Significance
Import of proteins to mitochondria and chloroplasts is essential for organelle biogenesis and organism survival. Proteins to be imported contain an N-terminal peptide targeting the protein to the correct organelle. The targeting peptides are cleaved off after the completed import. Because the free targeting peptides are potentially toxic to organellar activities, they must be removed. Here we report the identification and characterization of a unique mitochondrial and chloroplastic oligopeptidase, organellar oligopeptidase, that provides a complementary pathway for the degradation of targeting peptides and also participates in general organellar quality control mechanisms degrading the peptides produced from complete protein degradation.
Abstract
Both mitochondria and chloroplasts contain distinct proteolytic systems for precursor protein processing catalyzed by the mitochondrial and stromal processing peptidases and for the degradation of targeting peptides catalyzed by presequence protease. Here, we have identified and characterized a component of the organellar proteolytic systems in Arabidopsis thaliana, the organellar oligopeptidase, OOP (At5g65620). OOP belongs to the M3A family of peptide-degrading metalloproteases. Using two independent in vivo methods, we show that the protease is dually localized to mitochondria and chloroplasts. Furthermore, we localized the OPP homolog At5g10540 to the cytosol. Analysis of peptide degradation by OOP revealed substrate size restriction from 8 to 23 aa residues. Short mitochondrial targeting peptides (presequence of the ribosomal protein L29 and presequence of 1-aminocyclopropane-1-carboxylic acid deaminase 1) and N- and C-terminal fragments derived from the presequence of the ATPase beta subunit ranging in size from 11 to 20 aa could be degraded. MS analysis showed that OOP does not exhibit a strict cleavage pattern but shows a weak preference for hydrophobic residues (F/L) at the P1 position. The crystal structures of OOP, at 1.8–1.9 Å, exhibit an ellipsoidal shape consisting of two major domains enclosing the catalytic cavity of 3,000 Å3. The structural and biochemical data suggest that the protein undergoes conformational changes to allow peptide binding and proteolysis. Our results demonstrate the complementary role of OOP in targeting-peptide degradation in mitochondria and chloroplasts.
Protein turnover (degradation/biogenesis) in plant cells is a highly dynamic process, with as much as 50% of the total protein being replaced every 4–7 d (reviewed in ref. 1). In endosymbiotic organelles, mitochondria, and chloroplasts, the rate of protein degradation increases further during senescence and under conditions of environmental or developmental stress or insufficient carbon supply (reviewed in ref. 2). Highlighting the need for protein degradation is the fact that the genome of Arabidopsis thaliana encodes 723 putative proteases (3), the majority of which presently are uncharacterized.
The machineries involved in protein degradation in plant cytoplasm and endosymbiotic organelles are highly conserved and reflect their evolutionary origin. Proteins in the cytoplasm and the nucleus are cleaved mostly by the 20S/26S proteasome. In turn, chloroplastic and mitochondrial protein degradation is carried out by proteases of bacterial origin belonging to the serine (Lon, ClpP, Deg) or metalloprotease (FtsH) type families (reviewed in refs. 4–6). To date ∼35 different proteases in the chloroplasts and mitochondria of A. thaliana have been identified as being involved in the removal of misfolded or oxidatively damaged proteins and in the degradation of unassembled subunits of protein complexes.
Apart from the general degradation processes, chloroplasts and mitochondria also are sites of targeted proteolysis responsible for the maturation of precursor proteins. Approximately 3,000 chloroplastic and 1,000 mitochondrial proteins are synthesized in the cytosol as precursors carrying a targeting signal, allowing them to be imported posttranslationally to these organelles (7–9). For the proteins localized in the chloroplastic stroma or in the mitochondrial matrix, the targeting signal is an N-terminal extension termed “transit peptide” or “presequence,” respectively, which is cleaved off after import in a reaction termed “processing” (10). Most of the mitochondrial preproteins undergo processing by the mitochondrial processing peptidase (MPP), which in plants is integrated into the cytochrome bc1 complex of the respiratory chain (11). MPP is the only processing peptidase presently characterized in plant mitochondria. In some cases MPP cleavage is followed by additional trimming by octapeptidyl aminopeptidase 1 (Oct1), which cleaves off an N-terminal octapeptide (12, 13). Oct1 activity has been identified thus far only in yeast and mammals, but the gene coding for a putative ortholog also is present in the A. thaliana genome (5). Processing in the chloroplast stroma is carried out by the stromal processing peptidase (SPP), which cleaves off transit peptide and performs additional cleavage(s) within the transit peptide to generate shorter fragments (14, 15).
In A. thaliana, mitochondrial presequences range from 19 to 109 aa (16); chloroplastic transit peptides usually are longer, ranging from 26 to 146 aa (9).
Peptides generated as a result of preprotein processing can be degraded by the presequence protease (PreP), a metallopeptidase of the M16C family (reviewed in ref. 17). PreP initially was identified in plant mitochondria as the protease responsible for the degradation of the presequence of the ATPase beta subunit (pF1β) and the chloroplastic transit peptide of the small subunit of ribulose-1,5-bisphosphate carboxylase oxygenase (18, 19). Consistent with this result, A. thaliana PreP (AtPreP) was shown to have a dual localization in mitochondrial matrix and chloroplastic stroma (18, 20). In addition to targeting peptides generated by processing, AtPreP1 degrades a wide range of unstructured peptides ranging from 10 from 65 aa (18). The crystal structure of AtPreP1 revealed that the enzyme consists of two bowl-shaped halves connected by a hinge region, forming a catalytic chamber of 10,000 Å3, which is big enough to accommodate unstructured peptides up to 65 aa long; however, small folded proteins cannot be degraded. Residues localized in both halves of the enzyme contribute to proteolysis, suggesting that the structure must close for the cleavage to occur (21). Later studies led to the identification of AtPreP orthologs in yeast and mammals, both localized in mitochondrial matrix (22, 23). The human PreP also was shown to degrade amyloid-β peptides and has been associated with Alzheimer’s disease (23).
In addition to AtPreP and its orthologs, it has been suggested that other proteases of the M3A family take part in the degradation of signal sequences. For instance, in Escherichia coli, oligopeptidase A (OpdA) degrades the 20-aa signal peptide of prolipoprotein (24). Mitochondria-localized peptidases of M3A family were found in yeast (PRD1) and mammals (neurolysin) (25, 26). Their function in mitochondria remains unknown, although it has been suggested that PRD1 cooperates with Cym1 (the PreP ortholog in yeast) in the degradation of mitochondrial presequences (27).
The increase in abundance of peptides generated by either unspecific degradation or preprotein processing might influence organellar function and lead to various physiological consequences. For instance, in Caenorhabditis elegans peptides exported from mitochondria are involved in retrograde signaling as a part of the mitochondrial unfolded protein response (reviewed in ref. 28). Additionally, it has long been known that presequence peptides destabilize mitochondrial membranes, potentially leading to uncoupling of respiration or to dissipation of the membrane potential (29–32). Accumulation of presequences also was shown to impair mitochondrial precursor processing by direct inhibition of MPP activity (33). For these reasons, the turnover of peptides generated in mitochondria and chloroplasts must be regulated, either by active export or local degradation. In yeast, peptides up to 20 aa long can be exported out of mitochondria by an ABC-type transporter termed “Mdl1” (27, 34), but a similar system has not been identified in plants.
Here we describe the identification and characterization of organellar oligopeptidase (OOP), an M3 plant peptidase localized within the mitochondria and chloroplasts. We have characterized the evolutionary origin, the intracellular localization, and the structural and catalytic properties of OOP. Our results provide biochemical evidence of mitochondrial presequence degradation by an M3 peptidase and also suggest that this protein has a broader role in general peptide degradation in these endosymbiotic organelles.
Results
A. thaliana Contains Four M3A Peptidase Homologs.
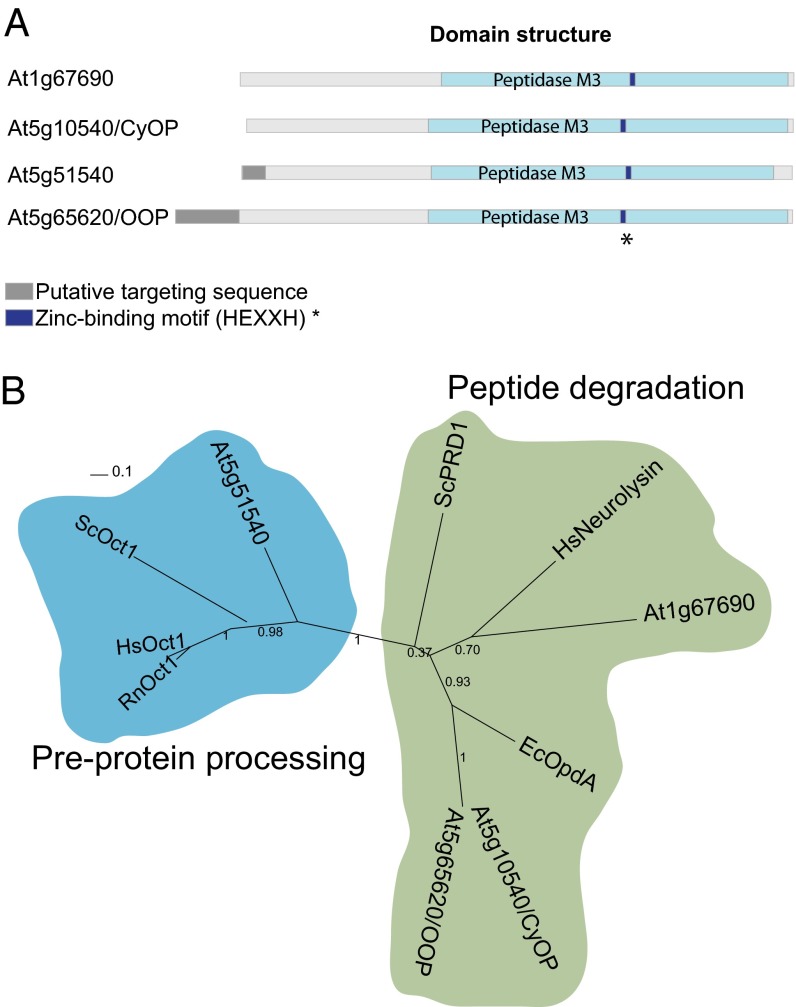
The M3A subfamily of metallopeptidases is widespread in prokaryotes and eukaryotes and includes the human neurolysin, the E. coli OpdA, and the yeast PRD1. This subfamily contains peptidases with two different functions. One group consists of oligopeptidases responsible for peptide degradation (OpdA and PRD1). The second group includes processing peptidases (Oct1) specialized in the removal of octapeptides from the N termini of MPP-processed precursor proteins. A survey of the Arabidopsis genome shows that it contains four genes encoding putative proteases of the M3A subfamily (Fig. 1A). None of these proteins has been characterized experimentally, but all possess a peptidase M3 domain carrying the zinc-binding motif HEXXH (where X is an uncharged residue) (Fig. S1A). To select putative organellar peptidases involved in peptide degradation, we analyzed their similarity to the currently known M3A-degrading peptidases and their possible intracellular localization. Based on these features, we selected two of the four A. thaliana M3A peptidases, At5g65620 and At5g10540, because both cluster with the degrading peptidases on the phylogenetic tree (Fig. 1B). Both were identified in chloroplasts using a high-throughput proteomic approach (9, 35), although only At5g65620 is predicted to possess a transit peptide (TargetP) directing it to chloroplasts. We excluded two other A. thaliana M3A peptidases from this study. Peptidase At5g51540 clusters along with Oct1 orthologs identified in yeast and mammals (Fig. 1B) and is likely to be involved in processing, not in peptide degradation, whereas peptidase At1g67690 does not show a discernible mitochondrial or chloroplastic targeting sequence.
Fig. 1.
M3A peptidase homologs in A. thaliana. (A) Schematic representation of the structural and functional elements in the primary structure of M3A family peptidases. The putative targeting sequence (TargetP; www.cbs.dtu.dk/services/TargetP/) is shown in gray, the peptidase M3 domain (Pfam; http://pfam.sanger.ac.uk/) in light blue, and the Zn-binding motif in dark blue. (B) Phylogenetic tree of M3A peptidase homologs from several organisms, showing the two distinct groups. The tree was built based on the alignment shown in Fig. S1A. Sequences used for comparison were A. thaliana At1g67690 (F4HTQ1), At5g65620 (Q94:00 AM1), At5g10540 (Q949P2), and At5g51540 (F4KDA5); S. cerevisiae ScOct1 (P35999) and ScPRD1 (P25375); Homo sapiens HsOct1 (Q99797) and HsNeurolysin (Q9BYT8); Rattus norvegicus RnOct1 (Q01992); and E. coli EcOpdA (P27298). (The UniProt accession number for each homolog is given in parentheses.)
A. thaliana Organellar Oligopeptidase and Cytosolic Oligopeptidase.
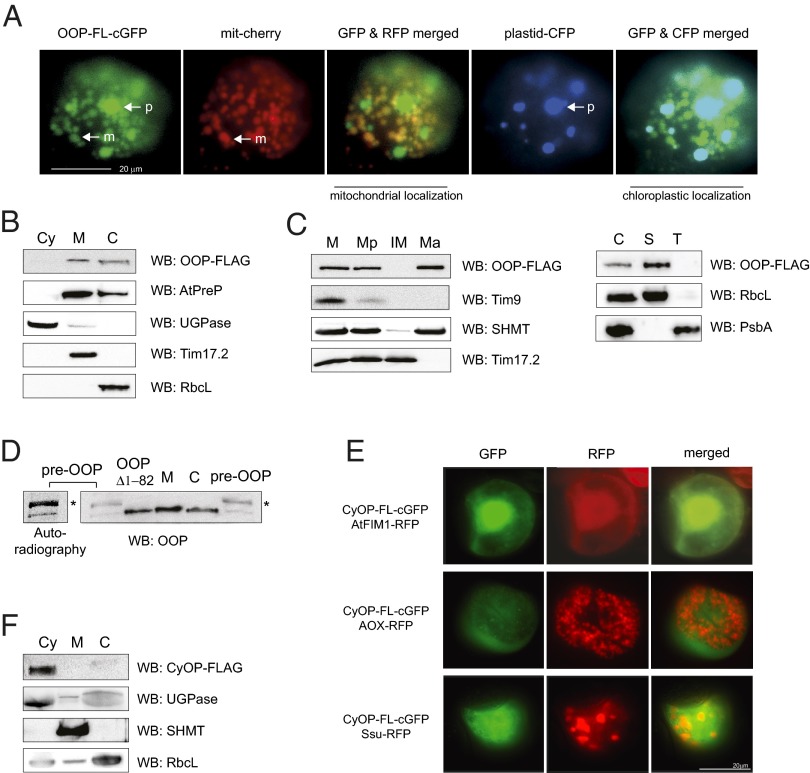
We investigated the subcellular localization of At5g65620 and At5g10540 using two independent in vivo approaches, transient expression in Arabidopsis cell suspensions and immunodetection. Detection in Arabidopsis suspension cells showed dual localization in mitochondria and chloroplasts for GFP fused with full-length At5g65620 (Fig. 2A) and a cytosolic localization for GFP fused with full-length At5g10540 (Fig. 2E). Because of the high sequence identity between At5g65620 and At5g10540 and thus the difficulty of producing specific antibodies, we generated A. thaliana lines expressing C-terminal FLAG-tagged full-length variants of At5g65620 and At5g10540. Detection of the FLAG tag in mitochondria, chloroplasts, and cytosol isolated from plants expressing tagged At5g65620 and At5g10540, respectively, confirmed the dual localization of At5g65620 in mitochondria and chloroplasts (Fig. 2B) and the cytosolic localization of At5g10540 (Fig. 2F). Based on these results, we termed At5g65620 the organellar oligopeptidase (OOP) and At5g10540 the cytosolic oligopeptidase (CyOP). Further subfractionation of mitochondria and chloroplasts showed that OOP is a soluble protein, localized in the matrix and stroma, respectively (Fig. 2C), having the same intracellular localization as the dual-localized presequence protease, AtPreP (Fig. 2B) (18, 36). According to TargetP (Fig. 1A and Fig. S1B), the precursor of OOP contains a transit peptide of 82 aa that is putatively cleaved off after import. To verify this result, we compared the in-gel mobility of the OOP precursor (pre-OOP) with that of an OOP variant lacking first 82 amino acids overexpressed in bacteria (OOPΔ1−82) and the native mature OOP forms in mitochondria and chloroplasts (Fig. 2D). The mature OOP forms detected in the isolated organelles migrate at a level similar to that of the overexpressed OOPΔ1−82 variant and at a lower level than the precursor form, confirming that the precursor of OOP undergoes processing.
Fig. 2.
Intracellular localization of OOP (At5g65620) and CyOP (At5g10540). (A) Fluorescence imaging of the full-length OOP fused to GFP with a mitochondrial cherry (mit-cherry) and chloroplastic cyan blue (chloro-CFP) marker in A. thaliana suspension-cultured cells. Mitochondria (m) and plastids (p) are indicated by arrows. (Scale bar: 20 µm.) (B) Detection of FLAG-tagged OOP in cytosol (Cy), mitochondria (M), and chloroplasts (C) isolated from stable A. thaliana transformants. (C) Fractionation of mitochondria and chloroplasts isolated from stable A. thaliana transformants expressing OOP-FLAG. Presence of FLAG was detected in total mitochondria (M), mitoplasts (Mp), inner membrane (IM), and matrix (Ma) fractions as well as in total chloroplasts (C), stroma (S), and thylakoids (T). (D) Analysis of proteolytic maturation of OOP in mitochondria and chloroplasts. In vitro synthesized and radioactively labeled precursor of OOP (pre-OOP) was resolved on SDS/PAGE alongside purified OOPΔ1–82 and total mitochondria, (M) and chloroplasts (C) isolated from wild-type A. thaliana plants. OOP was detected either by autoradiography or by immunodetection. The asterisk indicates the precursor form. (E) Fluorescence imaging of full-length CyOP fused to GFP and cytosolic, mitochondrial, and chloroplastic marker proteins fused to RFP in A. thaliana suspension-cultured cells. (Scale bar: 20 µm.) (F) Detection of FLAG-tagged CyOP in cytosol (Cy), mitochondria (M), and chloroplasts (C) isolated from stable A. thaliana transformants. Antibodies used in B–D and F were anti-FLAG, anti-AtPreP (mitochondrial matrix and chloroplastic stroma), anti-SHMT (serine hydroxymethyltransferase, mitochondrial matrix), anti-Tim17.2 (mitochondrial inner membrane), anti-PsbA (D1 protein, thylakoid membrane), anti-Tim9 (mitochondrial intermembrane space), anti-RbcL (chloroplastic stroma), anti-OOP, and anti-UGPase (cytosolic marker). WB, Western blot analysis.
OOP Can Degrade Short Presequences and Other Peptide Substrates.
Previous reports suggested that M3A peptidases may be involved in the degradation of targeting peptides and even may cooperate with M16C peptidases (as Cym1 or PreP) in this function (27).
We approached this idea by testing the activity of the mitochondria- and chloroplast-resident OOP against the Nicotiana plumbaginifolia pF1β, a well-characterized substrate of AtPreP. For activity assays we used the recombinant OOPΔ1−82 variant produced in bacteria (hereafter referred to as OOP).
Full-length pF1β 2-54 (53 aa) is not degraded by OOP, but it was degraded completely by the control, AtPreP1 (Fig. 3A and Fig. S2A). However, OOP was able to degrade both N- and C-terminal fragments derived from pF1β (12 and 11 aa, respectively) (Fig. 3A and Fig. S2A). The degradation of these fragments was analyzed by SDS/PAGE and confirmed by MS (Fig. 3A, Inset, Table 1, and Dataset S1). As a control for these experiments, we used a catalytically inactive OOP variant in which glutamine replaces the glutamate residue in the Zn-binding site (OOPE572Q) (Fig. 4 C and E).
Fig. 3.
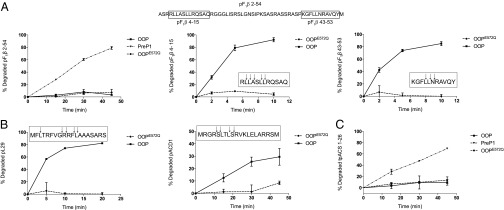
Degradation activity of the OOP peptidase. (A) Degradation of pF1β-derived peptides by OOP. Degradation of pF1β 2-54 by PreP1 is used as a positive control, and OOPE572Q is used a negative control. Insets in the graphs show the degradation pattern of the peptide substrates as analyzed by MS. The sequence of the tested pF1β peptides is shown for clarity. (B) Degradation of the presequences of L29 and ACD1 by OOP. Insets show pattern of cleavage analyzed by MS. (C) Degradation of the transit peptide of ACS (tpACS 1-26) by PreP1, OOP, and OOPE572Q.
Table 1.
Degradation of peptide substrates by OOP and PreP1 analyzed by MS
| Peptide | Description/source | Length, aa | Isoelectric point* | Cleavage by OOP | Cleavage by PreP1 |
| pF1β 4–15 | Presequence fragment | 12 | 12 | RL↓L↓A↓SL↓L↓RQSAQ | R↓L↓LASL↓L↓R↓QSAQ |
| N. plumbaginifolia | |||||
| pF1β 43–53 | Presequence fragment | 11 | 9.99 | KGFL↓L↓N↓RAVQY | KG↓FLL↓NR↓AVQY |
| N. plumbaginifolia | |||||
| pL29 | Presequence | 19 | 12.48 | MFL↓TRFVG↓R↓RF↓L↓AAASARS | MFLT↓R↓FVGRR↓FLAAAS↓ARS |
| A. thaliana | |||||
| pACD1 | Presequence | 20 | 12.18 | MRGR↓S↓LTL↓S↓RVKLELARRSM | MRGR↓SLTLS↓R↓VKLEL↓A↓R↓R↓SM |
| A. thaliana | |||||
| tpACS 1–11 | Transit peptide | 11 | 6.50 | MSS↓N↓SL↓R↓HVE↓S | MSSNS↓LRHVES |
| A. thaliana | |||||
| tpACS 1–19 | Transit peptide | 19 | 6.50 | MSSNSLRHVESM↓S↓Q↓LP↓S↓GA | MSSNS↓LR↓HV↓E↓S↓MSQLP↓SGA |
| A. thaliana | |||||
| tpACS 1–26 | Transit peptide | 26 | 8.52 | MSSNSLRHVESMSQLPSGAGKISQLN | MSSNS↓LRHVES↓MSQLPSGAG↓KISQLN |
| A. thaliana | |||||
| pThrRS 1–19 | Presequence | 19 | 8.52 | MASSH↓S↓LLF↓S↓SSFL↓SKPSS | MASSHS↓L↓L↓F↓S↓S↓S↓F↓L↓SKPSS |
| A. thaliana | |||||
| SytII 40–60 | Non-presequence – synaptotagmin | 21 | 4.94 | GESQE↓DMF↓AKLK↓DKFF↓N↓EINK | GESQEDMFAKLKD↓K↓FFNE↓IN↓K |
| Rattus norvegicus | |||||
| octMdh1 | Oct1 generated octapeptide | 8 | 5.52 | FS↓S↓TVA↓NP | FSSTVANP |
| S. cerevisiae | |||||
| octSdh1 | Oct1 generated octapeptide | 8 | 9.75 | FTSSAL↓VR | FTSSALVR |
| S. cerevisiae | |||||
| Cyt1p 294–309 | Non-presequence | 16 | 12.03 | GIKTRK↓FVF↓N↓PPKPRK | GIKTR↓K↓FV↓FNPPKP↓R↓K |
| S. cerevisiae | |||||
| pCox4 | Presequence | 23 | 12.01 | MLSLRQ↓S↓I↓RF↓F↓KPATRTLSSSRY | MLSLRQS↓I↓R↓F↓FKPAT↓RTLSSSRY |
| S. cerevisiae |
The cleavage sites were inferred from the data in Dataset S1.
Calculated using the ExPASy compute Mw/pI tool (http://web.expasy.org/compute_pi/)
Fig. 4.
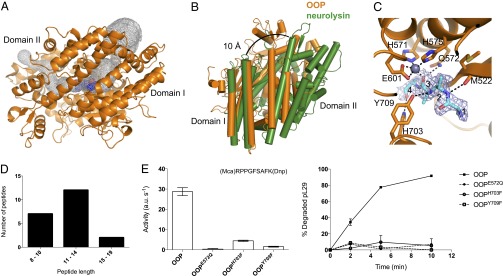
Structural features of OOP. (A) Overview of the OOP structure (orange) and its two domains, with the bound substrate shown as spheres (blue). The internal cavity in OOP is outlined in a gray mesh. (B) Comparison of OOP (orange) and neurolysin (green), superimposed on domain I, shows the possible path of opening. The neurolysin structure is opened by 10 Å at the top of the cleft. (C) Representation of the active site of OOP with the substrate bound in the structure of OOPE572Q modeled as a polyalanine tetrapeptide (cyan), with its electron density (2FO-FC, contoured at 1σ) shown in a blue mesh. The Zn ion is shown as a gray sphere, and the relevant residues in the active site are shown as sticks. Possible hydrogen bonds between the peptide and residues on the OOP active site are shown as black dashed lines. (D) Length distribution of the peptide ligands copurified with OOPE572Q (sequence of the peptides is shown in Dataset S2). (E) Activity of wild-type OOP and its variants carrying substitutions in the active site residues E572, H703, and Y709 analyzed by the degradation of the fluorogenic peptide substrate V and the presequence peptide pL29. Results are shown as average ± SEM.
Further analysis using other peptides of various sources showed that OOP can degrade several substrates and displays a substrate size restriction (Table 1 and Fig. S2B). The shortest peptides used in this study and degraded by OOP were two octapeptides (Table 1) generated by the yeast processing peptidase Oct1 from yeast Mdh1 and Sdh1 (12), and the longest one was the 23-aa presequence of the yeast Cox4 preprotein (Table 1 and Fig. S2B). OOP was unable to degrade a folded protein (insulin) or larger peptides including amyloid-β 1-28 and 1-40 and insulin-β chain (Fig. S2B), whereas AtPreP1 can degrade all these peptides except the short octapeptides (Table 1 and Fig. S2B).
Taken together, the set of experiments shown in Fig. 3, Table 1, and Fig. S2 suggest that OOP can degrade peptide fragments within the range of 8–23 aa.
Around 10% of the presequences identified in plants are shorter than or equal to 23 aa (6 of 62 for A. thaliana and 8 of 52 for Oryza sativa) (16) and thus might be potential natural substrates of OOP, considering solely the length criterion. On the other hand, the shortest identified transit peptide is 26 aa long (9) and thus is potentially too long to be degraded by OOP.
As a proof of principle, we selected two of the A. thaliana presequences within this size limit for degradation studies—the presequence of the ribosomal protein L29 (At1g07830; pL29; 19 aa) and the presequence of the 1-aminocyclopropane-1-carboxylic acid deaminase 1 (At1g48420; pACD1; 20 aa)—and the 26-aa transit peptide of acetyl-CoA synthetase (At5g36880; tpACS). Both presequences, pL29 and pACD1, were degraded upon incubation with purified OOP (Fig. 3B and Table 1) and also upon incubation with AtPreP1 (Table 1). In contrast, OOP could not degrade the full-length tpACS transit peptide but only its fragments of 11 aa and 19 aa (Fig. 3C and Table 1).
Taken together, all the data obtained from the MS analysis of the OOP cleavage products (13 peptide substrates) make it clear that OOP does not exhibit a strict cleavage pattern (Table 1). Although the reduced number of different substrates limits the certainty of our conclusions, the results indicate that OOP has a preference for hydrophobic residues (F/L) at the P1 position (in about 50% of the cleavages detected). Interestingly, in a similar set of substrates, PreP1 showed a preference for positively charged residues (K/R) at the P1 position (Table 1).
Structure of OOP with Bound Peptide Reveals the Molecular Basis for Substrate Length Restriction.
The biochemical analysis of peptide cleavage by OOP revealed a substrate length restriction and a relative promiscuity in the peptide cleavage sites. For a deeper insight into the molecular determinants of peptide degradation by OOP, we solved the crystal structure of OOP. Crystals of wild-type and the inactive variant OOPE572Q were obtained. The crystals belonged to the space group P212121, contained one molecule per asymmetric unit, and diffracted to 1.8–1.9 Å (Table S1).
OOP exhibits an overall ellipsoidal shape and consists of two major domains (domain I from residues 91–235 and domain II from residues 236–781) which enclose the catalytic cavity (Fig. 4A). The OOP homolog E. coli dipeptidyl carboxypeptidase has been crystallized in a closed conformation similar to the OOP structure presented here (rmsd 1.7 Å) (37). In contrast to the closed conformation of OOP, the mammalian homologs neurolysin and thimet oligopeptidase (TOP) were crystallized in open conformations (38, 39). The catalytic site of OOP is buried within a long internal cavity (Fig. 4A). The overall fold of OOP is the same as neurolysin (665 equivalent Cα atoms, rmsd of 3.0 Å) and TOP (654 equivalent Cα atoms, rmsd of 3.6 Å). The rmsd values are relatively large because OOP is in a closed conformation, whereas the other two are in an open conformation (Fig. 4B shows a comparison between OOP and neurolysin).
The total volume of the internal cavity in OOP is ∼3,000 Å3, which corresponds to the volume of a peptide with about 21 aa (40). This value agrees well with maximum peptide length that OOP can cleave (23 aa), as shown by our activity measurements (compare with Table 1). The internal cavity of OOP has two narrow openings to the outside solvent, but it is very unlikely that a peptide could enter the protein via these narrow openings. Supporting this idea is the fact that substitutions closing the openings to the outside (through the introduction of bulky tryptophan side chains) have little effect on OOP activity (Fig. S3).
Thus it is conceivable that substrate binding to OOP requires the opening of the structure between domains I and II, resembling the deep crevice observed in the neurolysin structure. The structural superposition shown in Fig. 4B between OOP (closed conformation) and neurolysin (open conformation) highlights the conformational flexibility needed to allow the peptide substrate access to the active site.
OOP contains the active site sequence motif HEXXH, characteristic for the whole MA clan of zinc metallopeptidases (41). The two histidines in this motif, H571 and H575, coordinate the Zn ion, with the more C-terminal residue E601 acting as the third zinc ligand (Fig. 4C). The nature and location of the ion was confirmed via anomalous data collected at the zinc edge on the OOPE572Q structure. The glutamate residue E572 is proposed to be important in coordinating a water molecule taking part in the nucleophilic attack of the scissile bond. In agreement with this notion, replacing this residue by glutamine completely inactivates the enzyme (Figs. 3 and 4E).
Despite being crystallized in the absence of added substrate, residual electron density became visible within the active site of OOPE572Q during refinement and accounted for a bound peptide. The electron density at the active site allowed the building of a tetrapeptide backbone trace but with no discernible side chains (Fig. 4C). This absence of side chains is a consequence of the conformational ensembles of multiple ligands and is typical for a mixture of endogenous peptides, as seen before in other examples of peptide-binding proteins (42). We have crystallized a trapped uncleaved peptide ensemble likely similar to the initial peptide-binding intermediate in the wild-type enzyme. The peptide substrate is buried within the catalytic cavity and bound via hydrogen bonds to five residues in the active site: A520, M522, Q572, H703, and Y709 (Fig. 4C). Interactions between the peptide backbone and these residues likely determine the correct positioning of the substrate for catalysis. In support of this idea, substitution of H703 or Y709 for phenylalanine abolishes the activity of the enzyme (Fig. 4E). It is noteworthy that the peptide main chain does not seem to be anchored, because no salt bridges are formed to the peptide termini. Also, the weak electron density suggests that the peptide continues into the cavities on both sides of the active site (Fig. 4C). The pockets that can accommodate the side chains of the peptide provide both polar and hydrophobic binding surfaces and therefore do not impose strict sequence specificity on the substrate. This lack of strict sequence specificity is in agreement with the observed promiscuity in the cleavage of the tested peptide substrates (Table 1), with only a weak preference for hydrophobic residues at the P1 position. To identify the endogenous peptides, MS analysis of the peptides extracted from OOPE572Q was performed. We detected a mixture of at least 21 different peptides originating from 10 different proteins (Fig. 4D and Dataset S2). The peptides range from 8 to 19 residues in length and most probably correspond only to a subset of the peptides bound to OOPE572Q. The sizes are fully consistent with the internal volume of the OOP cavity and the analyzed peptide substrates.
The structure of wild-type OOP was virtually identical to that of OOPE572Q (rmsd of 0.3 Å), with the major exception that no substrate was bound in the active site. This lack of a bound substrate was expected because the protein is still active, and any endogenous substrate would have been cleaved already.
Discussion
The cellular functions of mitochondria and chloroplasts are strictly dependent on the import of several hundred nuclear-encoded polypeptides into the correct organellar subcompartments. Most of the proteins imported to the mitochondrial matrix and the chloroplast stroma carry N-terminal targeting peptides that are removed within the organelles by targeted proteolysis. A consequence of this process is the generation of free presequences and transit peptides that either are exported from the organelles or are degraded in situ.
In the present report we explored the possibility of presequence degradation by proteases of the M3A family in plants.
Dual Targeting of OOP to Mitochondria and Chloroplasts.
The genome of A. thaliana encodes four putative M3A homologs, two of which have possible peptide degradation activity and potential organellar localization (Fig. 1). Using two independent in vivo approaches, we showed that peptidase At5g65620 is dually targeted to the mitochondrial matrix and the chloroplastic stroma, whereas At5g10540 is localized in the cytosol (Fig. 2). Based on this localization, we termed them “organellar oligopeptidase” (OOP) and “cytosolic oligopeptidase” (CyOP), respectively. OOP and CyOP display 92% sequence identity (Fig. S1B), differing mostly in the N terminus, with OOP containing a targeting peptide that is cleaved off upon import to mitochondria and chloroplasts (Fig. 2D). The high similarity between OOP and CyOP suggests that they arose from a recent gene duplication followed by acquisition of mitochondrial and chloroplastic localization by OOP.
OOP is one of the ∼100 proteins that presently are known to be dually targeted to both mitochondria and chloroplasts (43). Dual targeting is believed to occur in proteins taking part in processes and reactions common to both organelles, such as gene expression, protein synthesis, degradation, and stress response (43). Dual localization is a common feature of proteases, with several examples previously reported, including AtPreP, AtFtsH11, Lon1, and Lon4 (reviewed in ref. 5).
Molecular Constraints to Peptide Degradation by OOP.
Having assigned the intracellular localization of OOP to mitochondria and chloroplasts, we then evaluated its peptide degradation activity. After analyzing the cleavage of 13 different peptides, we concluded that OOP could degrade fragments in the range of 8–23 aa (Fig. 3, Table 1, and Fig. S2). Additionally, MS analysis of the cleavage fragments revealed a promiscuous cleavage pattern by OOP, although it showed a weak preference for hydrophobic residues at the P1 position of the substrate (Table 1).
Structural analysis of OOP provided several clues for the molecular determinants of substrate length and sequence restriction. OOP was crystallized in a closed conformation, with the structure showing an inner cavity of about 3,000 Å3 (Fig. 4A). This internal cavity is large enough to accommodate a peptide with about 21 aa, a value that agrees well with our biochemical analysis showing the cleavage of the longest peptide of 23 aa. Thus, it is likely that the volume of the internal cavity restricts the substrates of OOP. The structure of OOP (inactive variant OOPE572Q) also revealed the mode of binding of the peptide substrate, highlighting the fundamental role of residues H703 and Y709 for substrate stabilization through hydrogen bonding to the peptide backbone (Fig. 4 C and E). Furthermore, the substrate-binding pockets around the active site allow both hydrophobic and polar interactions, further supporting the idea that OOP has substrate promiscuity.
Presequence Degradation by OOP.
Having established that OOP is an active peptidase dually localized in mitochondria and in chloroplasts, we aimed to identify potential presequence peptide substrates, considering the observed size limitation.
Analysis of experimentally determined mitochondrial presequences in plants (A. thaliana and O. sativa), based on the identification of the N terminus of the mature mitochondrial proteins, showed ∼10% of presequences had a length ≤23 aa (16). From the group of mitochondrial presequences ≤23 aa, we tested pL29 (19 aa) and pACD1 (20 aa) as substrates for OOP. Indeed, both presequence peptides were degraded by OOP (Fig. 3B), supporting the hypothesis that M3 peptidases can degrade presequence peptides if they fit the size restriction (Fig. 5A).
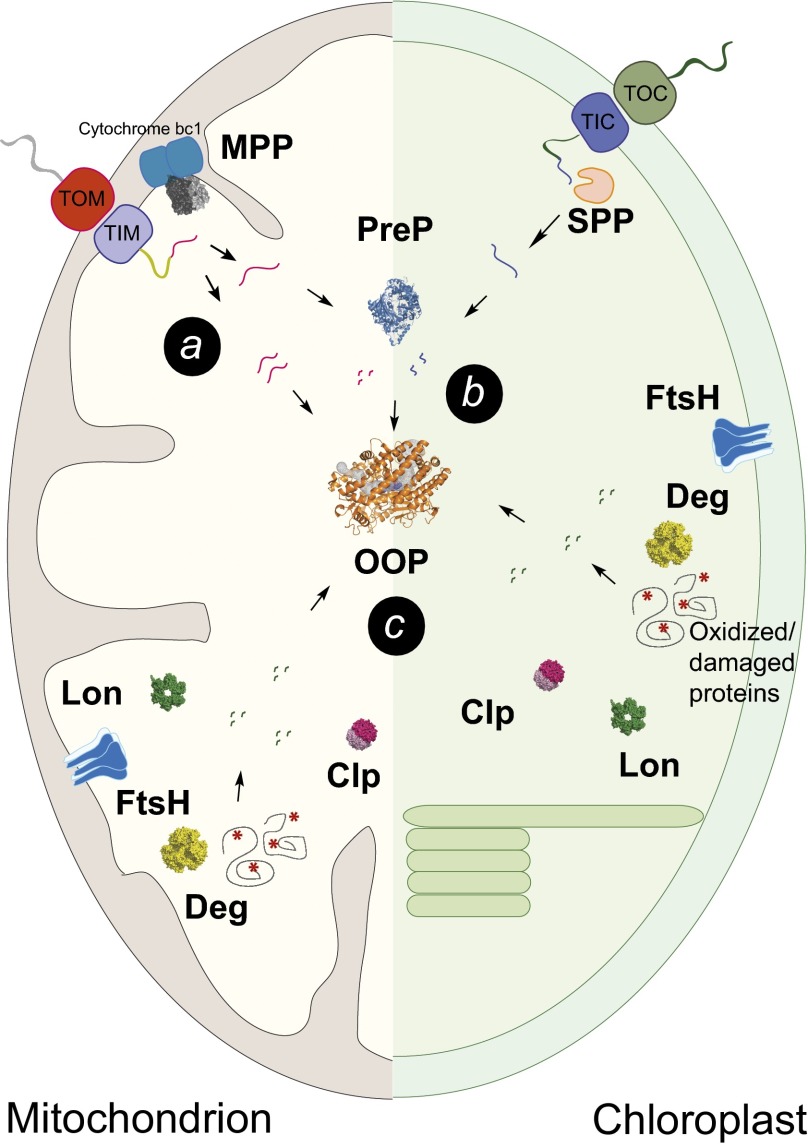
Fig. 5.
Cartoon representation of the proteolytic systems in mitochondria and in chloroplasts highlighting the possible roles of OOP in presequence degradation (a), degradation of presequence/transit peptide fragments (b), and degradation of peptides resulting from cleavage of damaged/oxidized proteins by proteases as FtsH, Clp, Lon, and Deg (c). TIC, translocase of the inner envelope of chloroplasts; TIM, translocase of the inner mitochondrial membrane; TOC, translocase of the outer chloroplast envelope; TOM, translocase of the outer mitochondrial membrane.
The chloroplastic transit peptides are longer than mitochondrial presequences in general, and even the shortest one presently determined in A. thaliana (tpACS, 26 aa) exceeds the OOP size limit. However, OOP degrades short tpACS fragments of 11 and 19 aa. It is worth noting that pL29 and pACD1, as well as tpACS, also were degraded by AtPreP1 (Fig. S2). In total, of 12 peptides degraded by OOP, 10 also were substrates for PreP. The apparent overlap in the substrate pool between PreP and OOP and the same subcellular localization suggests that these two peptidases might cooperate in vivo for peptide degradation in endosymbiotic organelles, a possibility supported by previous studies in yeast (27).
This functional connection between OOP and PreP may explain why we did not observe any phenotypic changes in oop-knockout plants (Fig. S4). Analysis of the seed germination rate, growth, and development progression as well as basic mitochondrial and chloroplastic functions (respiration and photosynthesis rate) in two independent oop-knockout lines revealed no difference between oop and wild-type plants (Fig. S4). However, crossing the oop plant with the double prep1 prep2 knockout results in a more severe phenotype. The oop prep1 prep2 triple-knockout plants display reduced growth rate as compared with the oop-knockout and prep1prep2 double-knockout plants alone (Fig. S5A). In contrast, genetic cross of oop plants with cyop plants carrying a transfer DNA (T-DNA) insertion in the gene coding for the cytosolic M3A peptidase did not result in a more severe phenotype (Fig. S5B). The genetic interaction between OOP and PreP agrees well with the functional overlap in peptide degradation observed for these two peptidases. However, it is unlikely that OOP and PreP would form a stable peptide-degrading complex, because immunoprecipitation of OOP-FLAG failed to show a physical interaction between the two peptidases in either the mitochondrial matrix or in the chloroplastic stroma (Fig. S5C).
Although the number of plant presequences that fit the OOP size restriction presently is rather small, the pool of OOP substrates among presequences might in fact be larger, because MPP can perform additional cleavages, generating shorter fragments (44). Also, the stromal processing peptidase can trim transit peptides upon cleavage from the mature protein, potentially making them accessible to OOP degradation (Fig. 5A) (14). OOP also could take part in targeting peptide degradation by acting not only in parallel but also downstream of PreP, cleaving peptides partially degraded by PreP (Fig. 5B). In support of this idea, incubation of pF1β 2−54 in the presence of both PreP1 and OOP yielded smaller fragments than incubation with PreP1 alone, suggesting that OOP can further degrade PreP1-generated products (Dataset S1).
Interestingly, one group of peptides tested in vitro turned out to be specific substrates for OOP (not degraded by AtPreP1). These substrates are octapeptides derived from a second step of preprotein processing by Oct1 (12). The octapeptides used in this study correspond to the fragments cleaved from the yeast proteins malate dehydrogenase (octMdh1) and succinate dehydrogenase (octSdh1) and were specifically degraded by OOP (Table 1). Even though processing by Oct1 and octapeptide generation has not been demonstrated in plants, these results open the interesting possibility that OOP (and possibly OOP orthologs in other organisms) are responsible for the degradation of octapeptides produced in mitochondria.
OOP Functions Beyond Targeting Peptide Degradation.
The function of OOP in the endosymbiotic organelles might not be restricted to the degradation of targeting peptides. In fact, several lines of evidence suggest that OOP might be involved in the degradation of peptides derived from other proteolytic events. In our assays OOP cleaved peptides not related to presequences (SytII and Cyt1p; Table 1). Moreover, 21 peptides generated by breakdown of 10 different bacterial proteins were bound to the active site of OOP overexpressed in bacteria and identified by MS (Fig. 4D). The lack of a strict sequence requirement, as revealed by the OOP structure, allows the cleavage of a wide range of substrates, constrained only by the peptide length.
Within mitochondria and chloroplasts, a network of proteases (Lon, Deg, ClpP, and FtsH) is involved in the degradation of unfolded and damaged proteins, working as a quality control system (45). Proteases such as ClpP and Deg were shown to generate short peptide fragments up to 25 aa in length, with the majority being 5–12 and 9–20 aa, respectively (46, 47). Considering the size restriction for OOP substrates, it is conceivable that OOP also acts downstream of such peptidases, cleaving the resulting fragments into smaller peptides (Fig. 5C).
The network of proteases acting within the endosymbiotic organelles is functionally analogous to the 20S/26S proteasome complex in the cytosol. In Arabidopsis, the proteasome generates short peptides 3–24 aa in length (48). It is possible that CyOP (the cytosolic paralog of OOP) is engaged in the degradation of proteasome-generated peptides as shown for cytosol-localized M3A orthologs in mammals (49).
Experimental Procedures
Plant Material and Cellular Fractionation.
A. thaliana plants (Columbia ecotype) were grown in a climate-controlled chamber at 23 °C and 130 μmol⋅m−2⋅s−1 light with a 16-h light/8-h dark photoperiod. In all cases seeds were stratified for 3 d at 4 °C after sowing before transfer to the light. Total protein extract was isolated from A. thaliana leaves as described in ref. 50. Mitochondria and chloroplasts were isolated from 14-d-old A. thaliana plants, as described in refs. 51 and 52, respectively. Mitoplasts were obtained by osmotic swelling of the intact mitochondria (22). Chloroplasts were fractionated as described in ref. 53. Cytosol fraction was isolated as follows. Fourteen-day-old A. thaliana plants were homogenized in a mortar in a grinding buffer [30 mM Hepes-KOH (pH 7.5), 2 mM EDTA, 300 mM sorbitol, and 1% (wt/vol) PVP 40 (Sigma)]. The homogenate was passed through Miracloth, and the resulting filtrate was centrifuged for 5 min at 2,500 × g. The pellet was discarded, and the supernatant was centrifuged for 20 min at 20,000 × g to pellet the organelles. The supernatant was collected and centrifuged again for 1 h at 60,000 × g. The cytosolic proteins were precipitated using trichloroacetic acid.
Biolistic Transformation and Microscopy.
Biolistic cotransformation of the vectors encoding GFP and RFP fusions was performed on Arabidopsis cell suspensions as previously described (54). For more detailed descriptions, see SI Experimental Procedures.
Protease Activity Measurements.
For the analysis of peptide degradation, samples of the different peptides used (1 μg pF1β2-54 and Aβ1-40; 8 μg pF1β4-15 and pF1β43-53; 4 μg of other peptides) were incubated with 0.5 μg of purified OOP or PreP1 for the indicated time frame in degradation buffer [50 mM Hepes-KOH (pH 7.4), with addition of 10 mM MgCl2 for the PreP1 assays] at 30 °C. After incubation the reactions were stopped by the addition of SDS peptide sample buffer [150 mM Tris⋅HCl (pH 7.0), 4% (wt/vol) SDS, 6% (vol/vol) β-mercaptoethanol, 30% (vol/vol) glycerol, 0.05% Coomassie blue G250] and subsequently were loaded on appropriate SDS polyacrylamide gels. In all cases gels were subjected to formaldehyde fixation for 1 h before staining with Coomassie Brilliant Blue (Sigma). The bands were quantified using MultiGauge software, and experiments were performed in duplicate. Amyloid-β 1-28, amyloid-β 1-40, insulin-β, and insulin were purchased from Sigma; pF1β 2-54 was produced as described in ref. 55, and the production of pF1β 4-15, pF1β 43-53, pL29, pACD1, tpACS 1-11, and tpACS 1-19 is described in SI Experimental Procedures. The peptides pCox4, Cyt1p 294–309, octMdh1, and octSdh1 were a kind gift from Chris Meisinger (University of Freiburg, Freiburg, Germany). Syt-II (residues 40–60) was synthesized by JPT Peptide Technologies Gmgh, and the peptides pThrRS 1-19 and tpACS 1-26 were synthesized by GenicBio. In the Substrate V assay, OOP (wild type or variants, 0.02 μg per assay) was mixed with 1 μg substrate V (R&D Systems) in degradation buffer (50 mM Hepes pH 7.4, reaction volume of 100 μL), and the increase in fluorescence (excitation, 327 nm; emission, 395 nm) was recorded immediately in a plate reader (SpectraMax Gemini). Results shown are the average of three independent measurements.
Supplementary Material
Acknowledgments
We thank Prof. Chris Meisinger (University of Freiburg, Germany) for providing the peptides octMdh1, octSdh1, Cyt1p, and pCox4 and the beamline scientists at the MAX IV Laboratory (Lund University), the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung, and the Swiss Light Source, Switzerland, for their support. This study was supported by grants from the Swedish Research Council (to E.G., P.S., J.L. and Ü.L.), the Wenner-Gren Foundation, the Swedish Foundation for Strategic Research, and by Magnus Bergvall (P.S.), and the Australian Research Council Centre of Excellence (J.W.). M.W.M. was supported by a Postdoctoral Fellowship from the Australian Research Council. R.P.-A.B. was supported by an EMBO Long-Term Fellowship and Marie Curie Actions. Cooperation between the E.G. and J.W. laboratories was supported by the Swedish Foundation for International Cooperation in Research and Higher Education.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4KA7 (E572Q mutant) and 4KA8 (wild-type organellar oligopeptidase)]. The raw mass spectrometry data have been deposited in the ProteomeXchange Consortium, http://proteomecentral.proteomexchange.org/cgi/GetDataset, via the PRIDE partner repository (dataset identifier PXD000444).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307637110/-/DCSupplemental.
References
- 1.Vierstra RD. Proteolysis in plants: Mechanisms and functions. Plant Mol Biol. 1996;32(1-2):275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 2.Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011;16(9):489–498. doi: 10.1016/j.tplants.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 3.García-Lorenzo M, Sjödin A, Jansson S, Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adam Z, Clarke AK. Cutting edge of chloroplast proteolysis. Trends Plant Sci. 2002;7(10):451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- 5.Kwasniak M, Pogorzelec L, Migdal I, Smakowska E, Janska H. Proteolytic system of plant mitochondria. Physiol Plant. 2012;145(1):187–195. doi: 10.1111/j.1399-3054.2011.01542.x. [DOI] [PubMed] [Google Scholar]
- 6.Schuhmann H, Adamska I. Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol Plant. 2012;145(1):224–234. doi: 10.1111/j.1399-3054.2011.01533.x. [DOI] [PubMed] [Google Scholar]
- 7.Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: From inventory to function. Cell. 2008;134(1):22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 8.Millar AH, Heazlewood JL, Kristensen BK, Braun HP, Møller IM. The plant mitochondrial proteome. Trends Plant Sci. 2005;10(1):36–43. doi: 10.1016/j.tplants.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 9. Zybailov B, et al. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3(4):e1994. [DOI] [PMC free article] [PubMed]
- 10.Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833(2):360–370. doi: 10.1016/j.bbamcr.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson AC, Sjoling S, Glaser E. The ubiquinol cytochrome c oxidoreductase complex of spinach leaf mitochondria is involved in both respiration and protein processing. Biochim Biophys Acta. 1994;1186(3):221–231. [Google Scholar]
- 12.Vögtle FN, et al. Mitochondrial protein turnover: Role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell. 2011;22(13):2135–2143. doi: 10.1091/mbc.E11-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalousek F, Isaya G, Rosenberg LE. Rat liver mitochondrial intermediate peptidase (MIP): Purification and initial characterization. EMBO J. 1992;11(8):2803–2809. doi: 10.1002/j.1460-2075.1992.tb05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter S, Lamppa GK. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol. 1999;147(1):33–44. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter S, Lamppa GK. Determinants for removal and degradation of transit peptides of chloroplast precursor proteins. J Biol Chem. 2002;277(46):43888–43894. doi: 10.1074/jbc.M206020200. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, Taylor NL, Whelan J, Millar AH. Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol. 2009;150(3):1272–1285. doi: 10.1104/pp.109.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmiec B, Glaser E. A novel mitochondrial and chloroplast peptidasome, PreP. Physiol Plant. 2012;145(1):180–186. doi: 10.1111/j.1399-3054.2011.01531.x. [DOI] [PubMed] [Google Scholar]
- 18.Moberg P, et al. Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J. 2003;36(5):616–628. doi: 10.1046/j.1365-313x.2003.01904.x. [DOI] [PubMed] [Google Scholar]
- 19.Stahl A, et al. Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. J Biol Chem. 2002;277(44):41931–41939. doi: 10.1074/jbc.M205500200. [DOI] [PubMed] [Google Scholar]
- 20.Bhushan S, et al. Catalysis, subcellular localization, expression and evolution of the targeting peptides degrading protease, AtPreP2. Plant Cell Physiol. 2005;46(6):985–996. doi: 10.1093/pcp/pci107. [DOI] [PubMed] [Google Scholar]
- 21.Johnson KA, et al. The closed structure of presequence protease PreP forms a unique 10,000 Angstroms3 chamber for proteolysis. EMBO J. 2006;25(9):1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alikhani N, et al. Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J Mol Biol. 2011;410(3):400–410. doi: 10.1016/j.jmb.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Falkevall A, et al. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281(39):29096–29104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- 24.Novak P, Dev IK. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol. 1988;170(11):5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Büchler M, Tisljar U, Wolf DH. Proteinase yscD (oligopeptidase yscD). Structure, function and relationship of the yeast enzyme with mammalian thimet oligopeptidase (metalloendopeptidase, EP 24.15) Eur J Biochem. 1994;219(1-2):627–639. doi: 10.1111/j.1432-1033.1994.tb19978.x. [DOI] [PubMed] [Google Scholar]
- 26.Kato A, et al. Targeting of endopeptidase 24.16 to different subcellular compartments by alternative promoter usage. J Biol Chem. 1997;272(24):15313–15322. doi: 10.1074/jbc.272.24.15313. [DOI] [PubMed] [Google Scholar]
- 27.Kambacheld M, Augustin S, Tatsuta T, Müller S, Langer T. Role of the novel metallopeptidase Mop112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J Biol Chem. 2005;280(20):20132–20139. doi: 10.1074/jbc.M500398200. [DOI] [PubMed] [Google Scholar]
- 28.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123(Pt 22):3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 29.Hugosson M, Andreu D, Boman HG, Glaser E. Antibacterial peptides and mitochondrial presequences affect mitochondrial coupling, respiration and protein import. Eur J Biochem. 1994;223(3):1027–1033. doi: 10.1111/j.1432-1033.1994.tb19081.x. [DOI] [PubMed] [Google Scholar]
- 30.Nicolay K, Laterveer FD, van Heerde WL. Effects of amphipathic peptides, including presequences, on the functional integrity of rat liver mitochondrial membranes. J Bioenerg Biomembr. 1994;26(3):327–334. doi: 10.1007/BF00763104. [DOI] [PubMed] [Google Scholar]
- 31.van ’t Hof R, Demel RA, Keegstra K, de Kruijff B. Lipid-peptide interactions between fragments of the transit peptide of ribulose-1,5-bisphosphate carboxylase/oxygenase and chloroplast membrane lipids. FEBS Lett. 1991;291(2):350–354. doi: 10.1016/0014-5793(91)81318-3. [DOI] [PubMed] [Google Scholar]
- 32.Zardeneta G, Horowitz PM. Analysis of the perturbation of phospholipid model membranes by rhodanese and its presequence. J Biol Chem. 1992;267(34):24193–24198. [PubMed] [Google Scholar]
- 33.Yang MJ, et al. The MAS-encoded processing protease of yeast mitochondria. Interaction of the purified enzyme with signal peptides and a purified precursor protein. J Biol Chem. 1991;266(10):6416–6423. [PubMed] [Google Scholar]
- 34.Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291(5511):2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- 35.Kleffmann T, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14(5):354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 36.Bhushan S, et al. Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep. 2003;4(11):1073–1078. doi: 10.1038/sj.embor.7400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comellas-Bigler M, Lang R, Bode W, Maskos K. Crystal structure of the E. coli dipeptidyl carboxypeptidase Dcp: Further indication of a ligand-dependent hinge movement mechanism. J Mol Biol. 2005;349(1):99–112. doi: 10.1016/j.jmb.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Brown CK, et al. Structure of neurolysin reveals a deep channel that limits substrate access. Proc Natl Acad Sci USA. 2001;98(6):3127–3132. doi: 10.1073/pnas.051633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray K, Hines CS, Coll-Rodriguez J, Rodgers DW. Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization. J Biol Chem. 2004;279(19):20480–20489. doi: 10.1074/jbc.M400795200. [DOI] [PubMed] [Google Scholar]
- 40. Creighton T (1992) Proteins: Structures and Molecular Properties (W. H. Freeman, New York), 2nd Ed.
- 41.Gomis-Rüth FX. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol. 2008;43(5):319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- 42.Berntsson RP, et al. The structural basis for peptide selection by the transport receptor OppA. EMBO J. 2009;28(9):1332–1340. doi: 10.1038/emboj.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrie C, Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833(2):253–259. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 44.Kmiec B, Urantowka A, Lech M, Janska H. Two-step processing of AtFtsH4 precursor by mitochondrial processing peptidase in Arabidopsis thaliana. Mol Plant. 2012;5(6):1417–1419. doi: 10.1093/mp/sss099. [DOI] [PubMed] [Google Scholar]
- 45.Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi KH, Licht S. Control of peptide product sizes by the energy-dependent protease ClpAP. Biochemistry. 2005;44(42):13921–13931. doi: 10.1021/bi0505060. [DOI] [PubMed] [Google Scholar]
- 47.Krojer T, et al. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci USA. 2008;105(22):7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polge C, et al. Evidence for the existence in Arabidopsis thaliana of the Proteasome Proteolytic Pathway: Activation in response to cadmium. J Biol Chem. 2009;284(51):35412–35424. doi: 10.1074/jbc.M109.035394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saric T, Graef CI, Goldberg AL. Pathway for degradation of peptides generated by proteasomes: A key role for thimet oligopeptidase and other metallopeptidases. J Biol Chem. 2004;279(45):46723–46732. doi: 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 50. Martinez-Garcia JF, Monte E, Quail PH (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20(2):251–257. [DOI] [PubMed]
- 51.Day DA, Neuburger M, Douce R. Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust J Plant Physiol. 1985;12:219–228. [Google Scholar]
- 52.Aronsson H, Jarvis P. A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 2002;529(2-3):215–220. doi: 10.1016/s0014-5793(02)03342-2. [DOI] [PubMed] [Google Scholar]
- 53.Hall M, Mishra Y, Schröder WP. Preparation of stroma, thylakoid membrane, and lumen fractions from Arabidopsis thaliana chloroplasts for proteomic analysis. Methods Mol Biol. 2011;775:207–222. doi: 10.1007/978-1-61779-237-3_11. [DOI] [PubMed] [Google Scholar]
- 54.Carrie C, et al. Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 2009;57(6):1128–1139. doi: 10.1111/j.1365-313X.2008.03745.x. [DOI] [PubMed] [Google Scholar]
- 55.Pavlov PF, Moberg P, Zhang XP, Glaser E. Chemical cleavage of the overexpressed mitochondrial F1beta precursor with CNBr: A new strategy to construct an import-competent preprotein. Biochem J. 1999;341(Pt 1):95–103. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.