Significance
Mammalian target of rapamycin complex 2 (mTORC2) controls a wide range of cellular and developmental processes and is a potential target for therapeutic strategies against a range of human diseases. Here we identify XPLN (exchange factor found in platelets, leukemic, and neuronal tissues) as an endogenous inhibitor of mTORC2 and show that XPLN negatively regulates cell survival and skeletal myoblast differentiation by inhibiting mTORC2 and subsequently the Ser/Thr kinase Akt. This XPLN action requires its N terminus and is independent of its canonical activity as a guanine nucleotide exchange factor. Our findings provide a molecular understanding of mTORC2 regulation and uncover XPLN as a potentially important player in many aspects of biology and diseases involving mTORC2 and Akt.
Abstract
Mammalian target of rapamycin complex 2 (mTORC2) controls a wide range of cellular and developmental processes, but its regulation remains incompletely understood. Through a yeast two-hybrid screen, we have identified XPLN (exchange factor found in platelets, leukemic, and neuronal tissues), a guanine nucleotide exchange factor (GEF) for Rho GTPases, as an interacting partner of mTOR. In mammalian cells, XPLN interacts with mTORC2 but not with mTORC1, and this interaction is dependent on rictor. Knockdown of XPLN enhances phosphorylation of the Ser/Thr kinase Akt, a target of mTORC2, whereas overexpression of XPLN suppresses it, suggesting that XPLN inhibits mTORC2 signaling to Akt. Consistent with Akt promoting cell survival and XPLN playing a negative role in this process, XPLN knockdown protects cells from starvation-induced apoptosis. Importantly, this effect of XPLN depletion is abolished by inhibition of Akt or mTOR kinase activity, as well as by rictor knockdown. In vitro, purified XPLN inhibits mTORC2 kinase activity toward Akt without affecting mTORC1 activity. Interestingly, the GEF activity of XPLN is dispensable for its regulation of mTORC2 and Akt in cells and in vitro, whereas an N-terminal 125-amino-acid fragment of XPLN is both necessary and sufficient for the inhibition of mTORC2. Finally, as a muscle-enriched protein, XPLN negatively regulates myoblast differentiation by suppressing mTORC2 and Akt, and this function is through the XPLN N terminus and independent of GEF activity. Our study identifies XPLN as an endogenous inhibitor of mTORC2 and delineates a noncanonical mechanism of XPLN action.
Mammalian target of rapamycin (mTOR) is an evolutionarily conserved Ser/Thr kinase that integrates signals from nutrient availability, growth factors, differentiation inducers, and various types of stress to control a wide range of cellular and developmental processes (1, 2). mTOR nucleates two distinct multiprotein complexes known as mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), characterized by the presence of raptor and rictor, respectively. Emerging evidence implicates the deregulation of mTOR signaling in a variety of diseases including cancer and diabetes (1), underscoring the importance of fully understanding the regulation of mTOR signaling.
mTORC1 regulates cell growth and proliferation by promoting biosynthesis of proteins, lipids, and organelles while inhibiting autophagy (1, 2). The best-characterized substrates for the mTORC1 kinase are S6 kinase 1 (S6K1) and eIF-4E–binding protein-1 (4E-BP1), both key regulators of protein synthesis (3). mTORC2 phosphorylates the hydrophobic motif site Ser473 on the Ser/Thr kinase Akt that is necessary for its activation (4), as well as the turn motif controlling the folding and stability of Akt (5, 6). The ribosome plays a direct role in activating mTORC2 (7), and association with the ribosome also allows mTORC2 to phosphorylate and stabilize Akt cotranslationally (8). Thus, mTORC2 is involved in a variety of processes that are regulated by Akt, including cell survival, glucose metabolism, and cellular differentiation (9–11). In addition, mTORC2 regulates cytoskeleton organization by promoting phosphorylation of protein kinase C (PKCα) (5, 6, 12, 13), and serum/glucocorticoid-regulated kinase 1 (SGK1) has also been identified as a substrate of mTORC2 (14). Compared with mTORC1, for which mechanisms of activation by upstream signals have been extensively studied, less is known about the regulation of mTORC2 signaling. Several endogenous inhibitors of mTOR have been reported. Although PRAS40 and FKBP38 are specific inhibitors of mTORC1, DEPTOR interacts with and inhibits both mTORC1 and mTORC2 (1, 2). Recently, the glucocorticoid-induced leucine zipper protein (GILZ) was reported to inhibit mTORC2 when overexpressed in BCR-ABL–expressing chronic myeloid leukemia (CML) cells (15).
XPLN (exchange factor found in platelets, leukemic, and neuronal tissues) is a guanine nucleotide exchange factor (GEF) for Rho GTPases (RhoGEF) selectively activating RhoA and RhoB in vitro (16). Like most RhoGEFs, XPLN contains a diffuse B-cell lymphoma (Dbl) homology (DH) domain followed by a pleckstrin homology (PH) domain. This protein is expressed in several human tissues, with the highest levels found in the skeletal muscle and brain (16). As expected for a protein with RhoGEF activity in vitro, overexpression of recombinant XPLN stimulates Rho-kinase–dependent assembly of stress fibers and focal adhesion and has cell-transforming activity (16). However, a biological function for the endogenous XPLN has not been reported.
Here we identify XPLN as an mTORC2-interacting protein. We find that XPLN inhibits mTORC2 kinase activity in vitro and activation of Akt in cells. Interestingly, this function of XPLN is independent of its GEF activity and is most likely mediated by a physical interaction between its N terminus and mTORC2. Furthermore, we show that XPLN negatively regulates cell survival and myoblast differentiation through inhibiting mTORC2 and Akt. These findings reveal XPLN as a regulator of mTORC2 signaling to Akt via a noncanonical mechanism.
Results
XPLN Interacts with mTORC2.
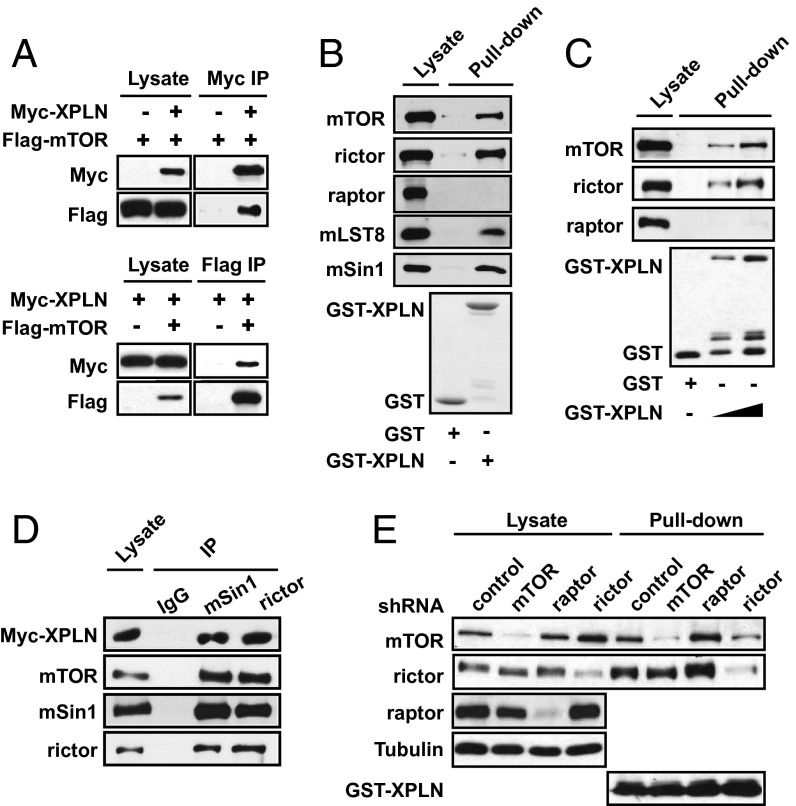
To explore interacting partners of mTOR, we carried out yeast two-hybrid screens using the C-terminal 1,188 amino acids of mTOR (a.a. 1362–2549) as bait against a HeLa cell cDNA prey library. XPLN cDNA emerged as a positive hit and was confirmed in secondary assays in yeast. The interaction between full-length mTOR and XPLN was then examined in mammalian cells. As shown in Fig. 1A, Flag-mTOR stably expressed in HEK293 cells coimmunoprecipitated with transiently expressed Myc-XPLN, and vice versa. In addition, bacterially purified GST-XPLN associated with endogenous mTOR in pull-down assays performed with both HEK293 (Fig. 1B) and mouse C2C12 cell lysates (Fig. 1C). Furthermore, GST-XPLN associated with endogenous rictor, and not raptor (Fig. 1 B and C), suggesting that the interaction may be specific for mTORC2. Indeed, both mSin1 and mLST8, the other components of mTORC2 (11), were found to associate with GST-XPLN (Fig. 1B), and immunoprecipitation of endogenous rictor and mSin1 brought down Myc-XPLN stably expressed in HEK293 cells (Fig. 1D).
Fig. 1.
XPLN interacts with mTORC2. (A) HEK293 cells stably expressing Flag-mTOR were transfected with Myc-XPLN. Anti-Myc or anti-Flag immunoprecipitation (IP) was followed by Western analysis. (B and C) GST pull-down assays were performed using purified GST-XPLN with GST as a negative control with HEK293 (B) or C2C12 (C) cell lysates and analyzed by Western blotting. Some degradation fragments were present in the GST-XPLN protein preparation. (D) Endogenous rictor and mSin1 were immunoprecipitated from HEK293 cells stably expressing Myc-XPLN, followed by Western analysis. (E) C2C12 cells were infected with lentiviruses expressing shRNAs for mTOR, raptor, rictor, or a scrambled sequence as control and then subjected to GST-XPLN pull-down assays.
The interaction between XPLN and mTOR complexes was further investigated in cells with lentivirus-delivered shRNA-mediated knockdown of mTOR, raptor, and rictor. Depletion of rictor significantly impaired the interaction between mTOR and XPLN, whereas removal of mTOR had no effect on rictor–XPLN interaction (Fig. 1E). On the other hand, raptor depletion slightly increased XPLN interaction with mTOR and rictor (Fig. 1E). Hence, the XPLN–mTOR interaction appears to be mediated by rictor, although we cannot rule out a possible involvement of mSin1, the presence of which is necessary for rictor association with mTOR (11). Our original two-hybrid result could be explained by the presence of orthologs of mTORC2 components in yeast (Avo3 and Avo1) (17, 18). Although the Avo3 (yeast rictor)-binding site (19) is in a region of TOR2 absent in our two-hybrid bait, the C-terminal 80 kDa of mTOR—included in the bait—binds rictor (20) and thus may also bind Avo3.
XPLN Negatively Regulates Akt and Cell Survival.
XPLN has been reported to stimulate the assembly of focal adhesions and stress fibers in a Rho-kinase–dependent manner (16). Because mTORC2 has been implicated in the regulation of actin cytoskeleton (12, 13), and in yeast TOR2 activates RHO1 and RHO2 through its GEF ROM2 (21), a plausible model would be that mTORC2 regulates GEF activity of XPLN toward RhoA proteins. However, knockdown of mTOR or rictor had no effect on RhoA-GTP levels in HEK293 cells, as assayed by pull-down of active RhoA with GST-RBD (Rho-binding domain of Rhotekin) (Fig. S1). RhoB expression was not detected in these cells. Although this lack of mTORC2 effect on RhoA did not completely rule out XPLN being a target of mTORC2 because of the existence of other RhoGEFs, we decided to examine the alternative possibility of XPLN being upstream of mTORC2. We used Akt as a readout because it was the best-characterized substrate of mTORC2.
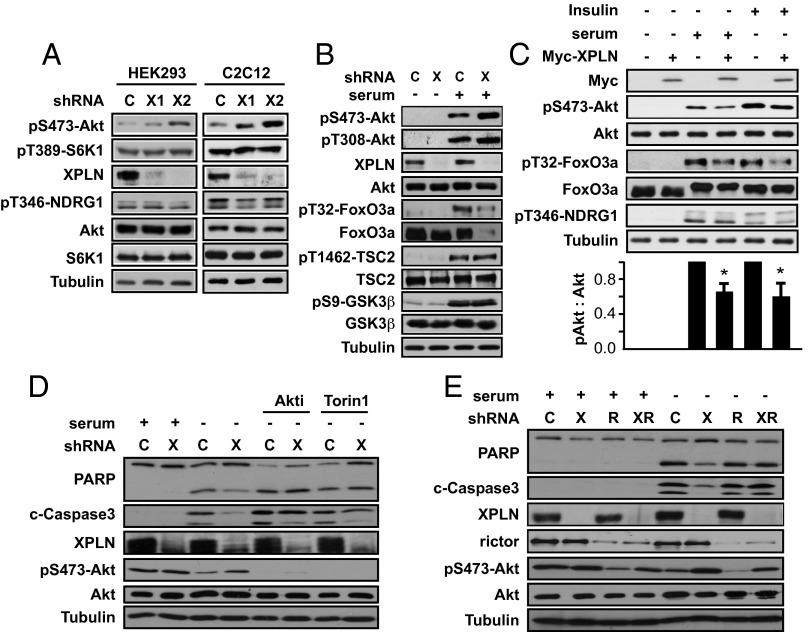
As shown in Fig. 2A, Akt phosphorylation on Ser473, the mTORC2 site, was markedly increased upon XPLN knockdown in both HEK293 and C2C12 cells, with two independent shRNAs for each cell line. On the other hand, XPLN knockdown did not affect the mTORC1 substrate S6K1 or the phosphorylation of NDRG1 (substrate of SGK1, another target of mTORC2) (Fig. 2A). The effect of XPLN depletion was observed on both steady-state and serum-stimulated pS473-Akt (Fig. 2 A and B). Phosphorylation of T308 on Akt was not markedly affected by XPLN knockdown (Fig. 2B), further confirming that XPLN acted through mTORC2. Several substrates of Akt are known to be differentially dependent on pS473-Akt; although FoxO3a phosphorylation requires pS473, phosphorylation of TSC2 and GSK3β can occur in the absence of pS473 (17, 22). Indeed, XPLN knockdown did not affect the levels of pT1462-TSC2 and pS9-GSK3β, both markedly stimulated by serum (Fig. 2B). The level of FoxO3a, on the other hand, was drastically reduced upon XPLN knockdown in serum-stimulated cells (Fig. 2B), most likely a consequence of enhanced phosphorylation by hyperactive Akt (23). Furthermore, overexpression of XPLN resulted in a modest, but nevertheless statistically significant, reduction of serum- and insulin-stimulated pSer473-Akt, with a similar effect on pT32-FoxO3a but no effect on pT346-NDRG1 (Fig. 2C). Taken together, these observations strongly suggest that XPLN is an endogenous inhibitor of Akt phosphorylation by mTORC2.
Fig. 2.
XPLN negatively regulates Akt phosphorylation and cell survival. (A) HEK293 or C2C12 cells were infected with lentiviruses expressing two independent XPLN shRNA (“X1,” “X2”) or a scrambled hairpin sequence as control (“C”), followed by Western analysis. Due to its lower abundance in HEK293 cells, XPLN was enriched by immunoprecipitation before Western blotting. (B) HEK293 cells were treated as in A and then serum-starved overnight, followed by stimulation with 10% (vol/vol) FBS for 30 min before Western analysis. (C) C2C12 cells were transfected with Myc-XPLN. After serum starvation overnight, the cells were stimulated with 10% (vol/vol) FBS or 100 nM insulin for 30 min followed by Western analysis. pS473-Akt and Akt bands were quantified by densitometry, and the relative ratios of pS473 versus total Akt were calculated with empty vector-transfected and stimulated samples designated as “1.” One-sample t test was performed to compare each data point to its respective control. *P < 0.05. (D) HeLa cells were infected with XPLN shRNA1 (“X”) or control and serum-starved for 48 h, followed by Western analysis. Some cells were treated with 1 µM Akti or 250 nM Torin1 for 3 h before cell lysis. (E) HeLa cells were infected with XPLN or rictor (“R”) shRNA or both and serum-starved for 48 h, followed by Western analysis.
Because one of the major functions of Akt is to support cell survival, at least partly through regulation of FoxO (24, 25), we tested whether XPLN might impact apoptosis. HeLa cells were serum-starved to induce apoptosis. As shown in Fig. 2D, XPLN knockdown decreased the levels of PARP cleavage as well as cleaved Caspase-3, both markers of apoptosis. Importantly, an Akt1/Akt2 inhibitor (Akti) and the mTOR kinase inhibitor Torin1 reversed the protective effect of XPLN depletion and enhanced apoptosis (Fig. 2D), suggesting that XPLN acts through Akt and mTOR. This is further confirmed by the reversal of the XPLN knockdown phenotype by the co-knockdown of rictor (Fig. 2E). It is noted that XPLN knockdown modestly enhanced pAkt even in rictor knockdown cells (Fig. 2E), most likely due to residual rictor protein especially in cells infected by both XPLN and rictor shRNA viruses, which had reduced rictor knockdown efficiency. In conclusion, XPLN negatively regulates cell survival by suppressing mTORC2 and Akt.
XPLN Regulation of Akt Is Independent of Its GEF Activity and Dependent on Its N Terminus.
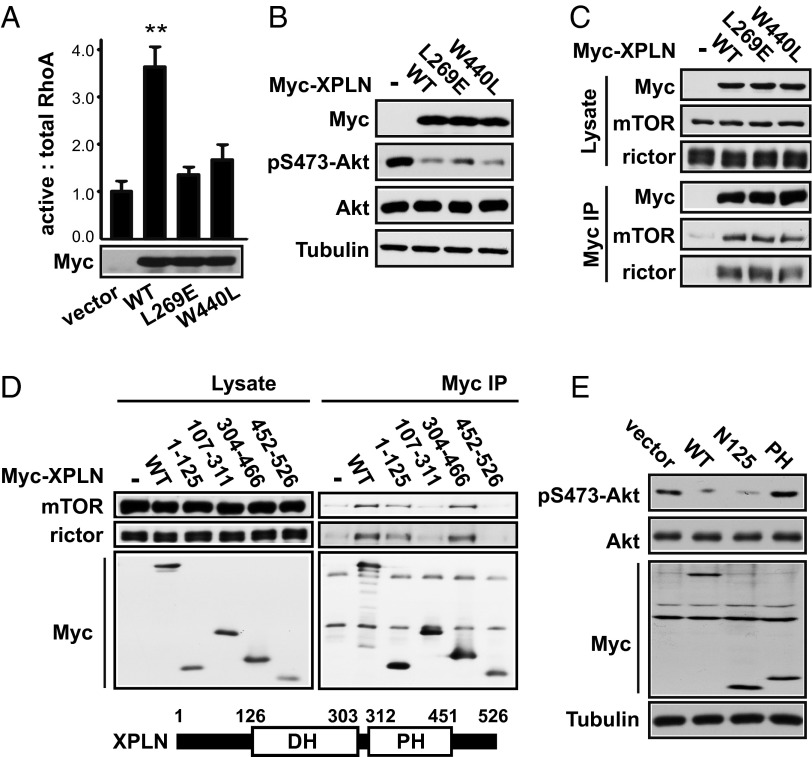
Because the only reported function of XPLN thus far is to act as a RhoGEF, we set out to test whether the GEF activity of XPLN was necessary for its regulation of Akt. It has been reported that two point mutations, L321E in the DH domain and W492L in the PH domain, are each sufficient to inactivate the GEF activity of NET1, the closest homolog of XPLN (26). We constructed analogous mutants of XPLN—L269E and W440L—and found that each mutation abolished the GEF activity of recombinant XPLN when overexpressed in cells (Fig. 3A). Strikingly, these XPLN mutants suppressed Akt phosphorylation in cells to the same degree as WT XPLN (Fig. 3B). Consistent with a GEF-independent, mTORC2-binding–dependent function of XPLN, both GEF-inactive XPLN mutants interacted with mTORC2 (Fig. 3C). These observations suggest that XPLN regulates phosphorylation of Akt in cells independently of its GEF activity.
Fig. 3.
XPLN regulation of Akt is independent of its GEF activity and dependent on its N terminus. (A) HEK293 cells were transfected with wild type, L269E, or W440L Myc-XPLN, followed by GST-RBD pull-down assays. The active RhoA (RhoA pulled down with GST-RBD) and total RhoA (RhoA in cell lysates) were quantified by densitometry. The ratios of active RhoA versus total RhoA were calculated and normalized against the control (empty vector). Paired t tests were performed to compare each data point to vector control. **P < 0.005. (B) C2C12 cells were transfected with various Myc-XPLN as indicated, followed by Western analysis. (C) HEK293 cells were transfected with various Myc-XPLN, followed by anti-Myc IP and then Western analysis. (D) HEK293 cells were transfected with fragments of Myc-XPLN as indicated, followed by anti-Myc IP and then Western analysis. (E) C2C12 cells were transfected with various Myc-XPLN fragments, followed by Western analysis.
To further examine the mechanism of XPLN action, we set out to map the region(s) of XPLN interacting with mTORC2. XPLN contains DH and PH domains with N- and C-terminal regions having no known sequence motifs. The DH domain of most GEFs confers the catalytic activity, whereas the PH domain may regulate protein localization by mediating protein–lipid or protein–protein interactions at least in some GEFs (27). As shown in Fig. 3D, the N-terminal region of XPLN (amino acids 1–125, designated N125) and the PH domain (amino acids 304–466) interacted with endogenous mTOR and rictor, suggesting that XPLN may have two independent binding sites for mTORC2. Importantly, overexpression of XPLN-N125 suppressed Akt phosphorylation to the same extent as overexpression of full-length XPLN (Fig. 3E). On the other hand, overexpression of the PH domain did not affect Akt (Fig. 3E) even though it interacted with rictor. Hence, we surmise that XPLN inhibits mTORC2 function via its N terminus interacting with rictor. A second interaction—between the PH domain and rictor—may serve to strengthen the interaction and enhance the inhibitory function of the N terminus of the full-length protein.
XPLN Inhibits mTORC2 Kinase Activity Toward Akt in Vitro.
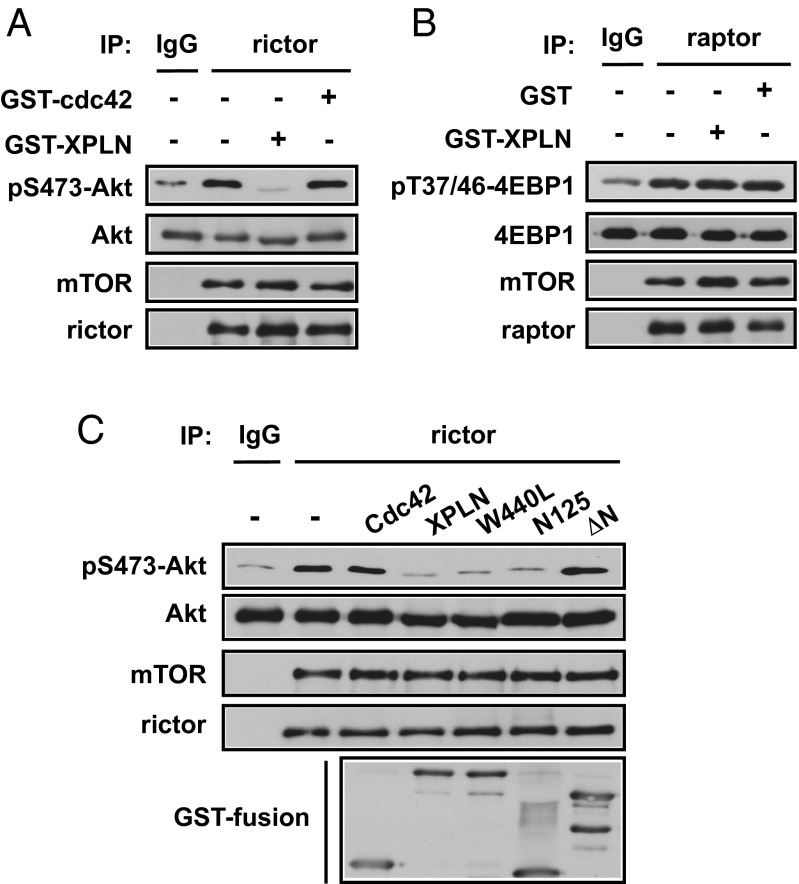
Given the physical interaction between XPLN and mTORC2, and the lack of detectable interaction between XPLN and Akt (Fig. S2), we reasoned that the simplest model explaining our observations thus far would be the inhibition of mTORC2 activity through XPLN binding. XPLN did not disrupt mTORC2 assembly, as overexpression of XPLN did not affect the amount of mTOR coimmunoprecipitated with rictor (Fig. S3). To examine if XPLN had a direct effect on mTORC2 kinase activity, we performed in vitro kinase assays with endogenous mTORC2 immunoprecipitated through rictor, using Akt as a substrate. Addition of bacterially purified GST-XPLN, but not GST-Cdc42, to the reaction markedly inhibited phosphorylation of Akt (Fig. 4A). At the same time, GST-XPLN had no effect on in vitro mTORC1 kinase activity using 4E-BP1 as a substrate (Fig. 4B).
Fig. 4.
XPLN inhibits mTORC2 phosphorylation of Akt in vitro. (A) Endogenous rictor was immunoprecipitated (IP) from HEK293 cells and subjected to in vitro kinase assays using His-Akt as the substrate and anti-pSer473 as the readout. Bacterially purified GST-XPLN or GST-Cdc42 (1 μg each) was added before kinase assays in the indicated samples. (B) Endogenous raptor was immunoprecipitated and subjected to in vitro kinase assays using GST-4EBP1 as the substrate and anti-pThr37/46 as the readout. GST-XPLN or GST (1 μg each) was added before kinase assays in the indicated samples. (C) Endogenous rictor immunoprecipitation and in vitro kinase assay were performed as described in A. Bacterially purified GST fusion protein of XPLN, W440L-XPLN, N125-XPLN, or ΔN-XPLN (1 μg each) was added before kinase assays.
Next, we examined XPLN mutants for their capacity to impact mTORC2 kinase activity. As shown in Fig. 4C, the GEF-inactive mutant, W440L, inhibited Akt phosphorylation in vitro as effectively as the WT XPLN. The N125 fragment was also inhibitory, whereas XPLN with the N-terminal 125 amino acids deleted (ΔN-XPLN) had no effect on the kinase activity (Fig. 4C), indicating that N125 is both necessary and sufficient for XPLN’s inhibition of mTORC2. Hence, in line with its regulation of Akt phosphorylation in cells, XPLN directly inhibits mTORC2 by a GEF-independent mechanism through its N terminus, most likely via physical interaction with the mTORC2 complex.
XPLN Negatively Regulates Myoblast Differentiation Through mTORC2 and Akt.
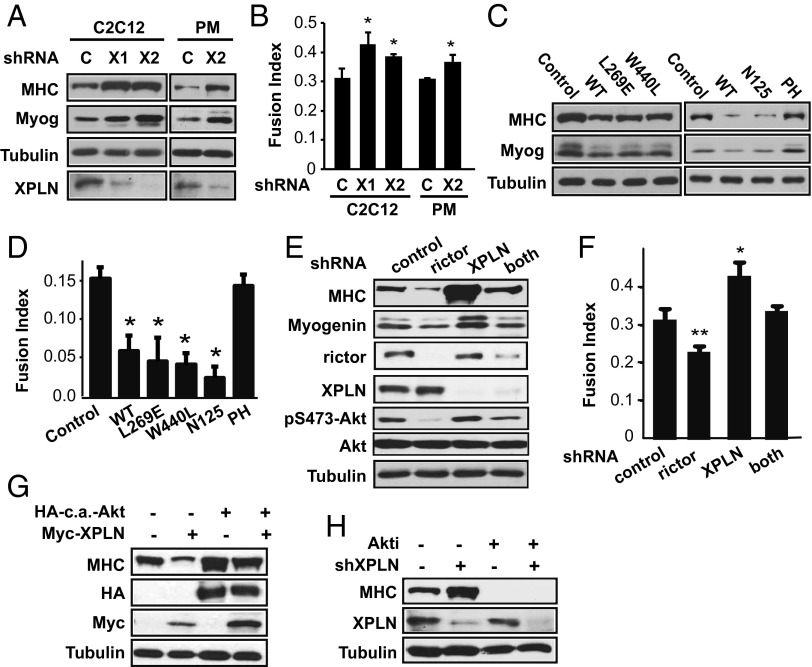
To further probe the biological relevance of this role of XPLN in regulating mTORC2 and Akt, we set out to examine a potential function of XPLN in myoblast differentiation because XPLN expression has been reported to be the highest in skeletal muscles among human tissues (16) and, additionally, Akt is a well-established regulator of skeletal myogenesis (28, 29). To that end, C2C12 myoblasts were induced to undergo myogenic differentiation by serum withdrawal. XPLN depletion led to a significant enhancement in myoblast differentiation compared with the control cells, as evidenced by elevated expression of the myogenic markers, myosin heavy chain (MHC) and myogenin (Fig. 5A), as well as increased fusion index (Fig. 5B). This XPLN knockdown phenotype was recapitulated in mouse primary myoblasts (Fig. 5 A and B). Meanwhile, XPLN overexpression decreased the degree of C2C12 differentiation (Fig. 5 C and D), which corroborated the knockdown phenotype. Interestingly, overexpression of the GEF-inactive mutants, as well as the N-terminal fragment of XPLN, had the same inhibitory effect on differentiation as WT XPLN (Fig. 5 C and D), indicating that the anti-myogenic function of XPLN is independent of its GEF activity and is conferred by its N terminus. This function closely correlates with the mode of XPLN action on Akt activity.
Fig. 5.
XPLN negatively regulates myoblast differentiation through mTORC2 and Akt. (A) C2C12 myoblasts and mouse primary myoblasts (PM) were infected with lentiviruses expressing XPLN shRNA (“X1,” “X2”) or the control hairpin (“C”). After differentiation for 3 d (C2C12) or 2 d (PM), the cells were subjected to Western analysis. (B) Cells as described in A were fixed and stained for MHC and DAPI and quantified for fusion index. (C) C2C12 cells were transfected with WT, mutants, or fragments of XPLN as indicated. After differentiation, the cells were subjected to Western analysis. (D) Cells as described in C were stained for MHC and DAPI and quantified for fusion index. (E) C2C12 cells were infected with lentiviruses expressing shRNA for XPLN, rictor, or both. After differentiation, the cells were subjected to Western analysis. (F) Cells as described in E were stained for MHC and DAPI and quantified for fusion index. (G) Cells were transfected with Myc-XPLN, c.a.-Akt, or both, followed by differentiation and then Western analysis. (H) Cells were infected with XPLN shRNA lentivirus, differentiated in 1 μM Akti or DMSO, followed by Western analysis. Paired t test was performed to compare each data point to control. *P < 0.05. **P < 0.01.
Furthermore, knockdown of rictor suppressed the increase in differentiation induced by XPLN knockdown (Fig. 5 E and F), confirming that XPLN acts through mTORC2. Similar to the observations in HeLa cells (Fig. 2E), co-knockdown led to less efficient depletion of rictor protein, which could explain the increased pAkt compared with rictor knockdown alone (Fig. 5E). The degree of differentiation correlated with the degree of pAkt (Fig. 5 E and F). In addition, a constitutively active (c.a.) Akt overcame the inhibition by overexpressed XPLN and restored differentiation (Fig. 5G). Conversely, Akti severely impaired differentiation in cells with XPLN knockdown (Fig. 5H). In aggregate, our observations strongly suggest that XPLN negatively regulates myoblast differentiation by inhibiting Akt activation through suppression of mTORC2.
Discussion
mTORC2 is critically involved in various cellular and developmental processes, but knowledge of its regulation has been scarce. Our studies have identified XPLN as a direct mTORC2 inhibitor in cells and in vitro. XPLN inhibits the kinase activity of mTORC2 toward Akt. This function of XPLN does not require its GEF activity and is most likely mediated by its direct interaction with mTORC2, revealing a noncanonical role of XPLN that is Rho-independent. Furthermore, we show that the endogenous XPLN negatively regulates cell survival and skeletal myoblast differentiation through inhibiting mTORC2 and Akt, validating the biological significance of this newly discovered regulatory mechanism. The only other mTORC2-specific inhibitor reported, GILZ, has been shown to inhibit mTORC2/Akt signaling when overexpressed in BCR-ABL–positive CML cells (15), but it is not known whether GILZ is an endogenous inhibitor of mTORC2 in normal physiological contexts.
Several RhoGEF proteins have been reported to have GEF activity-independent functions. For example, the GEF activity of Vav1 is not necessary for its ability to potentiate NF-AT activation in response to T-cell receptor signaling (30). Dbl binds and translocates Ezrin to the plasma membrane in a GEF-independent manner (31). The exact mechanisms by which such noncanonical functions are exerted by these GEFs are not clear, although protein–protein interactions mediated by modular domains outside of the catalytic region appear to be important. Interestingly, XPLN binds and inhibits mTORC2 via an N-terminal region that lacks sequence homology to any known modular domain. A second binding site is found in the PH domain of XPLN, but this domain is not sufficient to elicit an effect on mTORC2. It is possible that this additional interaction serves to increase the overall affinity between XPLN and mTORC2, in which case overexpression of the N terminus resulting in a high local concentration would be sufficient to exert an inhibitory effect without the need for the PH domain, as we have observed.
Interestingly, the action of XPLN not only is specific for mTORC2 but also may even be selective toward Akt. Another substrate of mTORC2, SGK1, is not regulated by XPLN in cells. This contrasts the observation with DEPTOR, which inhibits the phosphorylation of all mTORC1 and mTORC2 substrates tested (32). Overexpression of GILZ also inhibits all mTORC2 substrates in CML cells (15). It is possible that endogenous XPLN is localized in the cell where Akt, but not SGK1, is regulated. However, the lack of effect on SGK1 by XPLN overexpression (Fig. 2C), which would presumably override any requirement for subcellular localization, seems to argue against that possibility. An alternative mechanism is that XPLN binding to mTORC2 specifically blocks Akt as a substrate without affecting the other mTORC2 substrates. Future biochemical and structural studies should prove informative for the dissection of the exact mechanism by which XPLN inhibits mTORC2 phosphorylation of Akt.
Removal of XPLN inhibition alone is not sufficient to induce Akt phosphorylation in the absence of upstream stimuli when the basal activity of Akt is low. This is not surprising, as presumably activation of the kinase (mTORC2) requires positive inputs in addition to removal of XPLN suppression. Although very little is known about such inputs, PI3K activity and TSC1/2 have been reported to mediate mitogenic stimulation of mTORC2 kinase activity (33, 34). It is presently not known how or whether XPLN itself is regulated. Growth factor stimulation activates Akt in cells, but it does not affect XPLN levels (Fig. S4A) or the interaction between XPLN and mTORC2 (Fig. S4 B and C). DEPTOR is degraded by the proteasome in an mTOR-dependent fashion in response to serum stimulation, which forms a positive feedback loop to maximize mTOR activation (35–37). However, the slow kinetics of DEPTOR degradation (32) does not explain the well-known rapid activation of mTORC1 and mTORC2 substrates upon growth factor stimulation. It is possible that the inhibitors—DEPTOR and XPLN alike—may be overcome by a conformational change or modification of the kinase (mTORC2) without physical removal. In the case of XPLN in myoblast differentiation, there may be a simple mechanism of derepression: the level of XPLN does not change (Fig. S5), but mTOR levels increase drastically in the course of differentiation (38), which may allow mTORC2 to overcome XPLN stoichiometrically.
Akt regulates many physiological processes in addition to cell survival and myogenic differentiation, including cell proliferation, glucose metabolism, and other types of cellular differentiation. Regulation of Akt by XPLN in those processes warrants future investigation. A proto-oncogene, Akt is involved in tumorigenesis by promoting proliferation, survival, and motility of cancer cells (39). Evidence supports a direct role of mTORC2, at least partly through Akt, in driving tumorigenesis (1). Future examination of a role of XPLN in tumor suppression, especially in the context of hyperactive Akt, may prove informative for the understanding of and therapeutic strategy against cancer.
Materials and Methods
Antibodies and Other Reagents.
Rabbit polyclonal antibody against XPLN was generated by Proteintech Group Inc. using peptide REPQGETKLEQMDQSDSE as the antigen and affinity purified. All other antibodies were obtained from commercial sources, and the details are provided in SI Materials and Methods. All other reagents are also described in SI Materials and Methods.
Plasmids.
pCMV6-myristoylated-HA-Akt (c.a.-Akt) was previously described (40). pCMV-Myc-XPLN (human) and pGEX-4T-1-XPLN were generous gifts from Krister Wennerberg (University of Helsinki, Helsinki) (16). Various fragments and mutants of XPLN cDNA were generated by PCR or site-directed mutagenesis (Mutagenesis Kit, Stratagene).
Cell Culture.
The maintenance, differentiation, transfection, and lentiviral infection of HEK293 cells, HeLa cells, C2C12 myoblasts, and mouse primary myoblasts are described in detail in SI Materials and Methods.
Lentivirus-Mediated RNAi.
Lentivirus packaging and infection were performed as previously described (41). All shRNAs were from the MISSION TRC library (Sigma-Aldrich). The shRNAs for mouse mTOR, raptor, rictor, and negative control (scramble hairpin) have been reported (41). The clone identification numbers for XPLN shRNAs are provided in SI Materials and Methods.
Western Blotting, Immunoprecipitation, Purification of GST-Fusion Proteins, GST Pull-Down Assays, and in Vitro mTOR Kinase Assays.
These experimental procedures were performed as previously described (42) and also are provided in SI Materials and Methods.
RhoA Activity Assay.
GTP-bound RhoA was measured following the method described by Ren and Schwartz (43). Details are described in SI Materials and Methods.
Statistical Analysis.
All data are presented as mean ± SD, or representative blots, of at least three sets of independent experiments. Whenever necessary, statistical significance of the data was analyzed by performing one-sample or paired t tests.
Supplementary Material
Acknowledgments
We thank members of the J.C. laboratory for helpful discussions. This work was supported by National Institutes of Health Grants AR048914 and GM089771 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310434110/-/DCSupplemental.
References
- 1.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): Conducting the cellular signaling symphony. J Biol Chem. 2010;285(19):14071–14077. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 5.Facchinetti V, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27(14):1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan K-L. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27(14):1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144(5):757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Oh WJ, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29(23):3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge Y, Chen J. Mammalian target of rapamycin (mTOR) signaling network in skeletal myogenesis. J Biol Chem. 2012;287(52):43928–43935. doi: 10.1074/jbc.R112.406942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10(14):2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jacinto E, et al. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6(11):1122–1128. [DOI] [PubMed]
- 13.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 14.García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416(3):375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 15. Joha S, et al. (2012) GILZ inhibits the mTORC2/AKT pathway in BCR-ABL(+) cells. Oncogene. 31(11):1419–1430. [DOI] [PMC free article] [PubMed]
- 16.Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem. 2002;277(45):42964–42972. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- 17. Jacinto E, et al. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127(1):125–137. [DOI] [PubMed]
- 18.Frias MA, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16(18):1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 19. Wullschleger S, Loewith R, Oppliger W, Hall MN (2005) Molecular organization of target of rapamycin complex 2. J Biol Chem 280(35):30697–30704. [DOI] [PubMed]
- 20.Panasyuk G, et al. mTORbeta splicing isoform promotes cell proliferation and tumorigenesis. J Biol Chem. 2009;284(45):30807–30814. doi: 10.1074/jbc.M109.056085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88(4):531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 22.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 23. Plas DR, Thompson CB (2003) Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem 278(14):12361–12366. [DOI] [PubMed]
- 24.Lawlor MA, Alessi DR. PKB/Akt: A key mediator of cell proliferation, survival and insulin responses? J Cell Sci. 2001;114(Pt 16):2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- 25.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 26.Alberts AS, Treisman R. Activation of RhoA and SAPK/JNK signalling pathways by the RhoA-specific exchange factor mNET1. EMBO J. 1998;17(14):4075–4085. doi: 10.1093/emboj/17.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossman KL, Der CJ, Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 28.Jiang BH, Aoki M, Zheng JZ, Li J, Vogt PK. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc Natl Acad Sci USA. 1999;96(5):2077–2081. doi: 10.1073/pnas.96.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng XD, et al. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17(11):1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhne MR, Ku G, Weiss A. A guanine nucleotide exchange factor-independent function of Vav1 in transcriptional activation. J Biol Chem. 2000;275(3):2185–2190. doi: 10.1074/jbc.275.3.2185. [DOI] [PubMed] [Google Scholar]
- 31.Vanni C, et al. Phosphorylation-independent membrane relocalization of ezrin following association with Dbl in vivo. Oncogene. 2004;23(23):4098–4106. doi: 10.1038/sj.onc.1207509. [DOI] [PubMed] [Google Scholar]
- 32. Peterson TR, et al. (2009) DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137(5):873–886. [DOI] [PMC free article] [PubMed]
- 33.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28(12):4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gan X, Wang J, Su B, Wu D (2011) Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 286(13):10998–11002. [DOI] [PMC free article] [PubMed]
- 35.Duan S, et al. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell. 2011;44(2):317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao D, et al. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44(2):290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44(2):304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erbay E, Chen J. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J Biol Chem. 2001;276(39):36079–36082. doi: 10.1074/jbc.C100406200. [DOI] [PubMed] [Google Scholar]
- 39.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 40.Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003;163(5):931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge Y, Yoon MS, Chen J. Raptor and Rheb negatively regulate skeletal myogenesis through suppression of insulin receptor substrate 1 (IRS1) J Biol Chem. 2011;286(41):35675–35682. doi: 10.1074/jbc.M111.262881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon MS, Sun Y, Arauz E, Jiang Y, Chen J. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. J Biol Chem. 2011;286(34):29568–29574. doi: 10.1074/jbc.M111.262816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren XD, Schwartz MA. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/s0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.