Abstract
Adolescent idiopathic scoliosis (AIS) is one of the most common orthopedic disorders, affecting up to 4% of schoolchildren worldwide. We studied seven unrelated multiplex families of southern Chinese descent with AIS, consisting of 25 affected members. A genomewide scan with >400 fluorescent microsatellite markers was performed. Multipoint linkage analysis by GENEHUNTER revealed significant linkage of the abnormal phenotype to the distal short arm of chromosome 19, with both a maximum multipoint LOD score and a nonparametric LOD score of 4.93. Two-point linkage analysis by MLINK gave a LOD score of 3.63 (recombination fraction θ[m=f]=0.00) at D19S216. Further high-density mapping and informative recombinations defined the AIS critical region in the vicinity of D19S216, flanked by D19S894 and D19S1034, spanning 5.2 cM on the sex-averaged genetic map on chromosome 19p13.3.
Scoliosis is the most common form of spinal deformity. It can be either idiopathic or secondary to congenital abnormality or to neuromuscular, connective tissue, or bone disorders (Lonstein 1995). Adolescent idiopathic scoliosis (AIS), which has no known etiology and occurs in otherwise healthy adolescents, accounts for ∼80% of idiopathic scoliosis (IS [MIM 181800]). Its incidence among schoolchildren is 0.15%–4% (Lonstein 1995; Tse 1997; Reamy and Slakey 2001), with a third of cases (i.e., 5 per 10,000 schoolchildren) requiring immediate treatment with bracing or surgery (Lonstein 1995; Reamy and Slakey 2001). The identification of the disease gene or disease-susceptibility genes and the unraveling of the underlying defect may offer hopes for other therapeutic options.
AIS can be diagnosed clinically, by direct observation of the back or by the forward-bending Adams' test (Adams 1865). The extent of the curvature may be measured by calculation of the Cobb’s angle, on radiographic evaluation (Cobb 1948). If left untreated, AIS can result in cosmetic problems and can cause lower back pain or deformity, and pulmonary function can be impaired in advanced cases, requiring mechanical ventilation. For the early and milder cases, bracing may prevent further progression during pubertal growth. For severe cases, surgical correction of their spinal curve, with its inherent risks (Newton et al. 1977; Luk et al. 1996), is the only choice. However, the correction rate after surgery is only 50%–60% (Liu and Huang 1996).
The development of scoliosis may be due to the relative difference in length or growth rate between the anterior and posterior portions of the spinal column (Murray and Bulstrode 1996). This may explain why adolescents with a particularly high growth rate during puberty are more prone to develop scoliosis. Familial occurrences of IS have been noted (Garland 1934; Wynne-Davis 1968; Riseborough and Wynne-Davis 1973; Harrington 1977), and concordance for this condition in MZ twins further strengthens the case for a hereditary basis of the disease (Kesling and Reinker 1997). However, controversy exists as to the mode of inheritance; on the basis of surveys mostly conducted by questionnaire and/or physical examination, it has been proposed to be a multifactorial or complex trait (Fisher and De George 1967; Wynne-Davis 1968), an autosomal dominant trait (Garland 1934; Bell and Teebi 1995), or even an X-linked dominant trait (Cowell et al. 1972). With the advances that have been made in molecular genetics, it is possible to perform a genomewide search for the genetic locus/loci. In a study of four multiplex families with IS, Wise et al. (2000), using nonparametric linkage analysis, found evidence of allele sharing at three loci (chromosomes 6p, 10q, and 18q) in the affected individuals from two of these families. In the present article, we report our finding of a genetic locus responsible for AIS, mapped to chromosome 19p13 by linkage analysis.
We studied seven unrelated, multiplex families with AIS in Hong Kong, all originating from the Guangdong province of southern China. These families were identified through probands with severe AIS requiring surgery or bracing. All subjects, male and female, were ascertained by the same criteria. They were examined by the Adams' test and by a standing upright radiograph of the spine. The x-rays were read by two independent orthopedic surgeons who were blinded to the clinical state of the subjects. The Cobb’s angle was measured, and AIS was defined as >10° lateral curvature of the spine. Individuals >12 years of age and having a normal Adams' test and x-ray results were considered to be unaffected. Unaffected individuals <12 years of age were classified as “phenotype unknown” for the purpose of linkage analysis. From the seven kindreds, 25 affected members (5 male and 20 female), 25 unaffected members, and 2 members with phenotype unknown were available for study. Among the affected members, six had received surgical correction of spinal curvature and three had bracing (table 1). All phenotypes were assigned prospectively before genotyping. Each participating subject—or, in the case of minors, the responsible adult—gave informed consent as approved by the Ethics Committee of the Faculty of Medicine at the University of Hong Kong.
Table 1.
Clinical Data of Probands of the Families with AIS
| Family | Sex | Age atPresentation(years) | Location ofPrimary Curvea | Cobb’s Angleat Diagnosisb | SpinalSurgery | Bracing |
| 3 | F | 11 | T6–T11 | 55° | Yes | No |
| 8 | F | 13 | T10–L3 | 26° | No | Yes |
| 12 | F | 14 | T3–T10 | 42° | Yes | No |
| 13 | M | 11.5 | T5–T12 | 42° | No | Yes |
| 14 | F | 10 | T5–T11 | 20°c | Yes | Yes |
| 15 | F | 14 | T9–L3 | 48° | Yes | No |
| 18 | F | 10 | T5–T11 | 22° | No | Yes |
T = thoracic vertebra; L = lumbar vertebra.
A higher Cobb's angle value indicates a greater severity of AIS.
Curve continued to progress despite bracing, and surgery was performed when the Cobb’s angle reached 50°.
Genomic DNA, extracted from peripheral blood leukocytes and Epstein-Barr virus–transformed lymphoblastoid cell lines, was used for genotyping. Four hundred fluorescein-labeled microsatellite markers distributed at ∼10-cM intervals throughout the entire human genome (medium-density markers; ABI) were PCR amplified under conditions recommended by the manufacturer, and the products were analyzed on an automated sequencer (model 3700; ABI). Allele sizing and scoring were performed, with Genescan version 3.7 (ABI) software, by at least two persons unaware of the subjects’ clinical status at the time of scoring. Cyrillic version 2.1 (Cherwell Scientific) software was used for assignment of haplotypes.
Linkage analyses were performed, using parametric and nonparametric methods, by use of GENEHUNTER version 1.2 (Kruglyak et al. 1996). The allele frequency of each marker was calculated from our database or from married-in family members of the seven kindreds (Ott 1999). Two-point linkage analysis was performed with the MLINK software of the LINKAGE package version 5.2 (Lathrop et al. 1984). The map order and microsatellite marker locations were specified according to databases of the Center for Medical Genetics' Marshfield comprehensive human linkage map, the National Center for Biotechnology Information, the Whitehead Institute/MIT Center for Genome Research, the Genome Database, and the Stanford Human Genome Center Web sites.
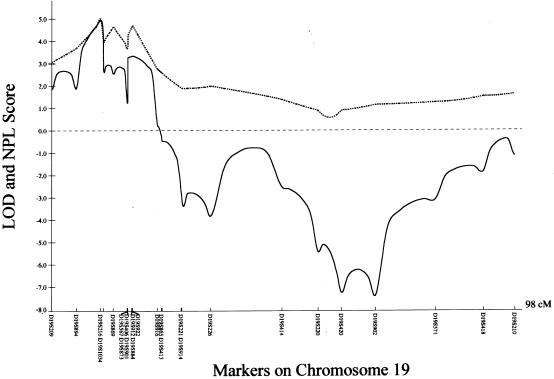
Since Wise et al. (2000) found evidence of allele sharing at three genetic loci in affected members of two families with IS, we initially screened a multiplex three-generation family (family 3) through use of polymorphic markers on these chromosomal regions—6p, 10q, and 18q—but failed to confirm linkage when a unified multipoint linkage approach was used (GENEHUNTER version 1.2). We next performed a genomewide scan for family 3 through use of 400 polymorphic microsatellite markers. Multipoint linkage analysis was used under a model of locus heterogeneity, specifying an autosomal dominant trait with a gene frequency of 0.01 and an estimated penetrance of 80%. Two intervals yielded initial multipoint LOD scores of >1. In particular, the locus at chromosome 19 yielded a multipoint LOD score of 1.75 and a nonparametric LOD (NPL) score of 6.71 (P=.00293). When we extended our study to six additional kindreds with AIS, multipoint linkage analysis of the seven families yielded a maximum LOD score of 4.48 and an NPL score of 5.36 (P=.00003) with markers on chromosome 19. We further genotyped 13 additional markers in the region flanked by D19S209 and D19S221, and the maximum multipoint LOD score and NPL score both were 4.93 at the region between D19S216 and D19S1034 (fig. 1). Two-point analysis with MLINK software yielded a maximum two-point LOD value of 3.63 at D19S216 (recombination fraction θ[m=f]=0.00). Further computation using hypothetical sets of different marker allele frequencies was performed, to exclude the possibility of reporting false-positive linkage (Ott 1992; Freimer et al. 1993). It did not significantly alter the multipoint LOD scores (which were 4.8–5.1) or two-point LOD values (which were 3.61–3.63 [θ[m=f]=0.00]) at D19S216. Other loci, D19S884 and D19S922, gave two-point LOD values of 4.02 and 4.09 (θ[m=f]=0.00), respectively (table 2).
Figure 1.
Multipoint linkage analysis of AIS to microsatellite markers on chromosome 19. The X-axis represents the relative location of these markers, and the Y-axis represents the multipoint LOD (solid line ) and NPL (broken line) scores from parametric and nonparametric linkage analyses, respectively, for all seven kindreds.
Table 2.
Two-Point LOD Scores for Linkage of Microsatellite Markers on Chromosome 19p13.3-13.2 to AIS at Different θ[m=f] Values in All Seven Kindreds, under an Autosomal Dominant Model
|
LOD Scorea at θ[m=f]= |
|||||
| Marker | 0 | .1 | .2 | .3 | .4 |
| D19S209 | −.888 | .819 | .782 | .569 | .295 |
| D19S894 | −1.179 | 1.822 | 1.561 | 1.052 | .482 |
| D19S216 | 3.625 |
2.934 | 2.136 | 1.301 | .541 |
| D19S1034 | −.026 | 1.217 | .918 | .507 | .148 |
| D19S869 | −.671 | 1.398 | 1.024 | .557 | .161 |
| D19S406 | −3.396 | 1.073 | .892 | .490 | .135 |
| D19S901 | 2.523 | 3.512 |
2.589 | 1.548 | .613 |
| D19S567 | .546 | 2.201 | 1.677 | 1.035 | .439 |
| D19S873 | −.752 | .910 | .702 | .387 | .113 |
| D19S912 | .491 | 1.825 | 1.349 | .784 | .294 |
| D19S884 | 4.020 |
3.184 |
2.254 | 1.310 | .495 |
| D19S922 | 4.087 |
3.150 |
2.179 | 1.239 | .450 |
| D19S916 | −2.287 | 2.137 | 1.672 | .992 | .378 |
| D19S413 | −1.607 | .822 | .692 | .409 | .127 |
| D19S865 | 1.180 | .876 | .555 | .270 | .071 |
| D19S914 | −1.990 | .015 | .169 | .135 | .046 |
| D19S221 | −3.606 | 1.169 | 1.074 | .715 | .307 |
Significant LOD scores are underlined.
A second chromosomal hotspot for AIS was located on chromosome 2, at D2S160-D2S347-D2S112-D2S151. A multipoint model-based linkage analysis showed a multipoint LOD score of 1.72, with an NPL score of 3.24 (P=.00357). No evidence of linkage was found with markers on the X chromosome.
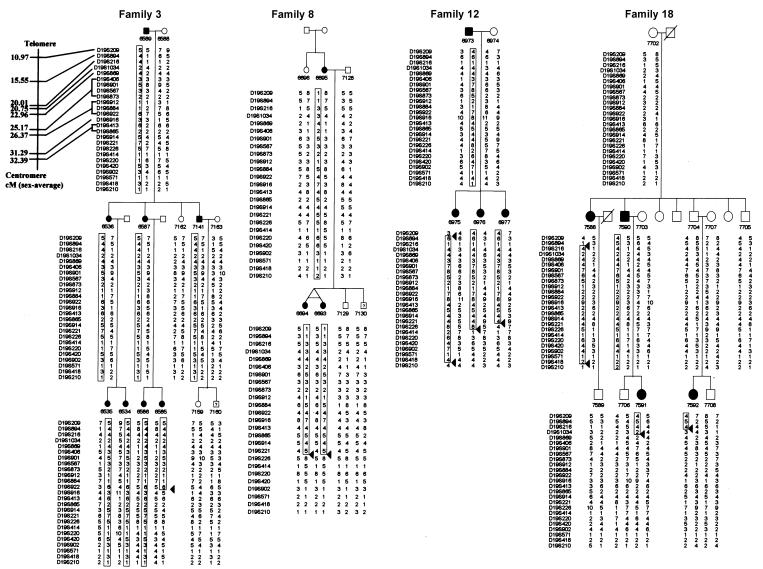
Examination of the chromosome 19 haplotypes of all family members in the seven kindreds revealed several informative recombinations. Families 3, 8, and 12 defined an ∼15-cM region for AIS (fig. 2). Examination of only affected members showed informative recombinations—for example, individual 6975 of family 12 defined the telomeric border at D19S209, and a recombination identified in individual 6585 of family 3 defined the centromeric border at D19S916. This finding was also supported by several recombinations identified in individuals of families 8 and 12.
Figure 2.
Abbreviated family pedigree and chromosome 19 haplotypes of four kindreds with AIS with informative recombinations. Affected individuals are denoted by blackened symbols, unaffected individuals are denoted by unblackened symbols, and individuals with unknown phenotype are indicated by a question mark. The identity of each microsatellite marker is shown to the left of each haplotype; their relative position along chromosome 19 is given in the top panel of family 3, and numbers on the far left are the sex-averaged chromosome distances (in cM) from the telomere of chromosome 19p. The individual number and haplotype is given under the symbol of each subject. The affected haplotype shared by relatives is boxed. An allele number represents a unique allele size across all families in the present study. Arrowheads indicate informative recombinations.
Further detailed examination of the haplotype of family 18 shows a recombination in individual 7588 that defined the telomeric boundary at D19S894. Other recombinations in individuals 7591 and 7592 further narrowed the centromeric border to D19S1034. This encompasses a region of 5.2 cM on the sex-averaged genetic map.
Wise et al. (2000) reported positive NPL scores of 1.42, 1.60, and 8.26 at hotspots on chromosomes 6p, distal 10q, and 18q, respectively, in a single family. Only the 10q site was revealed in a second family (Wise et al. 2000). In the present study, we failed to confirm linkage to any sites in these chromosomal regions. The variation may be ascribed to the different phenotypes of scoliosis in the two studies, since we have only recruited patients in whom scoliosis developed during adolescence. It is also generally accepted that AIS is a complex disease in which one or more genes may be responsible for the expressed phenotype and in which other modifying effects, such as age, sex, and environment, may play specific roles in the phenotypic variation between affected individuals (Miller 2000). The reduced penetrance (80%) assumed in the present study is conducive if the prospective candidate gene in this region is a disease-susceptibility gene and if the expression of the disease phenotype is dependent on the interaction of environmental and other genetic factors. The significant and similar LOD values for both parametric and nonparametric analysis of the seven kindreds is an indication that our model of a dominant mode of inheritance and locus heterogeneity is correct.
Informative recombinations observed in four of the families effectively narrowed the AIS critical region to around D19S216, between D19S894 and D19S1034, spanning 5.2 cM. This corresponds to a 1.9-Mbp region in the latest draft human genome sequence (Build 28). Within this region, there are ∼71 possible gene sequences identified. Genes expressed in chondrocytes, osteoclasts, muscles, or tendon could be potential candidate genes for AIS. Among these, the leucine-rich α-2-glycoprotein gene, which is similar to chondroadherin, expressed in chondrocytes, as well as the SH3-domain GRB2-like 1 gene, which is similar to endophilin 2, expressed in osteoclasts, are two strong candidates. Sequencing of these genes to identify the molecular defect, as well as demonstrating segregation of the defect in all affected members of a family, would be necessary to establish their candidacy.
Acknowledgments
The authors wish to thank the patients with scoliosis and their family members, for their participation. We are indebted to Prof. J. Ott of Rockefeller University, New York, for helpful comments on the manuscript; the Croucher Foundation and the University of Hong Kong Committee on Research and Conference Grants, for financial support of this project; and the Sir Robert Black Trust Fund of the Government of Hong Kong Special Administrative Region, for a training grant (to G.C.-Y.F.).
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for the Marshfield comprehensive human linkage map)
- Genome Database, The, http://www.gdb.org/
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for IS [MIM 181800]) [PubMed]
- Stanford Human Genome Center, http://shgc-www.stanford.edu/
- Whitehead Institute/MIT Center for Genome Research, http://www-genome.wi.mit.edu/
References
- Adams W (1865) Lectures on pathology and treatment of lateral and other forms of curvature of the spine. Churchill Livingstone, London [Google Scholar]
- Bell M, Teebi AS (1995) Autosomal dominant idiopathic scoliosis. Am J Med Genet 55:112 [DOI] [PubMed] [Google Scholar]
- Cobb JR (1948) Outline for the study of scoliosis. American Academy of Orthopaedic Surgeons Instructional Course Lectures 5:261–265 [Google Scholar]
- Cowell HR, Hall JN, MacEwen GD (1972) Genetic aspects of idiopathic scoliosis: a Nicholas Andry Award Essay, 1970. Clin Orthop 86:121–131 [DOI] [PubMed] [Google Scholar]
- Fisher RL, De George FV (1967) Idiopathic scoliosis: an investigation of genetic and environmental factors. J Bone Joint Surg 49:1005 [Google Scholar]
- Freimer NB, Sandkuijl LA, Blower SM (1993) Incorrect specification of marker allele frequencies: effects of linkage analysis. Am J Hum Genet 52:1102–1110 [PMC free article] [PubMed] [Google Scholar]
- Garland HG (1934) Hereditary scoliosis. Brit Med J 1:328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington PR (1977) The etiology of idiopathic scoliosis. Clin Orthop 126:17–25 [PubMed] [Google Scholar]
- Kesling K, Reinker KA (1997) Scoliosis in twins. Spine 22:2009–2015 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lauder ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-L, Huang D-S (1996) Scoliosis in China. A general review. Clin Orthop 323:113–118 [PubMed] [Google Scholar]
- Lonstein JE (1995) Idiopathic scoliosis. In: Lonstein JE, Bradfud DS, Winter RB, Ogilive JW (eds) Moe’s textbook of scoliosis and other spinal deformaties, 3rd ed. WB Saunders, Philadelphia, pp 219–256 [Google Scholar]
- Luk KDK, Cheung KMC, Chiu SW (1996) Thoracoscopic-assisted anterior release of the spine. J Orthop Surg 4:5–12 [Google Scholar]
- Miller NH (2000) Spine update. Spine 25:2416–2418 [DOI] [PubMed] [Google Scholar]
- Murray DW, Bulstrode CJ (1996) The development of adolescent idiopathic scoliosis. Eur Spine J 5:251–257 [DOI] [PubMed] [Google Scholar]
- Newton P, Wenger D, Mubarak S, Meyer R (1997) Anterior release and fusion in pediatric spinal deformity. A comparison of early outcome and cost of thoracoscopic and open thoracotomy approaches. Spine 22:1398–1406 [DOI] [PubMed] [Google Scholar]
- Ott J (1992) Strategies for characterizing highly polymorphic markers in human gene mapping. Am J Hum Genet 51:283–290 [PMC free article] [PubMed] [Google Scholar]
- ——— (1999) Analysis of human genetic linkage, 3rd ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Reamy BV, Slakey JB (2001) Adolescent idiopathic scoliosis: review and current concepts. Am Fam Physician 64:111–116 [PubMed] [Google Scholar]
- Riseborough EJ, Wynne-Davis R (1973) A genetic survey of idiopathic scoliosis in Boston, Massachusetts. J Bone Joint Surg 55A:974–982 [PubMed] [Google Scholar]
- Tse LY (1997) Student health service. In: Public Health and Epidemiology Bulletin. Department of Health, Hong Kong Government, Hong Kong, pp 9–10 [Google Scholar]
- Wise CA, Barnes R, Gillum J, Herring JA, Bowcock AM, Lovett M (2000) Localisation of susceptibility to familial idiopathic scoliosis. Spine 25:2372–2380 [DOI] [PubMed] [Google Scholar]
- Wynne-Davis, R (1968) Familial (idiopathic) scoliosis. A family survey. J Bone Joint Surg 50B:24–30 [PubMed] [Google Scholar]