Significance
Retinitis pigmentosa (RP) is a genetic disease that causes progressive blindness and that is caused by mutations in more than 50 genes. Conventional methods for identification of both RP mutations and novel RP genes involve the screening of DNA sequences spanning coding exons. In our work, we conversely test the use of whole genome sequencing, a technique that takes into account all variants from both the coding and noncoding regions of the human genome. In our approach, we identify a number of unique RP mutations, a previously undescribed disease gene, as well as pathogenic structural DNA rearrangements originating in introns.
Keywords: medical genetics, ophthalmology, ciliopathy, retinal blindness
Abstract
We performed whole genome sequencing in 16 unrelated patients with autosomal recessive retinitis pigmentosa (ARRP), a disease characterized by progressive retinal degeneration and caused by mutations in over 50 genes, in search of pathogenic DNA variants. Eight patients were from North America, whereas eight were Japanese, a population for which ARRP seems to have different genetic drivers. Using a specific workflow, we assessed both the coding and noncoding regions of the human genome, including the evaluation of highly polymorphic SNPs, structural and copy number variations, as well as 69 control genomes sequenced by the same procedures. We detected homozygous or compound heterozygous mutations in 7 genes associated with ARRP (USH2A, RDH12, CNGB1, EYS, PDE6B, DFNB31, and CERKL) in eight patients, three Japanese and five Americans. Fourteen of the 16 mutant alleles identified were previously unknown. Among these, there was a 2.3-kb deletion in USH2A and an inverted duplication of ∼446 kb in EYS, which would have likely escaped conventional screening techniques or exome sequencing. Moreover, in another Japanese patient, we identified a homozygous frameshift (p.L206fs), absent in more than 2,500 chromosomes from ethnically matched controls, in the ciliary gene NEK2, encoding a serine/threonine-protein kinase. Inactivation of this gene in zebrafish induced retinal photoreceptor defects that were rescued by human NEK2 mRNA. In addition to identifying a previously undescribed ARRP gene, our study highlights the importance of rare structural DNA variations in Mendelian diseases and advocates the need for screening approaches that transcend the analysis of the coding sequences of the human genome.
The identification of the genetic causes of rare Mendelian diseases is becoming increasingly important following some success with gene-based therapy, as recently reported for patients with a form of Leber congenital amaurosis (LCA), a severe autosomal recessive hereditary retinal dystrophy (1–3). The evidence that restoring a gene in the diseased retina could yield therapeutic effects has stimulated the pursuit of the genetic causes of other retinal dystrophies, including retinitis pigmentosa (RP).
RP is the name given to a group of hereditary retinal conditions in which degeneration of rod photoreceptors, responsible for vision under starlight or moonlight conditions, is more pronounced than that of cone photoreceptors, which mediate daylight vision. Individuals with RP typically experience night blindness at first, followed by progressive and unstoppable visual impairment in daytime conditions as well (4). Their visual fields become reduced gradually and sight is lost from the midperiphery to the periphery and then from the midperiphery to the center, resulting eventually in complete or near-complete blindness if left untreated. Most patients show intraretinal pigment in a bone spicule configuration around the fundus periphery, for which this condition was named. In addition, they typically show retinal arteriolar attenuation, elevated final dark adapted thresholds, and reduced and delayed electroretinograms (ERGs) (4). Vitamin A supplementation in combination with an omega-3 rich diet can slow the course of retinal degeneration and preserve visual acuity among adults with this condition (5, 6). Autosomal, recessively inherited RP (ARRP) is the most common form of hereditary retinal degeneration in humans. To date, over 50 genes have been associated with ARRP and allied disorders, among patients who are predominantly of European ancestry (RetNet; www.sph.uth.tmc.edu/retnet/home.htm). However, despite this high number of identified disease genes, ∼40–50% of all diagnosed cases have no mutations in recognized loci (7). Furthermore, genetic defects in RP are also population specific. For example, a screening of 193 unrelated Japanese patients with isolate or autosomal recessive RP for 30 disease genes identified commonly within North American or European patients revealed candidate pathogenic mutations in only 14% of the cohort (8).
Recent advances in massively parallel sequencing have enabled the analysis of large amounts of sequences (genes) at reasonable costs, revolutionizing the traditional approach of exon-by-exon Sanger sequencing (9). The two major forms of sequencing strategies allowing large-scale analyses are whole genome sequencing (WGS) and whole exome sequencing (WES). The former reads the entire genome with no distinction between exons and nonexonic regions. It allows the detection of intergenic variants, copy number variations (CNVs), and other structural rearrangements, as well as unrecognized exonic sequences. The latter technique relies on targeted DNA capture and focuses on the analysis of the known exonic content of the genome, performed according to the genomic annotation available at a given point in time.
In this study, we performed WGS as a method for mutation discovery in a highly genetically heterogeneous Mendelian disease; to this end, we evaluated 16 unrelated RP patients from diverse ethnic backgrounds.
Results
Genome Sequencing.
Genome sequencing in the 16 analyzed patients produced an average mapping yield of 200.8 ± 17.9 (mean ± SD) Gb and an average coverage of 66.1 ± 2.4 (mean ± SD) reads per base (SI Appendix, Table S1). This covered a genomic fraction of 0.968 ± 0.004, in which roughly 3.8 million putative variations were identified. Of these, ∼7.7% were not reported in dbSNP build 131 and were classified as novel variants. Variations present within transcripts were classified further as synonymous and nonsynonymous, and analyzed separately for the North American and Japanese sets of patients. Scoring of large structural variations (SVs) could be achieved only for seven genomes, as the remaining DNA samples, possibly because of their older age, did not produce reliable mate pair information (SI Appendix, Results S1).
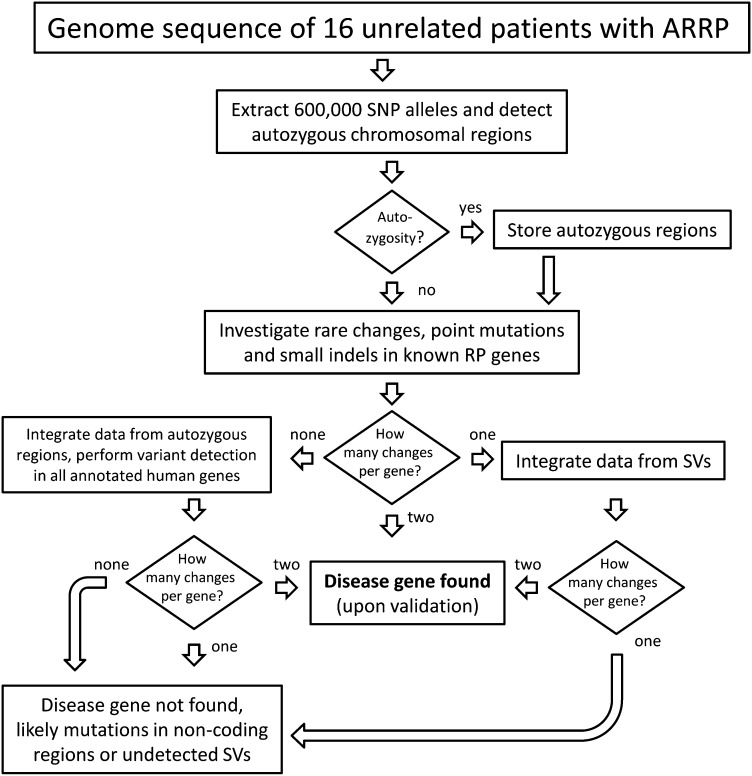
Assessment of pathogenic variants was performed by a series of filtering steps, summarized in Fig. 1.
Fig. 1.
Flowchart of the filtering process applied in this work.
Assessment of Autozygous Regions.
Each genome was evaluated for known or undocumented parental consanguinity as well as for possible founder mutation events by extracting genotypes of known polymorphic SNPs and by searching for long intervals with high degrees of homozygosity (at least 500 consecutive SNP markers, or ∼2.2 Mb on average), indicative of identity by descent (IBD). Significant genomic homozygosity was observed only in the five Japanese patients (individual IDs: R14, R15, R16, R18, and R19) who had documented parental consanguinity. The areas of IBD had essentially no overlap among these patients except for a 10-Mb interval on chromosome 1 shared by R15 and R19. Haplotype analysis indicated the shared intervals to be of different origins. No other patients carried genomic areas indicative of IBD; this was consistent with their family history reporting no parental consanguinity.
Sequence Analyses of Known RP Genes.
We first focused our analyses on genes known to be associated with ARRP. We investigated both small variants (from 1 to 50 bp) from the mapping of short reads and, whenever possible, large SVs. Our results are summarized in SI Appendix, Table S2; detailed results are provided in SI Appendix, Results S1, Figs. S1 and S3, and Table S3.
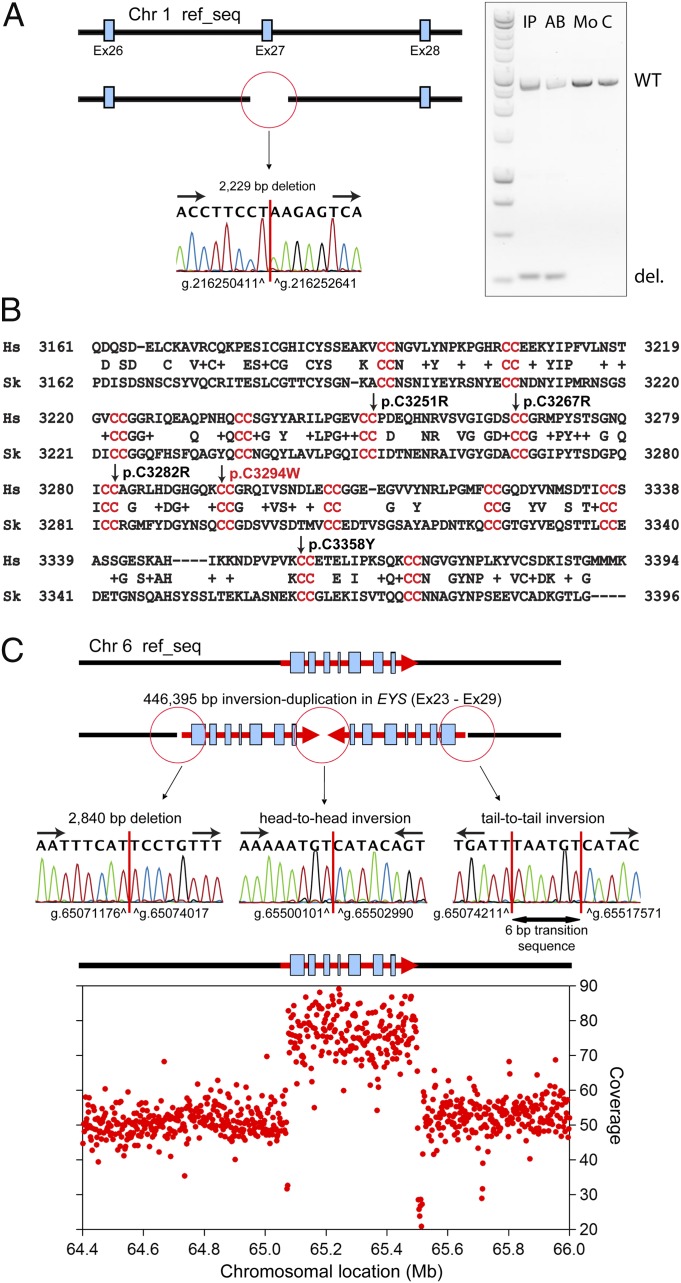
In addition to point mutations and short indels (insertion/deletions), we detected pathogenic SVs in USH2A and EYS in patients 003–019 and R9, respectively, by combining information from sequence coverage and abnormal junctions/mate pair distance. In the genome of patient 003–019, we identified a ∼2-kb deletion that removed exon 27 of USH2A, whereas patient R9 was found to carry a 446-kb head-to-head inverted duplication of the portion of chromosome 6 that included exons 23–29 of EYS (Fig. 2).
Fig. 2.
Pathogenic structural variations identified. (A) Sequence of the heterozygous USH2A 2,229-bp deletion in patient 003–019 (Left) and electrophoresis of the PCR fragments showing a smaller fragment carrying the deletion in the index patient (IP) and her affected brother (AB) but not in her mother (Mo) or a control DNA (C). del, deleted; WT, wild type. (B) Alignment of the USH2A protein from Homo sapiens (Hs) and Saccoglossus kowalevskii (Sk, acorn worm) showing the conservation of 13 CC repeat motifs (red) and the location of the mutation p.C3294W, newly identified in patient 003–019 and her sister. Four previously reported disease-associated missense changes (p.C3251R, p.C3267R, p.C3282R, and p.C3358Y) also affect neighboring CC repeats. (C) Schematic representation and DNA sequence of the junctions characterizing the chromosomal rearrangement detected in patient R9 and involving the EYS gene. Integration of the information obtained by Sanger DNA sequencing and WGS coverage of the region allows identifying an inverted duplication encompassing exons 23–29.
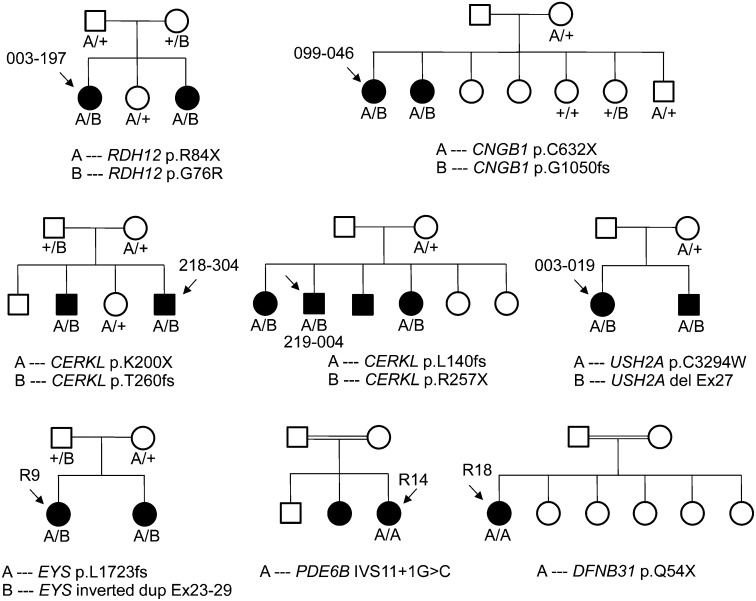
We found two pathogenic alleles, in either a homozygous or compound heterozygous state, in 8 of the 16 patients, 5 Americans and 3 Japanese, in seven different genes (SI Appendix, Table S2). Six patients carried mutations in one of the following genes: USH2A, RDH12, CNGB1, EYS, PDE6B, and DFNB31; 2 patients had mutations in CERKL. None of these mutations were found in the control cohorts of 95 healthy North American or 95 Japanese individuals. None of these mutations were reported previously, except p.R257X in CERKL and p.G76R in RDH12 (10, 11). All mutations cosegregated with RP as recessive, pathogenic alleles in all family members of the index patients for whom DNA samples were available (Fig. 3).
Fig. 3.
Cosegregation analyses. All mutations analyzed cosegregated with the disease according to an autosomal recessive pattern of inheritance.
Systematic Screening of All Genes.
Based on the data from the analysis of known RP genes, we adopted a pipeline to perform a systematic analysis targeting all annotated genes in the genomes of patients with unsolved genetic etiology (SI Appendix, Fig. S2). With the aim of selecting a restricted number of candidate genes, more aggressive filtering was adopted with respect to the one used for the screening of known disease genes. The major differences in the analytical pipeline included removal of all entries in dbSNP. We safely applied this filtering because, given the low frequency of individual mutations in ARRP genes (including undetected ones), the risk of eliminating pathogenic DNA variants that could be fortuitously included in dbSNP build 131 is negligible. Further, to validate this approach, we applied it again retrospectively to the genomes for which mutations in RP genes were already detected. All of identified RP mutations were present in the final list of variants, supporting the sensitivity of the strategy. Detailed results are provided in SI Appendix, Results S2 and are summarized in SI Appendix, Figs. S2 and S3 and Table S4.
In R19, in whom we did not find any clear-cut mutations in known ARRP genes, we found a homozygous frameshift variant (p.L206fs, c.617_624delTGTATGAGinsA) in the never in mitosis gene A (NIMA)-related kinase 2 (NEK2) gene. This variant was present within a highly homologous genomic stretch of 19.6 Mb of chromosome 1q32, predicted to be IBD (SI Appendix, Fig. S4). Similar to most frameshifts producing a premature termination codon, p.L206fs is predicted to result in an mRNA allele that is subject to nonsense-mediated mRNA decay, and therefore in no protein product. Targeted DNA screening revealed that c.617_624delTGTATGAGinsA was absent from 1,273 Japanese and 95 North American control individuals. The entire coding sequence of the NEK2 gene was then analyzed in a mixed cohort of 190 American patients with ARRP, in 64 Japanese patients with isolate RP, as well as in 13 patients found previously to show linkage between recessive retinal degeneration and the NEK2 region. However, other than known polymorphisms (rs1056729, rs12031285, and rs45623136), we found only a few isolated heterozygous missense variants (p.R26Q, c.77G>A; p.V137I, c.409G>A; p.I265V, c.793A>G; p.N189S, c.566A>G; and p.K103E, c.307A>G; none were present in dbSNP) insufficient to account for ARRP. Notably, an additional Japanese male with ARRP was found to carry the same frameshift variant p.L206fs, but heterozygously, with no other variants in the NEK2 coding sequence. This same patient (R51) was later found to carry the retinitis pigmentosa GTPase regulator (RPGR) mutation c.2405_2406delAG; p.E802fs (Human Gene Mutation Database entry: CD004115), described previously to be a sufficient cause of RP (12).
In light of a recent study reporting the involvement of noncoding RNA in the pathogenesis of retinal degeneration in mice (13), variants in noncoding RNA were also analyzed. After the removal of variants observed in 52 publicly available control genomes, only isolated heterozygous variants each with one entry per gene remained, insufficient to account for ARRP.
nek2 Inactivation and Rescue in Zebrafish.
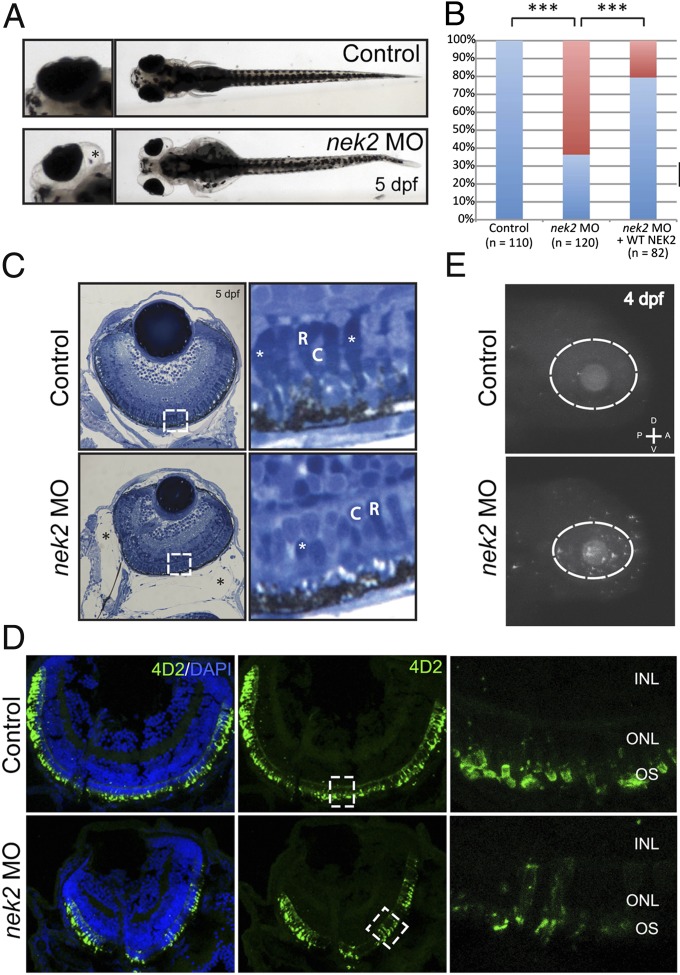
To validate the pathogenic role of NEK2 deficiency in RP, we suppressed the sole ortholog of NEK2 in zebrafish embryos and asked whether this manipulation might give rise to photoreceptor phenotypes. Upon injection of 6 ng of nek2 splice-blocking morpholino, we observed gross ocular defects, including microphthalmia and enlarged eye sockets in 5-d postfertilization (dpf) morphant (MO) embryos (Fig. 4A). Whereas 63% of MO embryos displayed such phenotypes, only 21% of embryos expressing both MO and wild-type human NEK2 mRNA did, suggesting that the ocular phenotypes are specific to the nek2 suppression (P < 0.001) (Fig. 4B).
Fig. 4.
In vivo functional evaluation of nek2 loss in zebrafish. (A) Bright-field representation of 5-dpf control and nek2 morphant zebrafish embryos. Magnified Insets highlight ocular phenotypes including microphthalmia and enlarged eye sockets (marked by the black asterisk). (B) Ocular phenotypes including microphthalmia and enlarged eye sockets vs. normal phenotypes (red bars and blue bars, respectively) are quantified in control and nek2 morphant embryos, as well as in morphant animals rescued with human WT NEK2 mRNA. Asterisks indicate statistically significant differences between groups (P < 0.001). (C) Histology of control and nek2 morphant embryos also show enlarged eye sockets (marked by black asterisks) and microphthalmia. Magnified Insets show a decrease in the number of photoreceptors with apparent changes in domains of condensed chromatin (white asterisks). C, cones; R, rods. (D) Immunohistochemical analyses of retinal cryosections from control and nek2 MO embryos, stained with DAPI (blue) and the 4D2 antibody against rhodopsin (green). Suppression of nek2 results in the depletion of rods and in the mislocalization of rod opsin from the outer segment (OS) of photoreceptors. INL, inner nuclear layer; ONL, outer nuclear layer. (E) TUNEL immunofluorescent images of 4-dpf embryos, showing an increase in the number of apoptotic cells in nek2 morphant embryos. The dotted ovals indicate the position of the eye. A, anterior; D, dorsal; P, posterior; V, ventral.
We next asked whether, in addition to overt structural abnormalities that may not directly inform the involvement of this gene to RP in humans, suppression of nek2 might also give rise to photoreceptor defects consistent with those of patients with ARRP. We therefore embedded and paraffin sectioned control and MO embryos. In addition to the small eye phenotype, we detected alterations in the photoreceptor layer. Specifically, after serial sectioning of 10–20 embryos injected with sham, MO, or MO + human NEK2 mRNA, we observed a persistent decrease in the number of photoreceptors with large central domains of condensed chromatin. This phenotype was seen in all nek2 MO embryos evaluated, but was absent from embryos injected with either sham or MO + human NEK2 mRNA, suggesting a loss of rod photoreceptors specific to the suppression of nek2 (Fig. 4C and SI Appendix, Fig. S5). To verify this observation, we used a rhodopsin (4D2) antibody to stain retinal cryosections from embryos injected with sham, MO, or MO + human NEK2 mRNA (Fig. 4D and SI Appendix, Fig. S5). Immunohistochemical analyses of cross-sections from each condition demonstrated that the suppression of nek2 resulted in the depletion of ∼24% of 4D2-positive rod photoreceptors. In addition, mislocalization of rod opsin throughout the photoreceptor cells was evident in the central retina of nek2 MO specimens, consistent with the hypothesis that nek2 is required for the appropriate trafficking of rhodopsin to the outer segments (Fig. 4D).
Further, to ask whether apoptosis, a major mechanism of photoreceptor loss in most known forms of RP (14), might account for some of the observed loss of photoreceptors, we performed TUNEL analysis. Masked scoring of embryos (∼50 embryos per injection mixture) revealed a sevenfold increase in the number of TUNEL-positive cells in the eye and head region of nek2 morphant embryos. By sharp contrast, we did not observe more than 1–10 TUNEL-positive cells in embryos injected with MO + NEK2 mRNA (Fig. 4E).
Finally, we were intrigued by the discovery of a heterozygous frameshift variant p.L206fs in NEK2 and the bona fide RPGR mutation p.E802fs in a patient with RP. We therefore asked whether the RPGR variant may interact genetically with the NEK2 locus. To test this possibility, we coinjected subeffective doses of the nek2 MO and rpgr MO and compared embryos with single or double MO (n > 100 at subeffective doses). Approximately 28% of embryos carrying subeffective doses of both nek2 and rpgr MO revealed ocular (P < 0.001) and rod photoreceptor phenotypes (serial sectioning of 10 embryos per genotype) that exceeded the number of affected embryos induced by either nek2 (3%) or rpgr (10%) MO alone, suggesting that the RPGR allele interacts in trans with the NEK2 locus to exacerbate photoreceptor defects (SI Appendix, Fig. S6).
Discussion
Massively parallel sequencing has proven to have a high potential to detect mutations in patients with rare Mendelian diseases (15). To date, most reports focus on monogenic conditions with no genetic heterogeneity, for which mutations can be recognized from benign variants since they invariantly affect the same gene in different patients.
In this study, we explored the efficacy of WGS in identifying mutations in unrelated patients from diverse ethnic backgrounds and presenting with a disease that is clinically the same but that has different genetic drivers. Whereas the small number of genomes analyzed in this study precludes an accurate analysis of quantitative measures, such as sensitivity of the WGS to detect mutations in known RP genes, we observed a few features that allowed us to make some valid comparisons between the different techniques currently available for genetic diagnosis. First, the majority of the pathogenic mutations identified were never reported before. This implies that tools that rely on systematic search for known pathogenic variants, both via mutation-centered resequencing and chip-based hybridization, may not be adequate for ARRP. Second, thanks to full-genome data, we detected complex structural variants whose junctions were located deep in noncoding regions. Because of their nature, these disease-causing variants would have been invisible to standard screening methods, or even to WES. Coverage-based analysis of CNV in exome sequencing has been attempted, with variable results. Limitations of this approach include the uneven efficiency of target DNA capture (and hence sequence coverage, on which assessment of number of copies is based) over different probes and, above all, the low probability of detecting junctions defining the SVs, which are more likely to be found in the nonexonic sequences composing ∼98% of our genome. Unambiguous detection of abnormal junctions and mate pair information are crucial parameters in defining a SV; for instance, they allow distinguishing a tandem duplication from an inverted one. Third, because we had access to the full wealth of genomic information, we could integrate many sources of information at once (e.g., SNP genotypes, phasing, etc.) that allowed us to accurately filter DNA variants that were related to the disease.
Genetic defects in EYS were proposed recently to be one of the major causes of ARRP in the Japanese population (16). We found that one of the pathogenic EYS alleles was a large SV (446 kb) with a complex genomic rearrangement. This finding supports the notion that SVs represent frequent pathogenic mutations in this gene (17). A homozygous nonsense mutation in exon 6 of DFNB31 was identified in R18, a patient with nonsyndromic ARRP. The DFNB31 gene encodes whirlin, a PDZ scaffold protein with expression in both hair cell stereocilia and retinal photoreceptor cells. Whirlin binds to the protein encoded by USH2A (18), a gene associated with both Usher syndrome type II (ARRP accompanied by hearing loss) and nonsyndromic ARRP (19). Whereas mutations in DFNB31 have been reported as rare causes of Usher syndrome type II (20, 21), no DNA changes in its sequence have yet been associated with nonsyndromic ARRP. However, at the age of 66, the past medical history of this patient was significant for only hyperlipidemia and she did not report any hearing loss. We could not perform an auditory examination because she was no longer reachable.
In patients from consanguineous families, regions of IBD allowed restricting the search for pathogenic mutations to only a fraction of the genome. However, these same regions were susceptible to carrying other rare but nonpathogenic homozygous changes as well. Indeed, a higher number of candidate genes/mutations remained among Japanese patients with parental consanguinity compared with those without it (SI Appendix, Table S4). These results suggest that even if the analysis should be restricted to areas of IBD, genomes with high homozygosity do not necessarily offer an extra advantage in mutation detection, when comprehensive genomic sequencing in single individuals is performed.
In three patients we identified clear-cut pathogenic but heterozygous mutations in known ARRP genes that could not be associated directly with the disease. This was particularly evident for patient R14, who carried a heterozygous frameshift in DFNB31 but was also homozygous for a mutation inactivating PDE6B (22). These findings are not surprising, given the elevated number of recessive ARRP mutations that are predicted to be present in the general population. Based both on theoretical assessments and on experimental data from control cohorts, we estimated that 1 in 3–7 individuals could be potential heterozygous carriers of an ARRP mutation (23, 24) or, as in the present case, 3 in 16.
The reasons why no candidate mutations of similar quality (i.e., two mutations, at least one of them being clearly deleterious in nature) to those revealed in known RP genes was uncovered in most of the unresolved genomes are unknown. Explanations for this observation may include the presence of variants or SVs that were undetected because of problems inherent in the mapping or sequencing procedure, or of less obvious pathogenic changes that alter splicing or transcription. These would include variants located in introns or in promoter regions, synonymous changes, or changes lying within important yet unannotated exons, genes, or genetic elements that have not been explored in the current study. Diseases caused by oligogenic modes of inheritance, or perhaps attributable to missense mutations for which efficient prioritization is difficult, is another possible explanation. De novo mutations in unknown dominant RP genes could also be evoked.
The search for mutations in unknown disease-causing genes revealed a number of genes with two nonsynonymous changes, which were mostly previously undescribed missenses. Application of more stringent filtering criteria by imposing the presence of at least one deleterious mutation followed by targeted annotation highlighted a single candidate, NEK2, in a Japanese patient who carried a homozygous frameshift in this gene. The serine/threonine-protein kinase NEK2 is known to play an important role in regulation of cell cycle progression through localization to the centrosomes and interaction with microtubules (25). The identified frameshift would result either in the creation of premature stop codon yielding a null allele or (less likely) a truncated protein lacking kinase activity and loss of microtubule binding. Importantly, defects in members of the Nek kinase family have been linked to impaired ciliogenesis and polycystic kidney disease (26). Recently, a role for Nek2 in the left–right patterning of vital organs (a phenotype associated with ciliary function) was established in Xenopus laevis (27). In the same work, in situ hybridization revealed the expression of nek2 transcripts in the eye (27). Furthermore, because NEK2 interacts with and can phosphorylate rootletin, a component of photoreceptor cilia (28, 29), NEK2 was considered to be an important candidate for ARRP.
Our zebrafish studies showed that lack of Nek2 induces microphthalmia as a gross morphological phenotype. More importantly, in nek2 morphants, we observed mistrafficking of rhodopsin, a hallmark of photoreceptor disease (30), and a reduced number of rod photoreceptors, likely via apoptotic processes. These phenotypes were rescued by injection of wild-type human NEK2 mRNA, validating the specificity of the induced defects. Microphthalmia is a phenotype that is difficult to interpret in the present context but that is not uncommon to zebrafish models of RP (31, 32). Meanwhile, photoreceptor death, mistrafficking of rhodospsin, and reduction of the outer retinal layers are classical features of RP in both patients and animal models (7, 14, 33). Indeed, no microphthalmia was noted in patient R19.
Intriguingly, the NEK2 frameshift identified in R19 was also present in R51, another patient with RP who had a deleterious mutation in RPGR. As the RPGR mutation in itself could explain the disease, an obvious question was whether the NEK2 mutation might in fact represent a common benign allele. We therefore searched for this variant in 1,273 control Japanese individuals and found that none carried it (allele frequency <3.9 × 10−4). The p.L206fs mutation in NEK2 is therefore exceedingly rare, such that its presence in a homozygous state in a patient is a strong argument in favor of its being an uncommon cause for ARRP. Although it is possible to attribute the presence of both NEK2 and RPGR mutations in R51 to chance, a more parsimonious explanation is that mutations in these two genes, both expressed in the connecting cilium, act synergistically to define a severe RP phenotype, due to the established principles of mutational load and oligogenic interactions of pathogenic alleles (34). In turn, this would increase the likelihood for the patient of being examined at earlier ages and analyzed genetically. Multiple genetic modifier genes have been reported for cilia-encoding genes and especially for RPGR (35). These modifiers may account in part for the wide phenotypic spectrum associated with genetic defects in this gene, ranging from localized macular atrophy to retinitis pigmentosa of variable severity. To investigate the possibility of the cooperative effect between deficiencies in these two ciliary genes, we performed in vivo genetic interaction studies and showed that loss of Rpgr function can exacerbate Nek2 ocular phenotypes, including defects comprising the trapping of rhodopsin in the inner segment. Taken together, our genetic and functional data indicate that NEK2 is a disease gene and that the retinal phenotype that results from its deficiency may represent a newly recognized ciliopathy.
To date, WGS has not been as widely explored as WES in the context of mutation detection. This can be attributed mainly to cost-related issues, because WGS is at least twice as expensive as WES procedures ensuring the same average coverage. We believe that the additional features displayed by WGS are worth the difference in price; however, this is a rather subjective matter that also depends on the disease that is being investigated. In the present case, WGS was essential to identify two pathogenic structural variations originating in introns. This is a significant finding, considering that only seven genomes could undergo SV analysis. Therefore, as a general rule, WGS is probably the strategy of choice when detection of structural variants or mutations in noncoding regions represents an important element of investigation. In the long term, considering that costs associated with massively parallel sequencing technology is expected to fall further and that analysis pipelines continue to evolve, it is probable that WGS would be just as workable economically and physically as WES. Limitations of WGS include the requirement of high-quality DNA to explore the full leverage of the mate-pair mapping and the lack of reliable pipelines to detect SVs ranging in size from 50 to a few hundred base pairs. Unexpectedly, the difficulty accompanied by handling the large amount of data produced by WGS was not a significant obstacle, given the power of desktop computers presently available on the market. Whereas samples with suitable quality could be obtained through careful preparation of fresh DNA samples, under detection of SVs may be a more problematic issue to solve. This occurs because current mapping is based on two steps: mapping of the short reads aimed at detecting variations between 1 and 50 bp and mate-pair mapping for detection of SVs larger than a few hundred bases; to our knowledge, a solution that could fill the gap between these two mapping approaches remains to be found.
In conclusion, in this study we identified clear-cut causative mutations among the overwhelming number of DNA variants present in the human genome, in single patients from genetically diverse populations. This happened without ambiguities in a highly heterogeneous disease, ARRP, and in more than 50% of the individuals analyzed. Furthermore, two cases presented mutations involving noncoding parts of the genome. Considering that the majority of patients referred for molecular genetics diagnosis are isolated individuals, our results are relevant not only to basic research, but also to future clinical genetic testing.
Methods
Our research protocol involving humans and animals was approved by the institutional review boards of our respective universities and organizations. Written informed consent for providing medical information and blood samples was obtained from each patient. Experimental procedures are described in detail in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Anna M. Siemiatkowska and Frans P. M. Cremers for sharing material from a person with RP, Adriana Ransijn for technical help, as well as Andrea Superti-Furga, and Luisa Bonafé for fruitful suggestions. Data storage was ensured by the Vital-IT Center for high-performance computing of the Swiss Institute of Bioinformatics. This work was supported by the Swiss National Science Foundation (Grant 310030_138346) and the Gebert Rüf Foundation, Switzerland (Rare Diseases-New Technologies Grant) (both to C.R.); a Center Grant from the Foundation Fighting Blindness (to E.L.B.); National Institutes of Health Grants DK072301 and MH-084018 (to N.K.); Ministry of Health, Labor and Welfare (MHLW) of Japan [Grant 23300101 (to S.I. and N. Matsumoto) and Grant 23300201 (to S.I.)]; MHLW, the Japan Science and Technology Agency, and the Strategic Research Program for Brain Sciences (N. Matsumoto); and a Grant-in-Aid for Scientific Research on Innovative Areas (transcription cycle) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Takeda Science Foundation (to N. Matsumoto).
Footnotes
Conflict of interest statement: R.G.T. is an employee and shareholder of Complete Genomics, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308243110/-/DCSupplemental.
References
- 1.Maguire AM, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainbridge JW, et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N Engl J Med. 2008;358(21):2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 3.Cideciyan AV, et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Natl Acad Sci USA. 2008;105(39):15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34(5):1659–1676. [PubMed] [Google Scholar]
- 5.Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Willett WC. ω-3 intake and visual acuity in patients with retinitis pigmentosa receiving vitamin A. Arch Ophthalmol. 2012;130(6):707–711. doi: 10.1001/archophthalmol.2011.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berson EL, et al. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch Ophthalmol. 1993;111(6):761–772. doi: 10.1001/archopht.1993.01090060049022. [DOI] [PubMed] [Google Scholar]
- 7.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 8.Jin ZB, et al. Identifying pathogenic genetic background of simplex or multiplex retinitis pigmentosa patients: A large scale mutation screening study. J Med Genet. 2008;45(7):465–472. doi: 10.1136/jmg.2007.056416. [DOI] [PubMed] [Google Scholar]
- 9.Tucker T, Marra M, Friedman JM. Massively parallel sequencing: The next big thing in genetic medicine. Am J Hum Genet. 2009;85(2):142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuson M, Marfany G, Gonzàlez-Duarte R. Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26) Am J Hum Genet. 2004;74(1):128–138. doi: 10.1086/381055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldahmesh MA, et al. Molecular characterization of retinitis pigmentosa in Saudi Arabia. Mol Vis. 2009;15:2464–2469. [PMC free article] [PubMed] [Google Scholar]
- 12.Vervoort R, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25(4):462–466. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 13.Sanuki R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 14.Cottet S, Schorderet DF. Mechanisms of apoptosis in retinitis pigmentosa. Curr Mol Med. 2009;9(3):375–383. doi: 10.2174/156652409787847155. [DOI] [PubMed] [Google Scholar]
- 15.Rabbani B, Mahdieh N, Hosomichi K, Nakaoka H, Inoue I. Next-generation sequencing: Impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet. 2012;57(10):621–632. doi: 10.1038/jhg.2012.91. [DOI] [PubMed] [Google Scholar]
- 16.Hosono K, et al. Two novel mutations in the EYS gene are possible major causes of autosomal recessive retinitis pigmentosa in the Japanese population. PLoS ONE. 2012;7(2):e31036. doi: 10.1371/journal.pone.0031036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieras JI, et al. Copy-number variations in EYS: A significant event in the appearance of arRP. Invest Ophthalmol Vis Sci. 2011;52(8):5625–5631. doi: 10.1167/iovs.11-7292. [DOI] [PubMed] [Google Scholar]
- 18.van Wijk E, et al. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet. 2006;15(5):751–765. doi: 10.1093/hmg/ddi490. [DOI] [PubMed] [Google Scholar]
- 19.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Missense mutation in the USH2A gene: Association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet. 2000;66(6):1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebermann I, et al. A novel gene for Usher syndrome type 2: Mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet. 2007;121(2):203–211. doi: 10.1007/s00439-006-0304-0. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, et al. Ablation of whirlin long isoform disrupts the USH2 protein complex and causes vision and hearing loss. PLoS Genet. 2010;6(5):e1000955. doi: 10.1371/journal.pgen.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaughlin ME, Sandberg MA, Berson EL, Dryja TP. Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat Genet. 1993;4(2):130–134. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- 23.Rivolta C, Sharon D, DeAngelis MM, Dryja TP. Retinitis pigmentosa and allied diseases: Numerous diseases, genes, and inheritance patterns. Hum Mol Genet. 2002;11(10):1219–1227. doi: 10.1093/hmg/11.10.1219. [DOI] [PubMed] [Google Scholar]
- 24.Nishiguchi KM, Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS ONE. 2012;7(7):e41902. doi: 10.1371/journal.pone.0041902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17(2):470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quarmby LM, Mahjoub MR. Caught Nek-ing: Cilia and centrioles. J Cell Sci. 2005;118(Pt 22):5161–5169. doi: 10.1242/jcs.02681. [DOI] [PubMed] [Google Scholar]
- 27.Fakhro KA, et al. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108(7):2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171(1):27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol. 2002;159(3):431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollingsworth TJ, Gross AK. Defective trafficking of rhodopsin and its role in retinal degenerations. Int Rev Cell Mol Biol. 2012;293:1–44. doi: 10.1016/B978-0-12-394304-0.00006-3. [DOI] [PubMed] [Google Scholar]
- 31.Luo N, Lu J, Sun Y. Evidence of a role of inositol polyphosphate 5-phosphatase INPP5E in cilia formation in zebrafish. Vision Res. 2012;75:98–107. doi: 10.1016/j.visres.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patil SB, Hurd TW, Ghosh AK, Murga-Zamalloa CA, Khanna H. Functional analysis of retinitis pigmentosa 2 (RP2) protein reveals variable pathogenic potential of disease-associated missense variants. PLoS ONE. 2011;6(6):e21379. doi: 10.1371/journal.pone.0021379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang GQ, Hao Y, Wong F. Apoptosis: Final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron. 1993;11(4):595–605. doi: 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- 34.Davis EE, Katsanis N. The ciliopathies: A transitional model into systems biology of human genetic disease. Curr Opin Genet Dev. 2012;22(3):290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahim AT, et al. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS ONE. 2011;6(8):e23021. doi: 10.1371/journal.pone.0023021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.