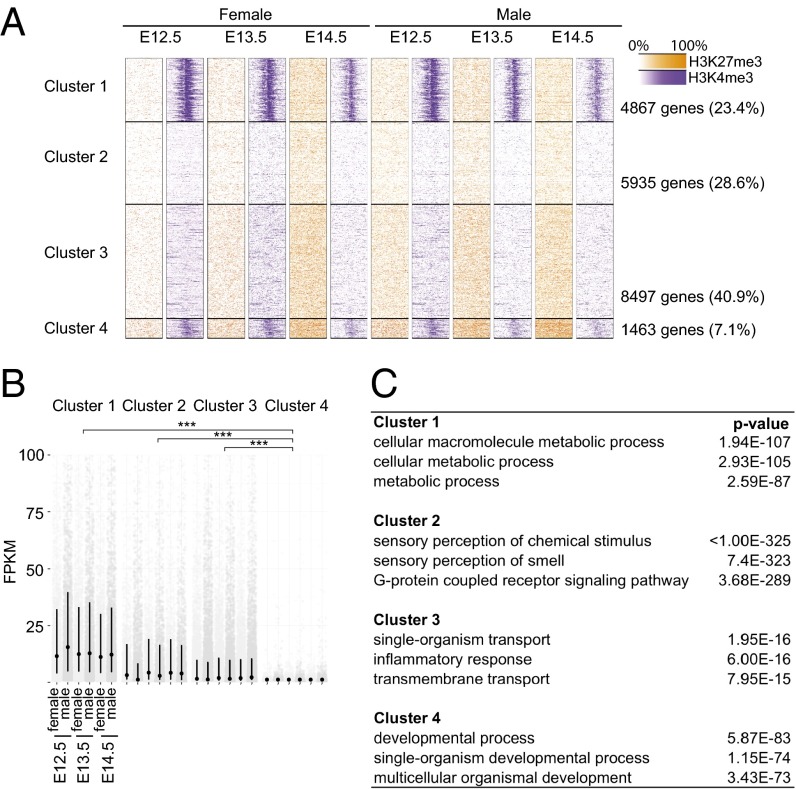
We next examined genome-wide chromatin states across sexes and developmental time points. First, to look for similarities across datasets, we treated all six conditions (two sexes and three time points) as equivalent, and used k-means clustering to group all genes according to similarity of H3K4me3 and H3K27me3 profile near their transcriptional start sites. In this analysis, no parameters regarding ChIP profile are initially specified to define the clusters; results therefore represent an unbiased description of common promoter chromatin states in the cell population. We observed that in both sexes and at all three time points, one cluster of genes was characterized by high H3K4me3 and high H3K27me3 signal, a characteristic of bivalent or poised genes in ESCs (Fig. 2A, cluster 4). Furthermore, this class of genes displayed markedly lower overall expression at all ages and in both sexes, compared with each of the other classes identified (Fig. 2B). Cluster 4 was significantly enriched for functional categories corresponding to development and organogenesis (Fig. 2C). The combination of high H3K4me3 signal, high H3K27me3 signal, low expression, and functional enrichment for developmental regulators led us to hypothesize that this class of genes exists in a poised state, similar to the poised state identified in ESCs (8).
Fig. 2.
Clustering of H3K4me3 and H3K27me3 profiles with associated gene expression and functional enrichment. (A) Clustering of genes by similarity of H3K4me3 and H3K27me3 profiles 2 kb upstream and downstream of the transcriptional start site. (B) RNA expression levels for genes in each of four clusters. Each gray point represents a single gene; black dots, median; black lines, interquartile range. ***P < 10−15 (two-sample t test). (C) Top three enriched GO categories for each cluster.
A Class of Poised Genes Does Not Resolve Between E12.5 and E14.5.
The presence of poised genes in E12.5 germ cells is consistent with current models for the function of poised genes in stem cells. In ESCs, poised genes are thought to retain H3K4me3, despite their transcriptionally repressed state, to facilitate transcriptional activation after receiving a differentiation signal (8). At E12.5, both male and female germ cells are thought to be relatively undifferentiated, and they can form pluripotent cell lines in culture. Between E12.5 and E14.5, however, germ cells of both sexes undergo major changes in cell state, as both transition to highly sex-specific transcriptional programs and begin the process of differentiation into mature sperm or oocytes (18). In particular, female germ cells at E14.5 have initiated meiotic prophase, a clear departure from an undifferentiated state. We therefore asked if genes that are poised in E12.5 or E13.5 germ cells tended to resolve toward H3K4me3 or H3K27me3 by E14.5.
Because the clustering described above treated all six conditions as equivalent, it did not account for changes in the state of each promoter over time. To track changes in poised promoter state over time at individual genes, we established a set of criteria that could be used to call a single promoter as poised or not at a given time point. We defined poised genes as having high H3K4me3 signal (>75th percentile of ChIP signal) and high H3K27me3 signal (>95th percentile of ChIP signal) within 2 kb upstream or downstream of the transcriptional start site, as well as absent transcription (fragments per kilobase per million reads ≤ 1).
To start, we analyzed data separately for male and female germ cells. We found 513 genes in male and 727 genes in female that met our criteria for the poised state at E12.5, and these numbers decreased to 291 and 270 genes at E14.5 in male and female, respectively (Fig. 3A). Among the genes meeting criteria for the poised state at E14.5, 247 in male (85%) and 254 in female (94%) were also poised at E12.5, E13.5, or both, indicating that genes that are poised at E14.5 had likely retained this state over time (Table S5). Of those genes that are poised in either sex at both E14.5 and earlier time points, 147 were shared between sexes (60% and 58% for male and female, respectively) (Fig. 3A and Table S5). In contrast, only 21 genes that resolved toward a repressed state (low H3K4me3, high H3K27me3, and low expression) were shared between males and females (28% and 18% of genes resolving toward the repressed state at E14.5 in males and females, respectively), and none of those that resolved toward an active state (high H3K4me3, low H3K27me3, and high expression) were shared between sexes. We conclude that a class of genes is maintained in a poised state in both male and female germ cells during the period encompassing sexual differentiation and female initiation of meiosis. This shared collection of germ-line–poised genes is a subset of cluster 4, the class of H3K4me3+/H3K27me3+ genes initially identified by genome-wide clustering of promoter ChIP signals (Fig. 3B). Notably, a subset of genes appeared poised in one sex but not the other. It is possible that these genes represent a true sex-specific set of germ-cell–poised genes; however, because our filtering criteria were designed to exclude false positives, it is likely that many of these genes are true positives in both sexes but met the filtering threshold in only one sex. The set of true shared, poised genes may therefore be larger than reported here.
Fig. 3.
A subset of poised genes is retained from E12.5 through E14.5. (A) Number of genes identified as “poised” at each fetal time point in male and female (gray bars), shared between time points in male and female (blue and red strips), and shared between time points and sexes (purple strip). (B) Distribution of poised genes identified in A among the four clusters identified in Fig. 2.
To see if this set of genes is also poised at earlier times in development, we compared our data to published H3K4me3 and H3K27me3 ChIP-seq data from mixed male and female germ cells at E11.5 (11). (The published dataset did not include corresponding transcriptional data.) We identified 805 genes at E11.5 with high H3K4me3 (>75th percentile) and high H3K27me3 (>95th percentile). Of the 147 poised genes that we identified as shared across fetal male and female germ cells between E12.5 and E14.5, 142 (97%) were included in this set of 805 genes at E11.5, supporting the continuity of the poised state at these genes in fetal germ cells.
Germ-Line–Poised Genes Include Critical Regulators of Early Development.
To better understand the biological function of this gene class, we looked for GO categories enriched in the set of 147 genes that is maintained in the poised state between E12.5/E13.5 and E14.5 and is shared between sexes, relative to the genome as a whole. We found that this gene set was overwhelmingly enriched for transcriptional regulators of somatic differentiation, including both general regulators of differentiation (“positive regulation of stem cell differentiation,” GO:2000738, fold-enrichment 58.72), and regulators of tissue-specific differentiation (including kidney, lung, heart, liver, pancreas, skeletal system, and brain; fold-enrichment 6.8–38.29) (Table 1). Of the 147 genes, 89 were associated with “developmental process” (GO:0032502, fold-enrichment 3.76). We found a similar set of enriched categories when the 147 poised genes were compared with all genes that lacked expression in fetal germ cells. The set of poised genes retained in male, female, or both during fetal development included essential regulators of ectoderm (Sox1, Olig2), mesoderm (T, Nkx2-5), and embryonic endoderm (Sox17, Foxa2), as well as primitive endoderm (Gata6, Pdgfra) and trophectoderm (Cdx2, Hand1) (Table S5). We evaluated expression of each of these genes during mouse embryogenesis, using publicly available data. Of the 147 genes, 127 are somatically expressed during mouse embryogenesis, and 52 are expressed before E8.75, implying that they are involved in early germ layer formation. Only 32 were expressed in mESCs. We conclude that the genes maintained in a transcriptionally poised state in fetal germ cells are developmental regulators. This class includes genes essential for specification of a complete conceptus, including the embryo proper, trophectoderm, and primitive endoderm (28).
Table 1.
GO categories enriched in genes maintained in a poised state in both male and female fetal germ cells
| Description | False-discovery rate q-value | Enrichment |
| Top five enriched GO categories | ||
| Developmental process | 2.35E-31 | 3.76 |
| Regulation of nucleobase compound metabolic process | 3.53E-29 | 3.97 |
| Regulation of transcription, DNA-dependent | 2.73E-29 | 4.4 |
| Regulation of RNA biosynthetic process | 2.28E-29 | 4.4 |
| Single-organism developmental process | 2.70E-29 | 4.45 |
| Selected enriched GO categories | ||
| Positive regulation of stem cell differentiation | 3.30E-05 | 58.72 |
| Formation of primary germ layer | 1.64E-03 | 13.85 |
| Retinoic acid receptor signaling pathway | 7.78E-03 | 27.52 |
| Nervous system development | 3.94E-08 | 7.63 |
| Brain development | 9.65E-06 | 11.29 |
| Sensory organ development | 7.83E-06 | 11.59 |
| Eye development | 2.12E-04 | 11.17 |
| Inner ear morphogenesis | 1.89E-06 | 17.27 |
| Cartilage development | 1.22E-05 | 13.5 |
| Skeletal system development | 1.42E-05 | 10.74 |
| Connective tissue development | 7.42E-05 | 10.48 |
| Kidney development | 2.66E-08 | 14.68 |
| Lung development | 3.05E-02 | 6.8 |
| Heart morphogenesis | 1.64E-02 | 11.51 |
| Liver development | 2.35E-02 | 10.3 |
| Pancreas development | 1.11E-03 | 24.46 |
| Endocrine pancreas development | 8.04E-07 | 38.29 |
| Limb morphogenesis | 4.86E-07 | 13.11 |
Germ-Line–Poised Genes Remain Poised During and After Meiosis in Males.
We next asked whether these germ-line–poised genes remain poised during and after meiosis. During meiosis, germ cells segregate homologous chromosomes to generate haploid gamete precursors, a highly specialized process requiring tight control of transcription and specialized chromosome packaging. Female germ cells initiate chromosome condensation and meiotic prophase at E14.5 and then arrest in late prophase until ovulation, whereas male germ cells first enter meiotic prophase only after birth (20). Both sexes undergo a second wave of epigenetic reprogramming after E15.5, including a wave of de novo DNA methylation (29). Therefore, retention of a transcriptionally poised state through meiosis would represent both temporal longevity, on a time scale of months, as well as preservation of the H3K4me3+/H3K27me3+ state during the large-scale chromatin changes associated with gamete differentiation, chromosome pairing, and homologous recombination.
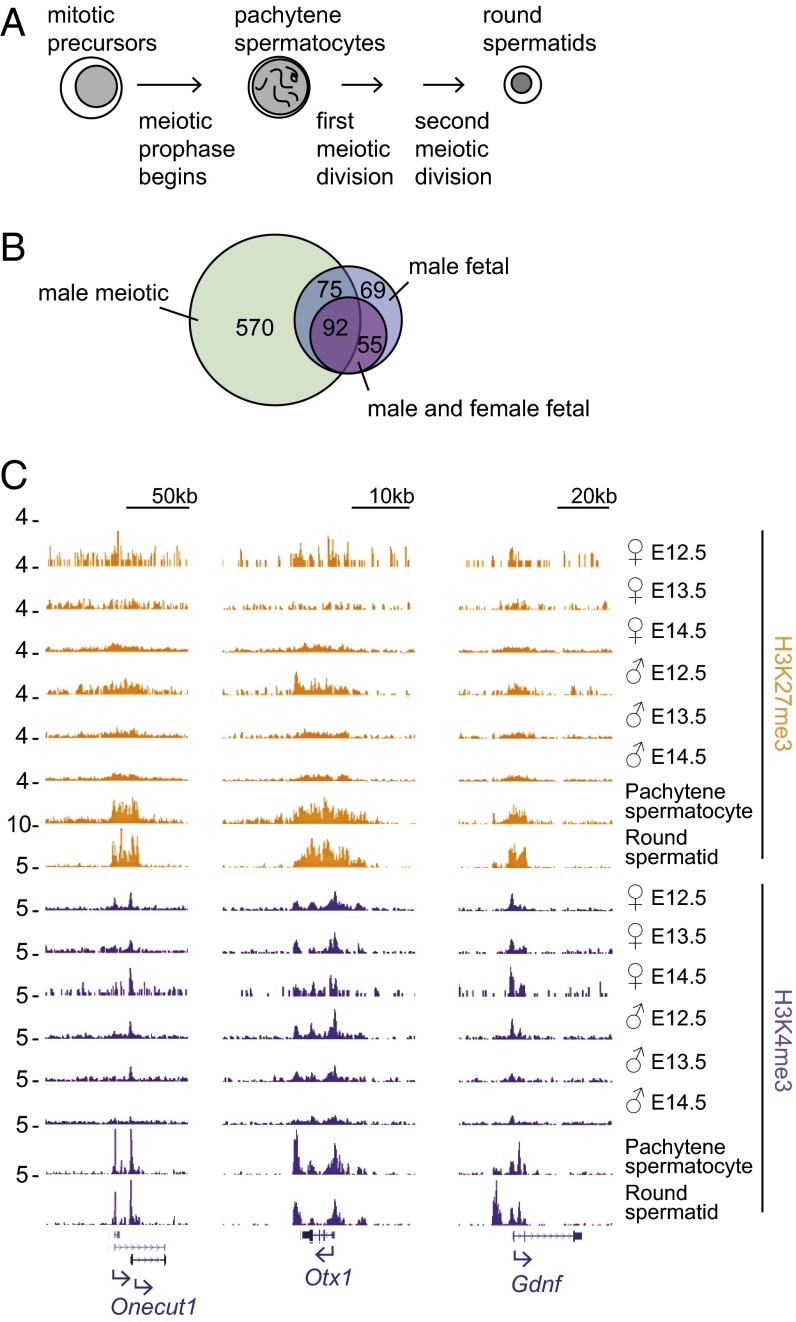
We performed RNA-seq and ChIP-seq on pachytene spermatocytes and on round spermatids. Pachytene spermatocytes are male germ cells in prophase I of meiosis that have completed homologous recombination but have not yet segregated their chromosomes, whereas round spermatids are haploid male germ cells that have completed meiosis but have not yet replaced histones with protamines (Fig. 4A). Using the same filtering criteria employed earlier, we found 737 promoters that are poised in both pachytene spermatocytes and round spermatids. Of these 737 genes, 167 were also among the 247 genes that are poised in male fetal germ cells at E12.5 and E14.5 (68% of male fetal poised genes), and 92 were among the 147 genes that are poised in both male and female fetal germ cells (63% of male and female fetal poised genes) (Fig. 4 B and C, and Table S5). As expected based on our fetal germ-cell data, these genes were strongly enriched for somatic, tissue-specific developmental regulators. We conclude that the class of poised genes maintained in male and female fetal germ cells is retained during and after meiosis, at least in the male germ line.
Fig. 4.
Promoters that are poised during fetal development remain poised during and after meiosis. (A) Schematic of meiosis in male germ cells. (B) Number of germ-line–poised genes retained across fetal time points in male only (blue) or in both male and female (purple), compared with genes identified as poised in both pachytene spermatocytes and round spermatids (“male meiotic,” green). (C) Sample gene tracks for three genes maintained in a poised state across germ cell development.
Discussion
We report here the presence of a class of essential developmental regulators that is maintained in a poised chromatin state throughout much of the life cycle in germ cells of both sexes. We collected high-throughput ChIP and RNA sequencing data from sorted male and female germ cells at three fetal time points surrounding sex differentiation and female meiotic initiation, as well as male meiotic and postmeiotic germ cells. Under each of these developmental conditions, we identified genes with high H3K4me3 and high H3K27me3 signal near their transcriptional start sites, as well as absent gene expression. A subset of poised genes, representing a range of critical developmental transcription factors and signaling molecules, was common to all germ-cell states examined; these genes were maintained in the poised state not only through the onset of sexual differentiation in fetal germ cells but also during and after meiosis in the adult male. The poised state has been well described in ESCs, where it is thought to allow more efficient entry into induced differentiation programs (8), and it has recently been suggested that its presence in the germ cells provides an in vivo correlate for this feature of pluripotent cells in vitro (11). Our data point to a broader biological function for the poised state: transmission through the germ line of the ability to generate a totipotent zygote at fertilization.
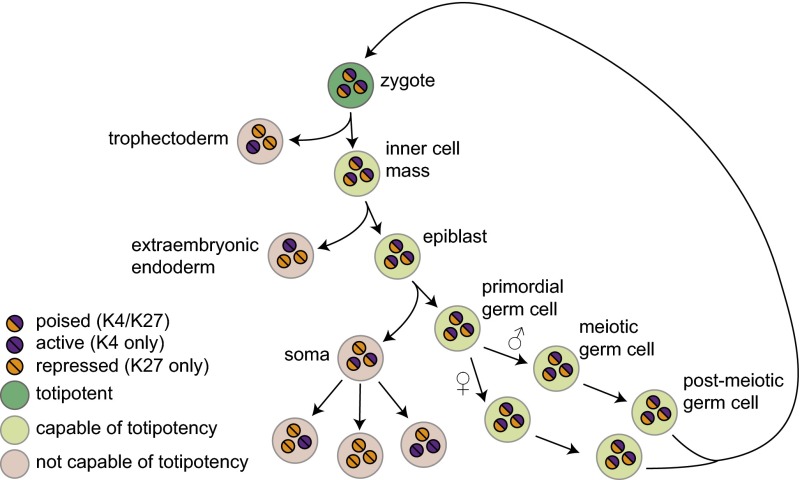
We propose a model wherein central regulators of embryonic and extraembryonic development are maintained in a poised state throughout the germ-cell life cycle, conferring a continuous germ-cell identity and permitting reestablishment of totipotency following fertilization (Fig. 5). Loss of the poised state at some or all of these genes corresponds to differentiation, and to the transition from immortal germ line to mortal soma or extraembryonic tissue. The concept of “latent pluripotency,” or continuous retention of a state conducive to establishment of tissues belonging to the three germ layers, has recently been proposed as a unifying feature of the germ line (6). We now suggest that the germ line is unified by a poised nuclear state that enables establishment not just of pluripotency, but of totipotency following fertilization. One implication of this model is that assaying the presence of poised genes will help to determine the potential of a cell to function as a gamete, for example during in vitro differentiation. In addition, this model raises several questions central to understanding the nature and regulation of the germ line.
Fig. 5.
Germ-line–poised genes as a foundation for totipotency in the germ line. Critical developmental genes are continuously maintained in a poised state (purple and orange circles), with transcription repressed. Loss of either H3K4me3 (resolution toward the active state, with associated increases in gene expression) or H3K27me3 (resolution toward the repressed state) at specific promoters results in terminal differentiation. Those cells that maintain a complete set of poised developmental regulators throughout development constitute the germ cells.
First, whether the poised state is maintained in mature sperm or oocytes, during fertilization, and in the mammalian early embryo and epiblast, remains an area of active investigation. Even if a core set of poised genes is required in the germ line for generation of a totipotent zygote, loss and reestablishment of that state might occur immediately following fertilization. In mammals, cooccurrence of H3K4me3 and H3K27me3 at promoters of select developmental regulators has been reported in epiblast cells, but genome-wide chromatin data are not available (30).
In mature mammalian sperm, histones are largely replaced by protamines, presumably eliminating information carried by histone modifications. However, a few modified histones are retained in mature sperm in mice (31, 32) and humans (31, 33). Of the 92 genes that we identified as poised at fetal through postmeiotic stages, 47 have been reported to retain histones marked by H3K4me3 and H3K27me3 after protamine replacement in mouse sperm (32). Notably, we observed that H3K4me3 signal in round spermatids sometimes spreads into broad domains around poised promoters, similar to the relatively broad H3K4me3 domains reported in mature sperm (31, 33). It will be intriguing to further explore this correlation.
Second, in addition to its association with poised or active gene promoters, the H3K4me3 histone modification is enriched at sites of recombination and crossing over during meiosis (34). In mammals, the methyltransferase PRDM9 (PR domain containing 9) directs recombination to defined hotspots, which are H3K4me3+ but are not associated with gene promoters; as a result, most recombination in wild-type individuals occurs in intergenic regions (35). In the absence of PRDM9, recombination is redirected toward H3K4me3+ promoter regions. This inappropriate recruitment of the recombination machinery to coding or regulatory regions might be expected to have deleterious effects, especially at developmentally critical genes. It is possible that the presence of the poised state in meiotic chromatin has an additional protective effect at gene promoters with which it is associated, acting as a failsafe for the targeting activity of PRDM9. In fact, we found a statistically significant depletion of recombination at germ-line–poised gene promoters in Prdm9 mutants, using previously published data (SI Text) (35). It will be interesting to determine whether a meiosis-specific set of histone modifications or chromatin factors accumulates around poised sites in meiotic germ cells.
Third, the factors that regulate maintenance of the poised state in the germ line remain unknown. Unlike ESCs (9), germ cells must fully differentiate into gametes while simultaneously protecting a complete set of poised genes and retaining the ability to give rise to a totipotent zygote. As a result, two independently targeted modes of regulation may be required. It was recently shown that two alternative components of Polycomb repressive complex 1, Ring1 and Rnf2, have genetically separable roles in female germ-cell maintenance and differentiation: both are required for survival of female fetal germ cells, but only Rnf2 is required to direct differentiation of these cells (36). If there are two classes of poised genes, those that resolve in germ cells and those that do not, then elucidation of how these classes are established and controlled could provide insight into modes of regulation of poised promoters both in vivo and in vitro. Furthermore, the role of factors involved in germ cell specification, such as Blimp1 and Prdm14, and differentiation, such as Dazl, can now be reevaluated in the context of their effects on regulation of germ-line–poised genes.
Finally, the fate of poised promoters in the meiotic and postmeiotic female germ line remains unknown. Analysis of germ-line–poised genes in the female compared with the male germ line will help to assess the universality of this gene class, and may illuminate sex-specific differences in regulation of the poised state. In both male and female gametes, the state of poised promoters during and after fertilization may be differentially regulated, with implications for maternal and paternal influences on inheritance and on development of offspring. In both sexes, a better understanding of the factors influencing maintenance of the poised state during and after fertilization will have broad implications for human development and disease.
Materials and Methods
Further details can be found in SI Materials and Methods.
Mice.
All experiments involving mice conformed to ethical principles and guidelines approved by the Committee on Animal Care at the Massachusetts Institute of Technology (Institutional Animal Care and Use Committee no. 0711–075-14). Germ cells were sorted from fetal gonads of mice carrying an Oct4-EGFP transgene, or sorted using a STA-PUT apparatus from wild-type adult mice to isolate pachytene spermatocytes and round spermatids.
ChIP-Seq.
For fetal germ cells, we used a small-scale ChIP protocol adapted from Adli et al. (22, 23). No additional amplification beyond the recommended number of cycles was used for either ChIP or input samples.
Data Analysis.
In brief, sequence data were aligned to mm9 using Bowtie, and enrichment peaks were called using MACSv1.4. Differential expression and clustering were evaluated using the R packages DESeq and Repitools, respectively. Enriched GO categories were identified using GOrilla. Custom R scripts were used for other analyses. Sequence data files are publically available under accession number SRA097278.
Supplementary Material
Acknowledgments
We thank Y.-C. Hu for back-crossing mice; H. Skaletsky for statistical advice; the Whitehead Bioinformatics and Research Computing Center for help with data analysis; the Whitehead Genome Technology Core for Illumina sequencing and library preparation; and D.W. Bellott, H.C. Christiansen, R. Desgraz, J. Mueller, and S. Soh for critical reading of the manuscript. This work was supported in part by a Hope Funds for Cancer Research Postdoctoral fellowship (to B.J.L.); National Institute of Child Health and Human Development Grant 1F32HD075591 (to B.J.L.); and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRA097278).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315204110/-/DCSupplemental.
References
- 1.Li R, Albertini DF. The road to maturation: Somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol. 2013;14(3):141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- 2.Jan SZ, et al. Molecular control of rodent spermatogenesis. Biochim Biophys Acta. 2012;1822(12):1838–1850. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Strome S, Wood WB. Immunofluorescence visualization of germ-line–specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1982;79(5):1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71(4):1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137(3):571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Leitch HG, Smith A. The mammalian germline as a pluripotency cycle. Development. 2013;140(12):2495–2501. doi: 10.1242/dev.091603. [DOI] [PubMed] [Google Scholar]
- 7.Seydoux G, Braun RE. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127(5):891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mochizuki K, Tachibana M, Saitou M, Tokitake Y, Matsui Y. Implication of DNA demethylation and bivalent histone modification for selective gene regulation in mouse primordial germ cells. PLoS ONE. 2012;7(9):e46036. doi: 10.1371/journal.pone.0046036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachs M, et al. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3(6):1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi H, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23(4):616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452(7189):877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyooka Y, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93(1–2):139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol. 2005;288(2):309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 17.McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262(1):1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 18.Jameson SA, et al. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8(3):e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labosky PA, Barlow DP, Hogan BL. Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development. 1994;120(11):3197–3204. doi: 10.1242/dev.120.11.3197. [DOI] [PubMed] [Google Scholar]
- 20.Hilscher B, et al. Kinetics of gametogenesis. I. Comparative histological and autoradiographic studies of oocytes and transitional prospermatogonia during oogenesis and prespermatogenesis. Cell Tissue Res. 1974;154(4):443–470. doi: 10.1007/BF00219667. [DOI] [PubMed] [Google Scholar]
- 21.Ng J-H, et al. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell. 2013;24(3):324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc. 2011;6(10):1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7(8):615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koubova J, et al. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci USA. 2006;103(8):2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowles J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312(5773):596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 26.Strickland S, Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978;15(2):393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- 27.Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226(2):322–330. doi: 10.1002/jcp.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139(1):3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- 29.Lees-Murdock DJ, De Felici M, Walsh CP. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics. 2003;82(2):230–237. doi: 10.1016/s0888-7543(03)00105-8. [DOI] [PubMed] [Google Scholar]
- 30.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci USA. 2010;107(24):10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17(6):679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 32.Erkek S, et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20(7):868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 33.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grey C, et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9(10):e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485(7400):642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokobayashi S, et al. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature. 2013;495(7440):236–240. doi: 10.1038/nature11918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.