Significance
For circadian clocks to modulate a daily cycle of metabolic and behavioral processes, temporal information must be transmitted to output pathways. In cyanobacteria, the circadian oscillator is composed of three Kai proteins that mediate cyclic phosphorylation of KaiC. We determined that a specific phosphostate of KaiC promotes genome-wide circadian output responses. A model that includes temporal feedback control of this KaiC phosphostate can accurately simulate key features of observed output responses of the two major contrasting promoter types. This study provides a unique perspective on the complex regulatory output responses of circadian oscillators.
Keywords: bioluminescence, chronobiology, transcription regulation
Abstract
The mechanisms by which cellular oscillators keep time and transmit temporal information are poorly understood. In cyanobacteria, the timekeeping aspect of the circadian oscillator, composed of the KaiA, KaiB, and KaiC proteins, involves a cyclic progression of phosphorylation states at Ser431 and Thr432 of KaiC. Elucidating the mechanism that uses this temporal information to modulate gene expression is complicated by unknowns regarding the number, structure, and regulatory effects of output components. To identify oscillator signaling states without a complete description of the output machinery, we defined a simple metric, Kai-complex output activity (KOA), that represents the difference in expression of reporter genes between strains that carry specific variants of KaiC and baseline strains that lack KaiC. In the absence of the oscillator, expression of the class 1 paradigm promoter PkaiBC was locked at its usual peak level; conversely, that of the class 2 paradigm promoter PpurF was locked at its trough level. However, for both classes of promoters, peak KOA in wild-type strains coincided late in the circadian cycle near subjective dawn, when KaiC-pST becomes most prevalent (Ser431 is phosphorylated and Thr432 is not). Analogously, peak KOA was detected specifically for the phosphomimetic of KaiC-pST (KaiC-ET). Notably, peak KOA required KaiB, indicating that a KaiBC complex is involved in the output activity. We also found evidence that phosphorylated RpaA (regulator of phycobilisome associated) represses an RpaA-independent output of KOA. A simple mathematical expression successfully simulated two key features of the oscillator—the time of peak KOA and the peak-to-trough amplitude changes.
Circadian biological clocks are recognized as endogenous 24-h timers that evolved through the selective fitness advantage they confer in anticipation of daily environmental variations and that generate rhythms in metabolic and behavioral processes (1–3). Both the ability to keep 24-h time and the mechanism by which such a clock regulates cellular processes are only partially understood in any organism. In the oxygenic photosynthetic bacteria known as cyanobacteria, the oscillator mechanism is a posttranslational protein interaction loop, and the nature of its temporal output signal is more easily addressable than in eukaryotic models. The recent report of a posttranslational circadian system that is shared among the kingdoms of life suggests a more universal role of posttranslational oscillators in nature (4, 5). Among the prokaryotic cyanobacteria, Synechococcus elongatus PCC 7942 is the prevalent model system for circadian studies due to its genetic manipulability and small (2.7 Mb) fully sequenced genome (6). The ability to monitor the circadian regulation of gene expression in vivo, achieved by fusing the promoter of a gene of interest to a bioluminescence reporter gene (7, 8), provides a tool for investigating the circadian clock and its connections with metabolism, cell division, and other fundamental cellular processes.
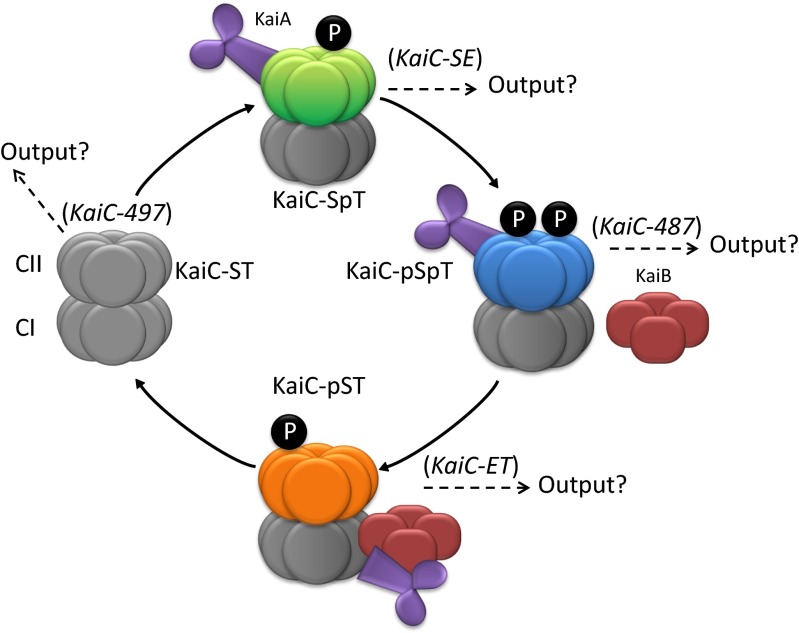
In S. elongatus, three key proteins, KaiA, KaiB, and KaiC, comprise the central circadian oscillator (Fig. 1). KaiC undergoes ordered autophosphorylation and autodephosphorylation events that signal the time of the day (oscillator timekeeping) for control of genetic expression patterns. Paramount to the circadian control of cellular responses is the ordered phosphorylation of two adjacent amino acid residues (Thr432 and Ser431) in the CII domain of KaiC; they become sequentially phosphorylated and then dephosphorylated during an ∼24-h cycle. As there are two phosphorylation sites, there are four possible states per KaiC monomer (ST, SpT, pSpT, and pST, in order, where pS and pT represent phosphorylated Ser431 and phosphorylated Thr432, respectively). KaiA facilitates the phosphorylation of Thr432 and then Ser431. Subsequently, KaiB antagonizes KaiA activity, and KaiC undergoes autodephosphorylation of Thr432 and then Ser431. The association/dissociation of all three Kai proteins controls the period, phase, and amplitude of the circadian oscillator. Great advances have been made in understanding the timekeeping functions of the central oscillator (for review, see ref. 6). However, mechanisms of the output activity have not been clearly established due to the complexity of positive and negative components and of the downstream networks. Thus, we developed a simple metric we call KOA (Kai-complex output activity) that compares the change in reporter gene expression between strains that carry wild-type or phosphomimetic-encoding alleles of KaiC and strains lacking a functional oscillator. KOA reflects the net activating or repressing action of an oscillator signal and the relative magnitude of this effect.
Fig. 1.
Simplified scheme of the central circadian oscillator system in S. elongatus. The central oscillator of the posttranslational circadian system of S. elongatus is composed of three proteins called KaiA, KaiB, and KaiC. KaiC, shown in its hexameric form, starting in the nonphosphorylated state (Left) coming out of the dark (gray). KaiC then proceeds through a series of phosphorylation steps (black filled P) involving the sequential phosphorylation of Thr432 and Ser431, returning back to the unphosphorylated state via dephosphorylation of p-Thr432 and p-Ser431 (6). The phosphorylation states of KaiC are indicated by different colors, with green CII domains representing the KaiC-SpT state and orange the KaiC-pST state. KaiA, shown in its dimeric form (purple), promotes the phosphorylation of KaiC whereas KaiB, shown in its tetrameric form (red), inhibits the effect of KaiA. The phosphorylation/dephosphorylation events of KaiC within the Kai complex determine timekeeping. The phosphomimetics used in this study are indicated in parentheses next to the state that each best mimics. Each state was queried for its potential participation in Kai complex output activity (designated as “Output?”).
Overall, determining the temporal signaling state(s) of KaiC that is/are active in KOA has been complicated by the lack of clarity regarding output mechanisms. The circadian clock modulates the promoter activity of most genes in the cyanobacterial genome (9); some of this rhythmicity may be attributable to an underlying rhythm of chromosomal compaction (10, 11). The transmission of circadian timekeeping information to transcriptional regulatory machinery has been proposed to occur through the phosphorylation state-dependent association of the circadian oscillator with output proteins such as the two-component regulatory system proteins SasA (Synechococcus adaptive sensor) and RpaA (Regulator of phycobilisome associated). The importance of SasA and RpaA in circadian gene expression has been demonstrated, and loss of RpaA causes arrhythmic gene expression (10, 12–14). In addition, the direct interaction of KaiC with DNA has been reported (15). Overexpression of KaiC suppresses expression from many genes (16), suggesting that the oscillator is a repressor. However, overexpression of KaiA, which stimulates KaiC phosphorylation, is associated with elevated expression from the kaiBC promoter, suggesting that “stimulated” KaiC is an activator or that KaiA represses the KaiC repressor.
In this work, we show that the absolute magnitude of reporter expression provides a quantifiable measure of KOA. The PkaiBC and PpurF promoters, used to drive luciferase expression, were chosen as the paradigms for class 1 and class 2 promoters, which display peak bioluminescence at dusk and dawn, respectively (17). Activity was tested both for WT KaiC, as a function of time as the oscillator cycles through the phosphorylation states, and for noncycling KaiC variants designed to mimic the four different phosphorylation states (Fig. 1). KOA provides a means to assess (i) the time in the circadian cycle in which the clock is most active in regulating gene expression, (ii) the influence of the oscillator on different classes of promoters, and (iii) the phosphorylation state(s) of KaiC that invokes the greatest output response. We found that, although class 1 and class 2 promoters show out-of-phase promoter activity, peak KOA for both classes of promoters occurs at the same phase of the circadian cycle (near anticipated dawn) and that both show peak KOA associated with the KaiC-pST state. Moreover, the absence of the transcription effector RpaA has (i) opposite effects on class 1 and class 2 promoters and (ii) opposite effects of those measured for kaiC deletion strains, suggesting that RpaA represses KOA. We also present evidence for an RpaA-independent output pathway. We developed a simple model for KOA involving those two key terms, the active KaiC-pST state and repression by phosphorylated RpaA. A mathematical description of KOA was developed and quantitatively compared with experimental measurements for both classes of promoters in WT strains containing native KaiA, KaiB, and KaiC. The model successfully describes the key features of the time of peak KOA and the peak-to-trough ratios.
Results
The Central Oscillator Represses the Class 1 Promoter PkaiBC.
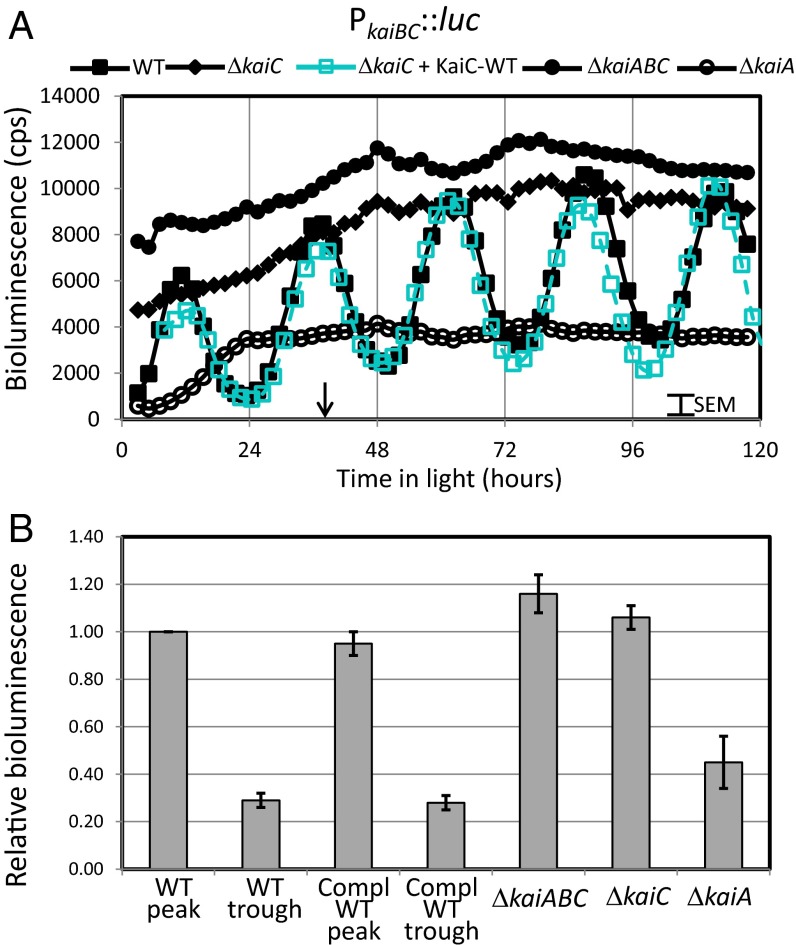
Models for the transmission of information from the circadian oscillator to the transcription machinery involve a direct link between the phosphorylation status of KaiC and Kai oscillator output. We first determined the default bioluminescence levels in the absence of key oscillator components as a baseline for testing the effects of specific KaiC variants. We hypothesized that transcription in the absence of KaiC would become arrhythmic and locked at a lower level, or midline level compared with the WT oscillation, as the knockout would remove any potential modulation of the output pathways, up or down, by (phosphorylated) KaiC protein. Strains were constructed in a WT firefly luciferase bioluminescent reporter background (AMC541, PkaiBC::luc) to assess the effects of knockouts on the promoter activity of PkaiBC, a class 1 paradigm promoter that is strongly influenced by the circadian clock (17). The bioluminescence levels of the WT and strains lacking KaiC or KaiABC were measured over several days (Fig. 2). Special care was taken to match the 750-nm optical density for all samples to assure that differences in bioluminescence intensity were not due to differences in cell number. As expected, the knockout strains displayed arrhythmic bioluminescence; specifically, the bioluminescence level in the absence of KaiC was constitutive at the upper end of the natural cycles (Fig. 2A). Thus, without KaiC present, the default state of the output pathway is high PkaiBC promoter activity, contrary to our expectations.
Fig. 2.
Default bioluminescence level is high for class 1 promoter PKaiBC in the absence of circadian oscillator. (A) Time dependence of bioluminescence of AMC541 (filled squares) and mutants with disruptions of KaiA, KaiC, or KaiABC measured using firefly luciferase driven by PkaiBC (open circles, filled circles, and filled diamonds, respectively). Following a 12-h dark entrainment, the WT AMC541 showed the expected ∼24.5-h circadian oscillations of bioluminescence, and knockouts of any of the Kai proteins eliminated the rhythms. Note that the bioluminescence levels of the KaiC and KaiABC knockouts are near the peaks of the oscillations for the WT strain. In contrast, the bioluminescence level of the KaiA knockout is near the trough. Upon introduction of KaiC-WT into NS1 of the kaiC deletion strain (open cyan squares, dashed line), the amplitudes and the rhythms were restored to those of the WT, showing the fidelity of the complementation system. The arrow indicates the time (at 36 h) used for relative bioluminescence comparisons described below. (B) Bar chart of the relative bioluminescence normalized to the peak level of the WT near the 36-h time. The data are the average of six to seven independent experiments with the error bars representing the observed SEM.
To assure that the bioluminescence levels are reflecting the genotype and not a consequence of the luciferase or detection system, we constructed an analogous set of strains starting from a different parental background that uses the luxAB bacterial luciferase system (Table S1 and Fig. S1). Detection of bioluminescence from this system differs in several respects, including the in vivo synthesis of a luciferase substrate that is not related to firefly luciferin, independence from ATP concentration, a different illumination setup for the solid medium cultures, and a different detection instrument (see SI Materials and Methods for more details). Regardless of differences in the experimental setups, the relative bioluminescence signal levels were similar—the KaiC and KaiABC knockout strains exhibited bioluminescence signal levels at the upper end of those measured in the WT strain (Fig. 1). Thus, under these conditions, changes in the relative bioluminescence levels are a consequence of the genotypic differences of the strains.
The means and SEMs are indicated for five to six independent experiments using the luc system described above, in which the bioluminescence levels were calculated relative to the peak WT level at 36 h (Fig. 2B), a time point at which variation of bioluminescence among independent experiments was minimal; the general scatter in the data is ∼7%. The results show that, in strains lacking an active central oscillator protein KaiC, the magnitude of the bioluminescence signals is locked at the peak values for the WT oscillations. Thus, it is more appropriate to view the influence of the KaiC oscillator as repressing the higher default promoter activity from the class 1 promoter PkaiBC. Notably, the kaiA deletion results in greater repression of the PkaiBC signal, suggesting that, in its absence, the repressive Kai-complex output is greater.
The Phosphomimetic KaiC-ET Signals Oscillator Output in the Presence of the Kai Complex.
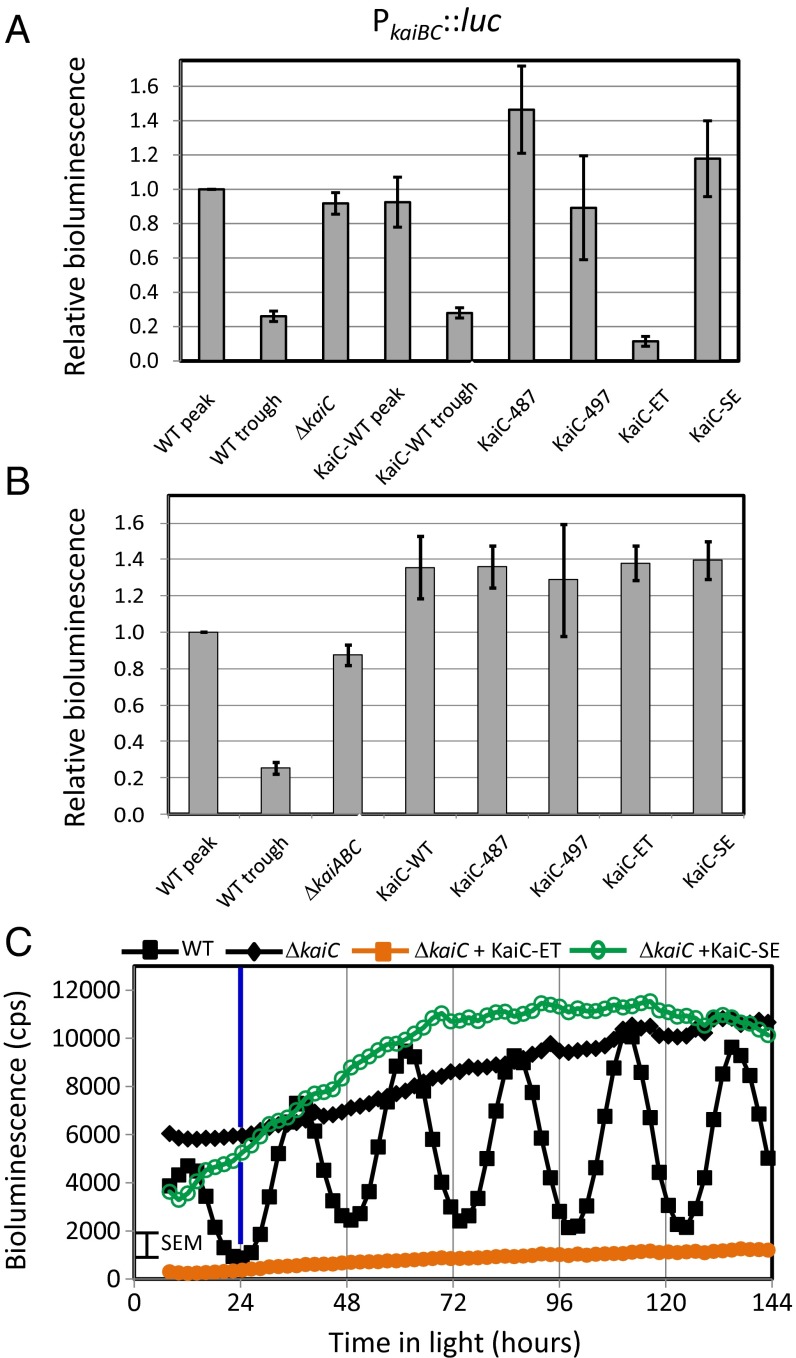
The sine wave of expression from different promoters whose magnitude peaks in different relative phases could reflect different quantities, or qualities, of output activity from different phosphostates of the oscillator (Fig. 1). What phosphostate(s) of KaiC is responsible for oscillator output, and does it differ for different promoters? Phosphomimetic variants of KaiC, designed to mimic the four possible phosphorylation states, ST, pST, SpT, and pSpT, were tested for their effects on PkaiBC reporter levels (Fig. 1). Two truncations that result in low and high ratios of phosphorylation of KaiC in vivo (18) (Table S1) and two point mutations constructed to mimic the pST and SpT states (19) were tested. WT or variant KaiC was reintroduced by ectopic expression from neutral site 1 (NS1) under control of the constitutive trc promoter in the KaiC and KaiABC knockout strains (Table S1), and the average and SEM bioluminescence signal was calculated (Fig. 3). All values were compared using the time corresponding to the peak WT bioluminescence at ∼36 h within each independent experiment, and the lower value of the WT bioluminescence rhythms was determined from the average of the troughs preceding and following the ∼36-h peak.
Fig. 3.
Bioluminescence response is specific for KaiC-ET variant for class 1 promoter PkaiBC. (A) Effect of introduction of KaiC variants in the KaiC-knockout background as reported from a class 1 promoter—PkaiBC. Shown in the first column is the peak level of the average bioluminescence of WT normalized to unity for reference, and the second column is the trough level of the WT. All data include five independent experiments and error bars reflecting the SEMs. Additional columns report values for ΔkaiC and ΔkaiC complemented with KaiC-WT, KaiC-C487, KaiC-C497, KaiC-ET, and KaiC-SE. Note that the only variant that causes a significant difference in the bioluminescence signal from that of ΔkaiC is KaiC-ET. (B) Effect of knockout and KaiC variants in a background lacking all of the Kai proteins (ΔkaiABC). Columns are similar to those in A. Note that all of the complemented deletion strains are similar to each other and to the deletion strain. This finding contrasts with those in A for the KaiC-ET variant, suggesting that KaiA and/or KaiB is required for the transmission of the KaiC phosphorylation information to the output proteins. (C) Plot of the bioluminescence as a function of time for representative strains involving the KaiC knockout. Shown are the WT (filled black squares), ΔkaiC (filled black diamonds), ΔkaiC + KaiC-ET (filled orange squares), and ΔkaiC + KaiC-SE (open green circles). Blue vertical line at 24 h highlights that bioluminescence from this class 1 promoter is at minimal levels at dawn.
In the KaiC knockout strain that retains kaiA and kaiB, introduction of WT kaiC in NS1 restored both rhythms and bioluminescence levels that were within 5% of the WT (Fig. 3A, KaiC-WT samples), confirming that the complementation system operates the same as the WT strain itself. Next, individual KaiC variants were introduced to complete the Kai complex. KaiC-487 and KaiC-497, truncation mutants that are fully phosphorylated and nonphosphorylated, respectively, had no effect on bioluminescence, nor did KaiC-SE, a phosphomimetic of the SpT state. Only the KaiC-ET variant, a phosphomimetic of KaiC-pST, caused bioluminescence to vary significantly from that of the knockout background strain, falling below the trough level observed in the WT rhythms (Fig. 3). Thus, KaiC-ET specifically alters expression of PkaiBC. Fig. 3C shows the time course of gene expression for select strains—WT, KaiC knockout, and the knockout engineered to express KaiC-ET or KaiC-SE.
In the KaiABC knockout strain, reintroduction of WT KaiC or any of the KaiC variants had no effect on the elevated and arrhythmic bioluminescence of the knockout host (Fig. 3B). The simplest interpretation is that the KaiC protein alone, regardless of its phosphostate, is inactive and/or incapable of invoking an output response. Thus, at least one of the other Kai proteins plays a greater role in regulating gene expression than merely coaxing KaiC to a specific signaling state.
Out-of-Phase Class 2 Promoter PpurF Displays Converse Behavior.
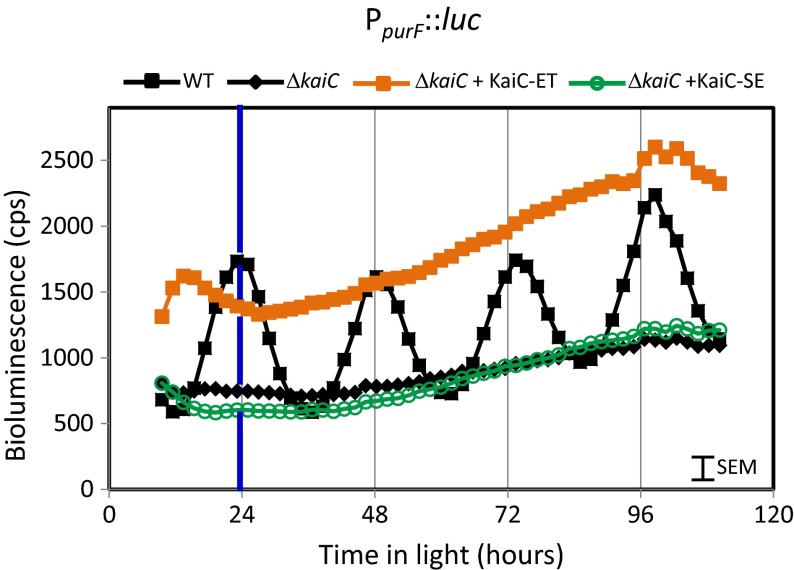
As the class 2 promoter PpurF displays promoter activity that is out-of-phase with PkaiBC, we asked whether different states of KaiC provide peak output response for different gene classes. Analogous constructs (Table S1) were tested for a PpurF::luc background. Notably, bioluminescence driven by PpurF was constant at trough levels in the KaiC knockout (Fig. 4), in contrast to the constitutive peak levels driven by PkaiBC::luc (Fig. 3C). Introduction of WT KaiC in NS1 restored rhythms as anticipated, increasing bioluminescence at nontrough times above that from the knockout strain (Fig. 4). As for the class 1 PkaiBC::luc reporter presented above, introduction of any KaiC variant other that KaiC-ET did not alter bioluminescence levels from the kaiC deletion reference level. PpurF::luc bioluminescence in the strain that expressed KaiC-ET remained at the upper end of the normal range of bioluminescence signals, and the strain carrying the KaiC-SE variant held at the lower end (Fig. 4). Thus, the effects of the knockout and complementation with KaiC variants were completely opposite to the results found for the class 1 promoter PkaiBC; however, the same variant, KaiC-ET, significantly altered expression levels for both classes of promoters.
Fig. 4.
KaiC-ET specifically elevates bioluminescence level for the class 2 promoter PpurF. Shown are representative traces for the strains ΔkaiC (filled black diamonds), ΔkaiC complemented with KaiC-WT (filled black squares), KaiC-ET (filled orange squares) or KaiC-SE (open green circles). Note that, in WT, the peak bioluminescence occurs at 24, 48, and 72 h, the KaiC knockout is at the lower end of the circadian range, and the knockout complemented with KaiC-ET runs high whereas the knockout complemented with KaiC-SE runs low. This behavior is opposite that observed with PkaiBC (Fig. 3). Blue vertical line at 24 h highlights that bioluminescence from this class 2 promoter is at maximal levels at dawn.
Contribution of the Response Regulator Protein RpaA to Circadian Gene Expression.
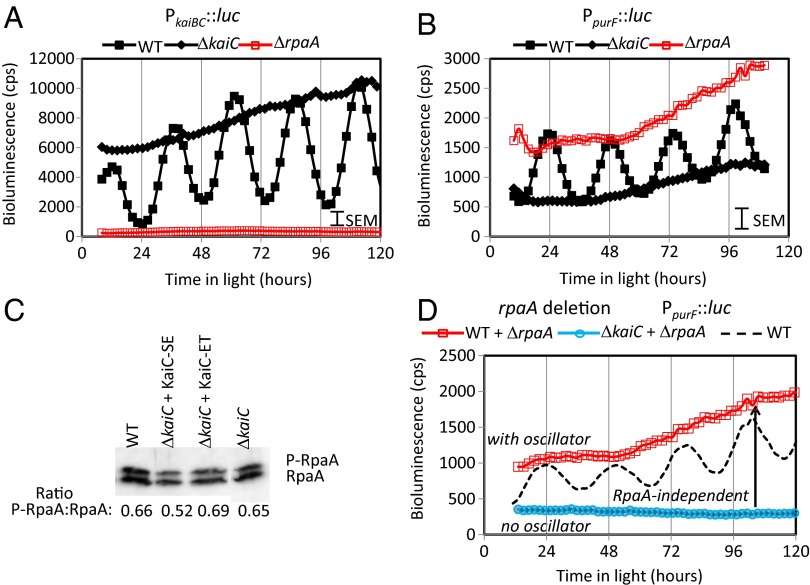
As RpaA is proposed to be a key conduit for oscillator output (14), we sought to address the contribution of RpaA to the oscillator’s influence on gene expression. We constructed a set of strains in which rpaA was deleted from the genome (Table S1). Removal of rpaA decreased bioluminescence from class 1 PkaiBC to near zero levels (Fig. 5A), consistent with previous reports and with a model that places RpaA as a master regulator of gene expression downstream of the oscillator. Furthermore, recent reports by Gutu and O’Shea (20) show a correlation between phosphorylated RpaA (P-RpaA) and the expression level from the class 1 PkaiBC reporter as the circadian oscillator progresses through its cycle. We similarly observe circadian cycling of phosphorylated RpaA (21). At 9 h into light following a dark period, a time when the difference in bioluminescence was near the maximal (Fig. 3), we measured the relative levels of P-RpaA in WT, kaiC deletions, and KaiC variant-substituted strains and were surprised to find that all showed essentially the same relative ratio of the phosphorylated (upper band) to the nonphosphorylated (lower band) RpaA (Fig. 5C). This result suggests an output pathway that is not as simple as modulation of RpaA or P-RpaA.
Fig. 5.
rpaA deletion has opposite effects on class 1 and class 2 promoters. Shown for comparison are the WT (filled black squares), ΔkaiC (filled black diamonds), and ΔrpaA (open red squares) for PkaiBC (A) and PpurF (B). Note that the RpaA knockouts have the opposite effect of the KaiC knockouts in both promoter classes. (C) Phospho-RpaA is present in all of the KaiC phosphomimetics. Samples were taken 9 h after lights on following a 12-h dark entrainment, and the ratio of P-RpaA to RpaA is provided. (D) Plot of bioluminescence reported by PpurF::luc in strains with (open red squares) and without (open cyan circles) the KaiC oscillator protein in an rpaA deletion background. The level measured in the strain containing the KaiC oscillator protein is approximately three- to fivefold greater than the strain lacking KaiC. This difference, which is comparable with the peak:trough ratio of the measured WT oscillations (shown B), indicates that there is a significant oscillator output that affects PpurF expression in the absence of RpaA.
Because we found that KaiC deletion strains and the KaiC variant-substituted strains show different effects for the class 2 promoter PpurF than for PkaiBC, we measured the bioluminescence of the rpaA deletion strains in PpurF reporter backgrounds. As shown in Fig. 5B, deletion of rpaA in the WT background had the opposite effect to that found for the class 1 PkaiBC promoter—the bioluminescence of the rpaA deletion strain was constitutive at the peak levels of the WT PpurF oscillator signals. However, this pattern was fully reversed by an additional kaiC deletion (Fig. 5D). Thus, in the absence of RpaA, the strain containing the KaiC oscillator had a level of promoter activity that was approximately three- to fivefold greater than that in the strain lacking the central KaiC oscillator. As this increase is comparable to the approximately threefold peak-to-trough ratio of the WT oscillations for PpurF (Fig. 5D, dotted line), we conclude that the oscillator can still transmit information to activate or repress PpurF expression levels independently of RpaA. Thus, we conclude that there is a significant RpaA-independent output from the Kai oscillator.
A Metric for KOA.
Because bioluminescence signals for knockout and complemented strains are opposite for class 1 and class 2 promoters, we sought to distinguish between high and low promoter activity and the influence of the central oscillator on promoter activity (KOA). To quantify the magnitude and direction (activating or repressing) of the oscillator on the output, it was essential to establish the basal level of expression in the absence of any oscillator output as a reference. This reference level was determined from a kaiC deletion strain that lacks a functional oscillator. Consequently, we can quantify KOA as the difference of the promoter activity (PA) with a functional oscillator (as measured by bioluminescence) and that measured in the absence of KaiC. By performing this transformation, the sign of KOA (positive or negative) directly indicates the effect of the oscillator on expression (activating or repressing, respectively) whereas the magnitude (absolute value) indicates the extent of the change. KOA reflects only the net effect of the oscillator on PA and is distinct from the timekeeping activity that results from phosporylation/dephosphorylation of Ser431 and Thr432. The simplest expression for KOA is:
where PA(WT) and PA(ΔkaiC) represent the promoter activity of the WT reporter and KaiC knockout strains, respectively. As the WT oscillator complex cycles through the phosphorylation states as a function of time, KOA is time-dependent and reflects the net output of the substates at that phase of the circadian cycle. For example, the substate that peaks at the time at which the magnitude of KOA is greatest is the most active substate. Correspondingly, KOA of the KaiC phosphomimetics was determined by replacing PA(WT) with PA(KaiC-mimetic). In this case, because the KaiC phosphomimetic cannot cycle, a time-dependent KOA is not expected. Correspondingly, the KaiC variant that has the greatest magnitude of KOA is the phosphomimetic that is most active. Although this calculation is merely a transformation of the reporter data, it more clearly reveals the following: (i) the influence of the circadian clock on the two predominant classes of promoters, class 1 and class 2; (ii) the point in the cycle when the magnitude of KOA is the greatest for a given promoter; and (iii) which KaiC variant/phosphostate invokes the greatest magnitude of KOA.
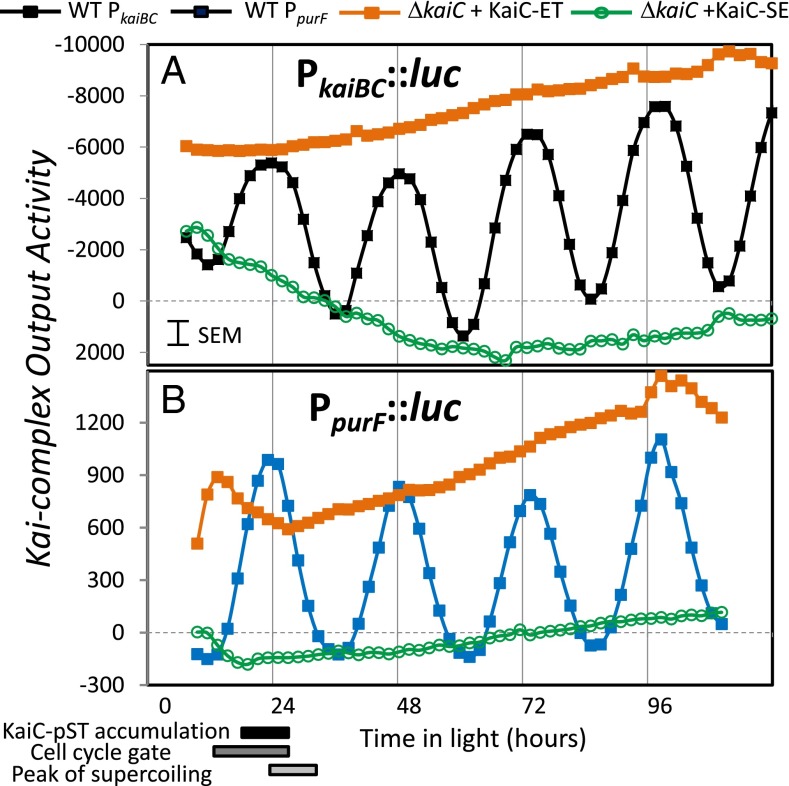
KOA was calculated for the PkaiBC::luc system as the paradigm for class 1 promoters (Fig. 6A). For this system, we find that (i) the native circadian clock represses promoter activity as shown by the negative sign of KOA; (ii) the KOA is most active at or near subjective dawn (24, 48, and 72 h) as shown by the time of greatest magnitude; and (iii) the KaiC-ET variant specifically invokes the greatest activity, whereas little KOA is invoked by any other KaiC variant (represented by KaiC-SE). Similarly, KOA was determined for the PpurF::luc system as representing class 2 promoters (Fig. 6B). For this system, the native circadian clock activates promoter activity as indicated by the positive KOA values; however, the timing of the peak magnitude of KOA and the activity of the KaiC-ET variant are the same as for PkaiBC::luc. No KOA was observed in the absence of the KaiC-pST or KaiC-ET states.
Fig. 6.
Kai-complex Output Activity (KOA) correlates with KaiC-pST or its phosphomimetic KaiC-ET for both class 1 (A) and class 2 (B) promoters. To more easily compare the results of the central circadian oscillator on class 1 and class 2 promoters, we defined KOA (Eq. 1) to better reflect the effect of the oscillator on the respective promoter outputs. Shown is a comparison of the KOA for the WT system (filled black and blue squares) and systems in which the KaiC-ET (filled orange squares) or KaiC-SE (open green circles) were the sole KaiC available in the Kai complex. Strikingly, the results show that the same state of KaiC that mimicked by KaiC-ET is the most active, resulting in the greatest magnitude of KOA for both class of promoters and that the activities of the circadian complex coincide at the times corresponding to dawn (24, 48, and 72 h). The difference is that the circadian clock increases promoter activity of PpurF but suppresses promoter activity of PkaiBC.
The concept of KOA makes it more apparent that these two promoter classes share more characteristics in common than initially thought. In particular, the greatest magnitude of KOA occurs simultaneously at or near dawn, and the KaiC-pST phosphomimetic (KaiC-ET variant) invokes the greatest KOA response. These findings support a model in which the same KaiC state alters both classes in opposing fashions, in contrast to a model in which different oscillator states stimulate (or repress) a particular subset of promoters.
A Modified Model That Includes an RpaA-Independent Output Pathway.
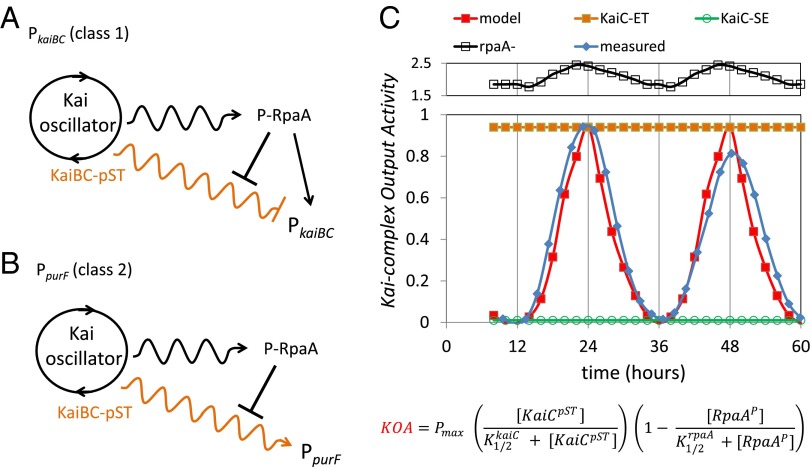
The standard model for the Kai oscillator output is that particular phosphostates of KaiC promote the phosphorylation of a “master output regulator,” RpaA, via SasA. Phosphorylated RpaA then acts to transmit the information to the transcription machinery (14). Recent work shows that P-RpaA accumulates in parallel with, and peaks at the same time as, bioluminescence for class 1 promoters, leading to an interpretation that P-RpaA activates promoter activity (20). Using KOA, we measured the net Kai output as it affects reporter gene expression, whether or not the pathway involves RpaA. Notably, the majority of our measurements for the class 1 promoter PkaiBC fit the standard model described above. However, the lack of correlation of the P-RpaA level and the bioluminescence level in the KaiC-phosphomimetic strains (Fig. 5) suggests that this model is incomplete. The simplest refinement of the model is to include a second output pathway from the oscillator to the transcription machinery that depends on the phosphostate of KaiC but not on the presence of P-RpaA (Fig. 7A), similar to models that include an oscilloid output (22). As our results also suggest a repressive effect of RpaA on KOA, we include the repressive link between P-RpaA and this output pathway. As our results suggest that peak KOA activity requires KaiB, we have also included KaiB in our model.
Fig. 7.
Simplified model of the central circadian oscillator and output activities. (A) Simplified scheme for the Kai oscillator output for the class 1 PkaiBC promoter. The standard model for the output response, involving modulation of P-RpaA via differential binding of SasA to KaiC dependent on phosphorylation state, can explain most of the observations. However, the discord between the level of P-RpaA and KOA necessitates the inclusion of a second, RpaA-independent output pathway as shown. This idea is similar in principle to a simplified version of the oscilloid model (22), in which we include an RpaA output pathway. For simplicity, we represent the KaiC phosphorylation cycle as a circle and highlight in orange only the KaiBC-pST state as the most active (see A above). Wavy arrows indicate that the “activity” is time-dependent in a circadian manner to distinguish it from a constant response indicated by straight arrows. (B) Simplified scheme for the Kai oscillator output for the class 2 PpurF promoter. In this case, the observations were not reconcilable with the standard model. Thus, we propose the unique scheme shown as a simple starting point to describe the behavior of the class 2 promoters. The model includes (i) a predominant RpaA-independent output pathway (orange wavy line) and (ii) suggests that P-RpaA represses this output pathway. It is somewhat satisfying that both of these effects were already brought into the revised model for class 1 promoter PkaiBC, indicating that behavior of class 1 and class 2 promoters likely shares much in common. Among those common characteristics are that the KaiBC-pST state is the most active in invoking KOA, which simultaneously suppresses PkaiBC (class 1) and activates PpurF (class 2). (C) Comparison of the relative values of KOA taken from Fig. 6 for PpurF (blue curve) and the simulation of the mathematical model using Eq. 2 (red curve). The equation for KOA is shown below. The simulation accurately predicts the time of peak KOA as well as the peak-to-trough ratio. In addition, by increasing the catalytic term or decreasing the repressive term, the higher levels for KOA found in the system with KaiC-ET (orange) and in the rpaA deletion (open black squares) were accurately described. By decreasing the catalytic term, the low values for KOA were reproduced for the KaiC-SE (open green circles).
Although we could rescue the standard model by inclusion of the second output pathway for the class 1 promoter PkaiBC, the behavior for the class 2 promoter PpurF is incompatible with this scheme. Most strikingly, the presence or absence of an intact Kai oscillator complex affects the PpurF reporter as much in an rpaA deletion background as the usual magnitude of PpurF reporter oscillation in a WT strain (Fig. 5). These data identify a predominantly RpaA-independent output pathway for the class 2 PpurF promoter. We indicate this component in the simplest manner in Fig. 7B. As P-RpaA represses KOA for the class 2 PpurF promoter as well, we include the repressive shunt in the figure. The necessity to introduce a unique output pathway to explain our data further supports the refined model for the class 1 promoter PkaiBC (Fig. 7A). Additional studies are necessary to determine whether this behavior is common to all class 2 promoters.
The unique aspects of the models include (i) a significant (predominant) RpaA-independent output from the central oscillator whose magnitude of KOA peaks upon accumulation of the KaiC-pST state and (ii) requirement for KaiB that coincides with the accumulation of the KaiC-pST state. P-RpaA remains a key player in controlling the observed oscillations in the bioluminescence; however, we propose a nonstandard role in which P-RpaA represses an alternative Kai output pathway. The timekeeping response of the output then results from the net effect of the temporal variation of KaiC-pST and the (time-dependent) repression of KOA by P-RpaA (which peaks at dusk at a time when KOA is minimized).
A Mathematical Description of KOA.
Implicitly we have assumed that a KaiC hexamer composed solely of KaiC-ET accurately reflects the behavior exhibited by the WT hexamer, which is never homogenously composed of the KaiC-pST state (23). Thus, we wanted to quantitatively test the validity of the model deduced above. We developed a simple mathematical description, based on the previous arguments, namely, that KaiC-pST is the most active state and P-RpaA is a repressor of the oscillator output (Fig. 7 A and B), to test whether the simple scheme can faithfully reproduce key features of the experimental observations (Figs. 5 and 6). The model includes one term for the output involving the concentration of KaiC-pST and a second term for the inhibition of output by P-RpaA. The simple mathematical expression is shown below:
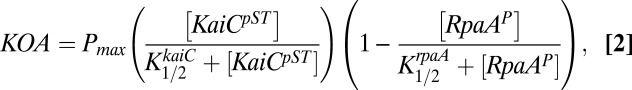 |
where 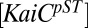 is the relative concentration of the KaiC-pST substate,
is the relative concentration of the KaiC-pST substate, 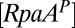 is the relative concentration of phosphorylated RpaA,
is the relative concentration of phosphorylated RpaA, 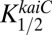 is the half saturation level for the relative level of KaiC-pST,
is the half saturation level for the relative level of KaiC-pST, 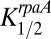 is the half saturation level for the relative level of P-RpaA, and Pmax is the maximum output level. The first term in the parentheses is a catalytic term reflecting the amount of the active KaiC-pST state present at any time. The second term is due to an inhibition of output reflecting the amount of the inhibitory P-RpaA present at any time. The time dependence of KOA comes from the time variations of
is the half saturation level for the relative level of P-RpaA, and Pmax is the maximum output level. The first term in the parentheses is a catalytic term reflecting the amount of the active KaiC-pST state present at any time. The second term is due to an inhibition of output reflecting the amount of the inhibitory P-RpaA present at any time. The time dependence of KOA comes from the time variations of 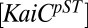 and
and 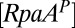 . We used the data from Rust et al. (23) on the time dependence of the KaiC-pST state for the catalytic term for the time window from 8 to 36 h. Comparison of the in vitro data with in vivo data can be done by aligning peak promoter activity of PkaiBC::luc with the peak of the total KaiC-P as suggested by Iwasaki et al. (24), allowing comparison with KOA via elapsed time in the circadian cycle. Because in vitro and in vivo experiments are performed under different conditions, this alignment is only approximate to within about an hour and is consistent with in vivo results presented by Johnson et al. (25). We used data from Gutu and O’Shea (20) on the time dependence of P-RpaA. As those data were obtained from in vivo measurements from entrained cells, we assumed no time shift to synchronize their data with our results. The calculated values were extended another 24 h to better compare with measured values over two complete cycles. Both
. We used the data from Rust et al. (23) on the time dependence of the KaiC-pST state for the catalytic term for the time window from 8 to 36 h. Comparison of the in vitro data with in vivo data can be done by aligning peak promoter activity of PkaiBC::luc with the peak of the total KaiC-P as suggested by Iwasaki et al. (24), allowing comparison with KOA via elapsed time in the circadian cycle. Because in vitro and in vivo experiments are performed under different conditions, this alignment is only approximate to within about an hour and is consistent with in vivo results presented by Johnson et al. (25). We used data from Gutu and O’Shea (20) on the time dependence of P-RpaA. As those data were obtained from in vivo measurements from entrained cells, we assumed no time shift to synchronize their data with our results. The calculated values were extended another 24 h to better compare with measured values over two complete cycles. Both 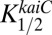 and
and 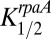 were assumed to be constant with time (i.e., not dependent on the phosphorylation state of KaiC) and were adjustable parameters to obtain a good simulation. For comparison, the measured data for the class 2 PpurF promoter were rescaled to range from zero to one. Pmax was adjusted to fit the calculated data to this rescaling for the WT strain, and the calculated values were shifted down to align the troughs of the oscillations (Fig. 7C). Values used for the simulation were Pmax = 4,
were assumed to be constant with time (i.e., not dependent on the phosphorylation state of KaiC) and were adjustable parameters to obtain a good simulation. For comparison, the measured data for the class 2 PpurF promoter were rescaled to range from zero to one. Pmax was adjusted to fit the calculated data to this rescaling for the WT strain, and the calculated values were shifted down to align the troughs of the oscillations (Fig. 7C). Values used for the simulation were Pmax = 4, 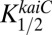 = 0.1, and
= 0.1, and 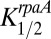 = 0.11. Quite satisfactorily, given the simplicity of the model, the calculated WT data accurately describe both the time dependence of KOA, including the time it peaks, and the extent of peak-to-trough values.
= 0.11. Quite satisfactorily, given the simplicity of the model, the calculated WT data accurately describe both the time dependence of KOA, including the time it peaks, and the extent of peak-to-trough values.
The behavior of the deletion strains and the KaiC phosphomimetics can also be understood in the context of this model. The low output for KaiC-SE comes from a zero value for the catalytic term. The high value for KOA in KaiC-ET comes from a maximal value for the catalytic term. The high value for KOA in the rpaA deletion comes from the lack of inhibition due to the lack of P-RpaA. Using the values from the simulation of the WT data, the expected values for the rpaA deletion are approximately twofold greater than observed. The lower observed value can be attributed to an inability of the transcription machinery to achieve such a high response. In other words, we attribute the smaller than predicted magnitude of KOA in the rpaA deletion strain to limitation of the transcription machinery as opposed to a direct reflection of KOA. Interestingly, the model predicts the small residual rhythms in the rpaA deletion, which is likely dampened by the limitation of the transcription machinery. Although such rhythms are not routinely observed in an rpaA deletion strain, it is worth noting that residual rhythms are observed in sasA deletion strains (14), which to first order would be modeled in a similar manner as eliminating P-RpaA (14). Thus, to first order, the simplified model can be fit to the observations for the WT as well as deletion strains and KaiC phosphomimetics, showing that it contains key ingredients for a description of KOA. Importantly, the equation shows how the peak magnitude of KOA and the extent of peak-to-trough oscillations can be modeled considering a single active KaiC-pST state and repression of output by P-RpaA. As the time dependence of KOA for both class 1 and class 2 promoters is very similar (Fig. 6), the model will successfully simulate the same key features for the class 1 promoter by simply adjusting Pmax. The success of this model to predict the observed behaviors lends support to the ideas that there is a single active state, KaiC-pST, that leads to an RpaA-independent output and that (at least in part) P-RpaA is repressive of the Kai oscillator output.
Discussion
In this study, we introduce the concept of Kai-complex output activity (KOA) to distinguish among the following properties of clock-controlled gene expression: promoter activity, the effect of the Kai oscillator on promoter activity, the most active output state of the oscillator, and the timekeeping activity that modulates the output activity. We chose PkaiBC and PpurF as paradigms for class 1 and class 2 promoters because their regulation is predominated by oscillator control. A key aspect of the experimental design was establishing conditions under which the bioluminescence levels could be quantitatively trusted and the inclusion of the opposing PkaiBC and PpurF reporters for all analyses, which revealed aspects of clock control of gene expression that have been missed previously. Even though the promoters show mutually out-of-phase promoter activity, the peak magnitude of KOA coincides for both near subjective dawn and with the same phosphomimetic, KaiC-ET (Fig. 6). Importantly, when KaiC is in the active KOA state, the output from the oscillator simultaneously represses PkaiBC (class 1) and activates PpurF (class 2). Furthermore, another Kai protein—we propose KaiB—is important for facilitating output. In addition, our results show that RpaA acts to repress the magnitude of KOA, and we find a significant RpaA-independent output activity that is evident with the PpurF reporter (Fig. 5D).
KaiC Requires KaiB for Peak KOA.
The results presented above show that KaiC alone is incapable of effecting an output response, even if a phosphomimetic is used to mimic an active KaiC state (Fig. 2). Furthermore, a KaiA knockout affects expression in an opposite manner compared with that of a KaiC knockout (Fig. 2), suggesting that the presence of KaiA represses the magnitude of KOA. Notably, KaiA is not necessary to achieve large magnitudes for KOA. The repressive effect of KaiA on the oscillator can be attributed to its propensity to promote formation of the KaiC-SpT and KaiC-pSpT states (Fig. 1). Neither of these states was active based on (i) data from analogous phosphomimetic variants and (ii) lower KOA at circadian times during which these states are most prominent. It should be noted that decreased levels of KaiC were observed upon insertional inactivation of KaiA, but decreased levels of KaiC cannot explain the opposing effects of the KaiA knockout compared with the KaiC knockout. Because KaiA represses the oscillator output (Fig. 2), it is most likely that KaiB is the required player for high KOA. The increase in the magnitude of KOA in the absence of KaiA is perhaps surprising as KaiC becomes predominantly nonphosphorylated (24). This result could suggest a possible direct role of KaiA in clock output, but further work is necessary to support this idea.
The proposal that KaiB is required for KaiC activity is intriguing because there are numerous cyanobacterial strains that contain one (or more) KaiBC operons without KaiA (26). The absence of KaiA suggests an additional oscillator-independent activity for the KaiBC complex. Furthermore, the sensitivity of the bioluminescence level to specific KaiC variants suggests a transcription-related activity for the KaiBC complex; at least in the case of PpurF, this activity cannot be accounted for by modulation of RpaA. Further studies are needed to elucidate this proposed activity.
Peak KaiC-pST Levels Coincide with Peak KOA in WT Strains.
Because the results show that the peak magnitude of KOA is near dawn and is promoted by the KaiC-pST mimetic (KaiC-ET), we hypothesized that peak levels of KaiC-pST should coincide with the peak magnitude of KOA. The composition of the KaiC hexamer during the circadian cycle has been determined previously both in vitro and in vivo (23, 25). Synchronization of the in vitro data to in vivo data was described above in the description of the mathematical model. The faithful simulation of the simple mathematical model to describe key features of KOA (Fig. 7) strongly supports the conclusion that KaiC-pST is the most active output state of KaiC in the native circadian system.
The KaiC-pST state, accumulating late in the circadian cycle, was also shown to be important for binding of KaiB to form the entire Kai complex (22, 25, 27–29). This result is in agreement with our finding that KaiB is required to reach peak KOA (Fig. 3).
Using SAXs and fluorescence, Murayama et al. (29) found that KaiC-pST is the only phosphorylation state of KaiC that remained at near constant levels at different temperatures, suggestive that the KaiC-pST state is key to the temperature compensation of the clock oscillator complex. From our perspective, keeping the levels of the active KaiC-pST state temperature-compensated assures a KOA that is temperature-compensated as our model is sensitive specifically to the KaiC-pST state (Eq. 2).
The KaiC-pST state does not accumulate to near 100% either in vivo or in vitro. Rather, peak accumulation appears to be in the range of 15–25%, suggesting that the Kai complex is composed of a mixed KaiC hexamer involving just one or two KaiC-pST components, with the remainder being nonphosphorylated (23, 29, 30). Although several crystal structures have been obtained that provide a wealth of data (31, 32), the structures show a greater extent of phosphorylation than expected for the KaiC hexamer at the time of greatest KOA. A structure more representative of the active state would yield important clues to the function of KaiC in cellular processes.
Relation of KOA and DNA Topology.
A link between DNA topology and the circadian system was shown by the finding that both chromosomal compaction (10) and supercoiling of endogenous plasmids (33) follow the circadian cycle. Both chromosomal compaction and supercoiling peak at subjective dawn at a time corresponding to the peak magnitude of KOA in this study. In addition, it was shown previously that a supercoiling-sensitive Escherichia coli promoter drives class 2 expression in S. elongatus whereas canonical E. coli promoters are class 1 in the cyanobacterium, supporting the idea that promoter response is a function of the local DNA topology (34). Furthermore, Vijayan et al. (11) showed that addition of inhibitors that affect DNA topology have different effects on expression from class 1 and 2 promoters. These results, combined with the knowledge that the crystal structure of KaiC resembles a molecular motor such as a helicase or ATPase (25, 31, 32), hint at a more direct role for KaiC in modulating promoter activity by, for example, binding to chromosomal and plasmid DNA. Previous results to test direct action of KaiC on DNA have shown weak binding of KaiC to forked DNA in support of this idea (15), but no conclusive evidence of KaiC possessing a biochemical activity directly on DNA was obtained.
Relation of Circadian KOA to Cell-Cycle Gating.
A Kai oscillator-based checkpoint for the cyanobacterial cell cycle has been identified (35). This “circadian gate” also occurs late in the circadian cycle at time between 14 and 17 h upon introduction back into light (35). During this time, the KaiC-pST ratio is increasing and the other phosphorylated KaiC states are decreasing (19, 23). Although these times are shifted from the peak magnitude of KOA, which is observed closer to 24 h upon introduction back into light, of all of the KaiC phosphostates, the KaiC-pST state best correlates with the cell-cycle gate. Any real differences in the timing may be attributed to the formation of a “gate complex” that could differ in other protein components from those involved in the circadian activity discussed above.
RpaA-Independent Output of the Central Oscillator.
Our results argue for a significant RpaA-independent output pathway, in particular for the class 2 PpurF promoter. Faithful reproduction of the circadian output oscillations using a simple model derived using this idea provides strong support for the proposal. However, what could be the RpaA-independent pathway? It could involve another protein such as RpaB, whose role in circadian gene expression was reported recently (36), or possibly other response regulator proteins. However, current models for the activity of RpaB invoke interactions with RpaA and would likely need modification to be compatible with the results presented here. Our results are consistent with a model in which the KaiBC complex regulates an alternative output process, such as through direct interaction of KaiC with DNA, an oscilloid model (33). Support for these ideas stems from the similarity in sequence and structure of KaiC to known DNA/RNA interacting proteins, specifically helicases (24, 31, 32). Experiments designed to more directly assess such interactions are necessary to determine the RpaA-independent activity of the KaiBC complex.
Materials and Methods
Cyanobacterial Strains, Media, and Culture Conditions.
All WT reporter and mutant cyanobacterial strains were created in S. elongatus PCC 7942. Cyanobacterial strains were grown in BG-11 medium (37) under continuous light (photosynthetically active radiation I ∼100–150 μmol photons⋅m−2⋅s−1) at 30 °C with appropriate antibiotics. The reporter strain AMC541 carries the firefly luciferase gene driven by the kaiBC promoter [PkaiBC::luc (pAM2105 NS2 Cmr)] (Table S2) (38) in the neutral site 2 (NS2) locus. Construction of kaiC and kaiABC knockouts, as well as the strains complemented with KaiC variants (Table S1), were constructed via homologous recombination using plasmids shown in Table S2. More details are described in SI Materials and Methods.
Bioluminescence Assay and Data Analysis.
The measurement of bioluminescence from the reporter strains was performed as described previously (39–43). Briefly, batch cultures for in vivo bioluminescence studies were grown in 25-mL sterile clear glass tubes containing 5 mL of BG-11 medium and the appropriate antibiotics. Cultures were synchronized by placement in a cycle of 12 h light and 12 h dark for 1 d and then returned to constant light conditions for bioluminescence sampling. To best match the number of cells for each culture, the OD750 was measured on a Beckman Coulter DU 640B Spectrophometer, and volumes were adjusted to give equivalent cell counts. More details are included in SI Materials and Methods.
Protein Sample Preparation, Gel Electrophoresis, and Phos-tag Acrylamide Gels.
Total protein sample preparation and immunoblot analysis for single-time-point experiments were performed as described previously (4). Samples prepared by adding one-tenth volume of 2× SDS loading dye to ∼5–6 μg of sample (for RpaA detection) were loaded onto SDS-polyacrylamide gels containing 25 μM Phos-tag reagent (Wako Pure Chemical Industries, Ltd.). Following separation and gel rinses (SI Materials and Methods), protein was electrophoretically transferred to a PVDF membrane. The filter was blocked by incubating in 5% nonfat dry milk/Tris-buffered saline with 0.1% Tween 20. The primary antibody was added and incubated for 1 h at room temperature. Following rinses, the secondary antibody was then incubated at 20 °C for 1 h (for RpaA, goat anti-rabbit-HRP). The blots were developed using SuperSignal West Fempto Maximum Sensitivity Substrate (Thermo Scientific) according to the manufacturer’s directions. Chemiluminescence was detected using an Alpha Innotech FluorChem HD2 Imaging System (Alpha Innotech).
Mathematical Simulations of KOA.
Eq. 2 was used to simulate the relative values of KOA determined from the experimental data measured in this work. Values for the ratio of KaiC-pST state and the P-RpaA were obtained from the literature (20, 23). The simulation was performed using Microsoft Excel, and the relative KOA and simulations were superimposed.
Supplementary Material
Acknowledgments
We thank S. Cohen, Y.-I. Kim, S. Brody, W. Bechtel, and R. Greenspan for helpful discussions; C. Chang, J. Bordowitz, M. B. Paddock, and R. Shultzaberger for technical assistance; and E. O’Shea for RpaA antiserum. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award R01GM062419 (to S.S.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315170110/-/DCSupplemental.
References
- 1.DeCoursey PJ. Effect of light on the circadian activity rhythm of the flying squirrel, Glaucomys volans. Z Vgl Physiol. 1961;44(4):331–354. [Google Scholar]
- 2.Johnson CH. Testing the adaptive value of circadian systems. Methods Enzymol. 2005;393:818–837. doi: 10.1016/S0076-6879(05)93043-7. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469(7331):554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine: genetics of the cyanobacterial circadian clock. Adv Genet. 2011;74:13–53. doi: 10.1016/B978-0-12-387690-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo T, et al. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90(12):5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Golden SS, Kondo T, Ishiura M, Johnson CH. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995;177(8):2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9(12):1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 10.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103(22):8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106(52):22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kageyama H, Kondo T, Iwasaki H. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J Biol Chem. 2003;278(4):2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki H, et al. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101(2):223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 14.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103(32):12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori T, et al. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA. 2002;99(26):17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi Y, et al. labA: A novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2007;21(1):60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clerico EM, Cassone VM, Golden SS. Stability and lability of circadian period of gene expression in the cyanobacterium Synechococcus elongatus. Microbiology. 2009;155(Pt 2):635–641. doi: 10.1099/mic.0.022343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YI, Dong GG, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105(35):12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang YG, Kuo NW, Tseng R, LiWang A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2011;108(35):14431–14436. doi: 10.1073/pnas.1104221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50(2):288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boyd JS, Bordowitz JR, Bree AC, Golden SS (2013) An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc Natl Acad Sci USA 110(34):13950–13955. [DOI] [PMC free article] [PubMed]
- 22.Pattanayek R, et al. Combined SAXS/EM based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS ONE. 2011;6(8):e23697. doi: 10.1371/journal.pone.0023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318(5851):809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99(24):15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: From biophysics to bioevolution. Annu Rev Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiegard A, et al. Biochemical analysis of three putative KaiC clock proteins from Synechocystis sp. PCC 6803 suggests their functional divergence. Microbiology. 2013;159(Pt 5):948–958. doi: 10.1099/mic.0.065425-0. [DOI] [PubMed] [Google Scholar]
- 27.Kageyama H, et al. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23(2):161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 28.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22(9):2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murayama Y, et al. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 2011;30(1):68–78. doi: 10.1038/emboj.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331(6014):220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattanayek R, et al. Structures of KaiC circadian clock mutant proteins: A new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS ONE. 2009;4(11):e7529. doi: 10.1371/journal.pone.0007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattanayek R, et al. Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol Cell. 2004;15(3):375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104(47):18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min HT, Liu Y, Johnson CH, Golden SS. Phase determination of circadian gene expression in Synechococcus elongatus PCC 7942. J Biol Rhythms. 2004;19(2):103–112. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- 35.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140(4):529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanaoka M, et al. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J Biol Chem. 2012;287(31):26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111(Mar):1–61. [Google Scholar]
- 38.Chen Y, et al. A novel allele of kaiA shortens the circadian period and strengthens interaction of oscillator components in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2009;191(13):4392–4400. doi: 10.1128/JB.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson CR, et al. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 40.Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu Rev Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 41.Holtman CK, et al. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 2005;12(2):103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 42.Kondo T, Ishiura M. Circadian rhythms of cyanobacteria: Monitoring the biological clocks of individual colonies by bioluminescence. J Bacteriol. 1994;176(7):1881–1885. doi: 10.1128/jb.176.7.1881-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.