Significance
AdoMet radical enzymes harness the power of radical-based chemistry to carry out complex chemical transformations. The structure of butirosin biosynthethic enzyme BtrN reveals both unforeseen differences and surprising similarities compared with other members of this rapidly expanding enzyme superfamily. In particular, variations in how BtrN binds S-adenosyl-L-methionine (AdoMet) warrant redefinition of the core fold responsible for adenosyl-radical generation whereas similarities in how BtrN binds an auxiliary iron–sulfur cluster provide the basis for assignment of a previously undescribed structural motif.
Keywords: radical SAM enzyme, iron-sulfur cluster fold, twitch domain
Abstract
The 2-deoxy-scyllo-inosamine (DOIA) dehydrogenases are key enzymes in the biosynthesis of 2-deoxystreptamine–containing aminoglycoside antibiotics. In contrast to most DOIA dehydrogenases, which are NAD-dependent, the DOIA dehydrogenase from Bacillus circulans (BtrN) is an S-adenosyl-l-methionine (AdoMet) radical enzyme. To examine how BtrN employs AdoMet radical chemistry, we have determined its structure with AdoMet and substrate to 1.56 Å resolution. We find a previously undescribed modification to the core AdoMet radical fold: instead of the canonical (β/α)6 architecture, BtrN displays a (β5/α4) motif. We further find that an auxiliary [4Fe-4S] cluster in BtrN, thought to bind substrate, is instead implicated in substrate–radical oxidation. High structural homology in the auxiliary cluster binding region between BtrN, fellow AdoMet radical dehydrogenase anSME, and molybdenum cofactor biosynthetic enzyme MoaA provides support for the establishment of an AdoMet radical structural motif that is likely common to ∼6,400 uncharacterized AdoMet radical enzymes.
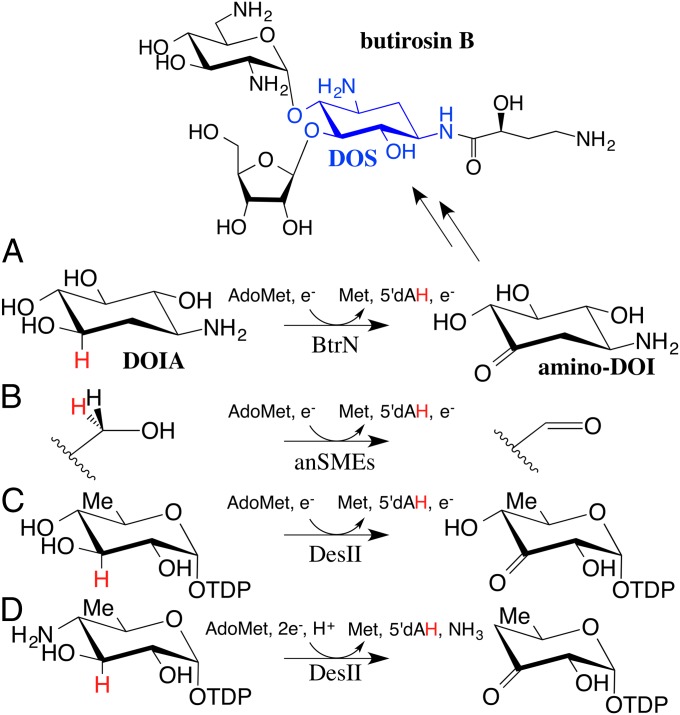
Due to mounting antibiotic resistance, the discovery and/or modification of antibiotic compounds to fight bacterial infection is a vital task of our time (1). Understanding antibiotic biosynthesis is an important first step in being able to engineer a new wave of therapeutic molecules. Aminoglycosides are a class of antibiotics that inhibit protein synthesis by interacting with the 30S subunit of the bacterial ribosome and have had considerable success in the clinical setting (2). Most FDA-approved aminoglycosides, including neomycin, gentamicin, and kanamycin, share a common glucose-6-phosphate–derived 2-deoxystreptamine (DOS) structural core (3) (blue in Scheme 1). Although the majority of aminoglycoside-producing bacteria use similar reaction sequences to biosynthesize this DOS core, including the penultimate two-electron oxidation of 2-deoxy-scyllo-inosamine (DOIA) to amino-dideoxy-scyllo-inosose (amino-DOI) (Scheme 1, A), identification of the enzymes responsible has not always been straightforward. For example, the btrE gene product in the butirosin B producing Bacillus circulans was proposed to be an NAD-dependent enzyme responsible for the generation of amino-DOI, as in other aminoglycoside pathways (4). This hypothesis proved incorrect upon further sequence and biochemical analysis (5, 6). Instead, Yokoyama et al. found that production of amino-DOI required another enzyme, BtrN, an S-adenosyl-l-methionine (AdoMet, SAM) radical dehydrogenase (5). Here, we report structures of catalytic and noncatalytic forms of BtrN, providing a structural basis for the unique reaction it performs.
Scheme 1.
Butirosin B biosynthesis and AdoMet radical dehydrogenase activity in (A) BtrN, (B) anSMEs, and (C) DesII; (D) the native DesII deaminase activity.
The AdoMet radical enzyme family catalyzes a diverse array of radical-based reactions, including sulfur insertions, complex chemical transformations and rearrangements, DNA and RNA modifications, and, in the case of BtrN, dehydrogenation (7). BtrN is one of two known members of the dehydrogenase subfamily of AdoMet radical enzymes, joining the anaerobic sulfatase maturating enzyme (anSME) family, which performs a two-electron oxidation of a serine or cysteine to generate a formylglycine residue on a sulfatase protein substrate (8, 9) (Scheme 1, B). BtrN and the anSMEs are thought to use identical reaction mechanisms (10). Unlike anSMEs, BtrN is an AdoMet radical carbohydrate-tailoring enzyme with a substrate similar to that of DesII, a deaminase involved in the biosynthesis of TDP-d-desosamine (11) (Scheme 1, D). BtrN, DesII, and anSMEs, like other AdoMet radical family members, require a [4Fe-4S] cluster, a molecule of AdoMet, and a reducing equivalent to initiate turnover (5). Three cysteines, arranged in a CX3CX2C motif, and AdoMet ligate this required [4Fe-4S] cluster (12–14). Electron transfer from the cluster to AdoMet causes homolysis of a carbon–sulfur bond in AdoMet, forming methionine and a 5′-deoxyadenosyl radical (5′dA•). This radical species subsequently abstracts a hydrogen atom from substrate, resulting in 5′-deoxyadenosine (5′dAH) and a substrate-based radical. Members of this superfamily display very similar structural features for cluster and AdoMet binding, leading to a description of an “AdoMet radical core” (15, 16). Outside of this core, however, very little structural similarity is observed, presumably leading to the diverse chemistry performed by members of this superfamily.
In BtrN, hydrogen atom abstraction by 5′dA• results in a substrate-based radical intermediate localized at position C3 (5, 17). Subsequent deprotonation and oxidation of this intermediate yields the amino-DOI product (Scheme 1, A). Thus, catalysis requires both a proton and an electron acceptor. Interestingly, both AdoMet radical dehydrogenases BtrN and anSMEs house [4Fe-4S] clusters in addition to the one required for AdoMet homolysis (10). BtrN binds one additional cluster (18) whereas the anSMEs bind two (9, 19), suggesting that at least one auxiliary (Aux) cluster is required for AdoMet radical dehydrogenase chemistry (18). Direct ligation of substrate to an Aux cluster became an attractive hypothesis for the role of these clusters, as it would aid in both deprotonation and oxidation of the substrate intermediate (9, 18). However, the recently determined structure of anSME from Clostridium perfringens (anSMEcpe) solved in complex with a peptide substrate shows that these Aux clusters are fully ligated by cysteine residues from the protein and do not play a direct role in substrate binding (20).
These two Aux clusters in anSMEs are surrounded by a C-terminal sequence motif, “CX9–15GX4C—gap—CX2CX5CX3C—gap—C” (21), that is referred to as a SPASM motif or domain. SPASM motifs are found in more than 280 AdoMet radical enzymes, all of which are putatively involved in the maturation of ribosomally translated peptides (21). The name SPASM derives from the biochemically characterized members of this subfamily, AlbA (22), PqqE (23), anSMEs (9, 19), and MtfC (24), involved in subtilosin A, pyrroloquinoline quinone, anaerobic sulfatase, and mycofactocin maturation, respectively. Surprisingly, the anSME structure revealed similarities between the SPASM domain (20) and the Aux cluster-binding domain of molybdopterin biosynthetic enzyme MoaA, an AdoMet radical enzyme with a nonpeptide substrate (25, 26). These conserved features include a β hairpin surrounded by iron ligating cysteine positions and followed by a helical region. Due to its partial-SPASM makeup, this substructure was named a “twitch” domain (20). Given that BtrN contains these sequence elements (CX9–15GX4CXn), it was hypothesized to contain its Aux cluster in a twitch domain similar to MoaA (20). Interestingly, the Aux cluster in MoaA has an available iron ligation site that, unlike in anSMEcpe, is used to bind substrate (27, 28). The structures of BtrN, presented below, address both the role and the structure of the Aux cluster-binding region and provide the structure of an AdoMet radical dehydrogenase that acts on a nonpeptide substrate.
Results
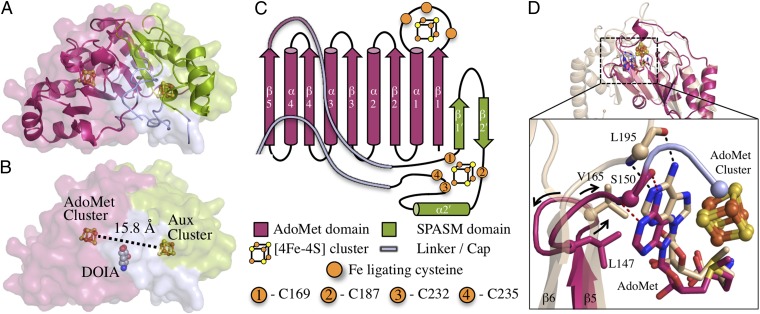
Anomalous signal from a SeMet-derivatized C-terminally His6-tagged BtrN dataset collected at the selenium edge was used to phase an initial structure of the enzyme (Table S1). This structure, solved to 2.02 Å resolution, represents an OPEN conformation of BtrN, as the C-terminal region is extended from the protein core, leaving the active site highly solvent-exposed (Fig. S1). In this OPEN conformation, residues in the α4 helix (121–134), a loop following the AdoMet cluster binding loop (28–32), and a linker region connecting the N-terminal AdoMet domain to the C-terminal auxiliary cluster domain (146–161) are disordered and not included in the model. In addition, electron density for BtrN substrates AdoMet and DOIA, which were included in the crystallization conditions, is not observed in this OPEN structure. To obtain a CLOSED structure of BtrN, a native (non-SeMet) N-terminally His6-tagged construct was used. A structure of this construct, in which the entire BtrN fold was apparent, was solved to 1.56 Å resolution. In this model, a CLOSED conformation of the protein is observed, with the C-terminal region capping the enzyme’s active site. This CLOSED structure includes the disordered regions not present in the OPEN structure and clear electron density for both BtrN substrates. Both the OPEN and CLOSED structures are constituted with two [4Fe-4S] clusters, and, in each case, the Aux cluster is fully protein-ligated. The overall fold of BtrN includes a partial AdoMet radical fold (residues 1–150), an auxiliary cluster binding motif (residues 169–235), and two loop regions—a linker joining the AdoMet and auxiliary cluster domains (residues 151–168) and the C-terminal cap (residues 236–250) (Fig. 1).
Fig. 1.
The BtrN fold. (A) BtrN includes an AdoMet domain (residues 1–150; magenta), an auxiliary cluster domain (residues 169–235; green), and linker regions (residues 151–168 and 236–250; light blue). (B) The BtrN substrate, DOIA (gray), binds between the two clusters. (C) Topology of BtrN (D) BtrN (colored as in A) lacks the α5- and α6-helices and the β6-strand found in all structurally characterized AdoMet radical proteins (15, 16) (represented by anSMEcpe, PDB ID code 4K36, in tan). The GXIXGXXE motif provides a hydrophobic contact to the AdoMet adenine and stabilization to the loop after β5 (V165, T167, and T170 in anSMEcpe). This hydrophobic contact is conserved in BtrN (L147) and stabilized by an upstream residue (H144, Fig. S3). Contacts provided by the backbone atoms of residues in the β6 motif of AdoMet radical enzymes (L195 in anSMEcpe) are conserved in BtrN (S150; Cα’s in spheres) but are found in a loop following the β5-strand. R152 (Cα in light blue sphere) is unique to BtrN (Fig. 2 and Fig. S3).
BtrN Has an Abridged AdoMet Radical Fold.
A typical AdoMet radical fold consists of a conserved (β/α)6 partial TIM barrel fold with a [4Fe-4S] cluster at the top of the barrel in a loop following β1 (15, 16). BtrN initiates in the same manner with C16, C20, and C23 providing three ligands to its AdoMet radical cluster. The full β6/α6 is not conserved in BtrN, which instead has a β5/α4 structure before ending the AdoMet radical domain (Fig. 1). Despite this difference in fold, interactions provided by all four previously described AdoMet binding motifs, including the “GGE” motif, the ribose motif, the “GXIXGXXE” motif, and the β6 motif (16), are for the most part conserved in BtrN (Figs. S2 and S3). In addition, the fold contains a basic residue, H117, that interacts with the carboxyl group of AdoMet, as seen in HemN (29), HydE (30), PylB (31), and anSMEcpe (20), and another hydrogen bond common to nearly all AdoMet radical members, between the N6 position of AdoMet and the carbonyl of Y22, the hydrophobic residue of the AdoMet radical cluster binding motif (16) (Fig. S3). The Y22 interaction is disrupted in the OPEN structure, as the conformation of W21 is altered due to crystal packing, distorting the AdoMet cluster binding loop and possibly contributing to the inability of the OPEN structure to bind AdoMet (Fig. S1).
Although BtrN contains all AdoMet binding motifs, the GXIXGXXE and β6 motifs are unique due to the abridged nature of the enzyme’s AdoMet radical fold. A traditional GXIXGXXE motif provides a hydrophobic contact at the end of β5 to the adenine ring of AdoMet and is stabilized by a backbone interaction with a polar residue in α5. BtrN has no α5, and its β5 strand terminates before reaching the adenine moiety. Following the β5 strand in BtrN, however, a loop containing L147 provides a similar hydrophobic interaction to the adenine as found in the GXIXGXXE motif of other AdoMet radical enzymes (15, 16). Instead of being stabilized by a polar residue from α5, this L147 position is stabilized by a helical turn that immediately follows β5 (Fig. 1D and Fig. S3). The β6 motif in most AdoMet radical enzymes involves two backbone positions of a residue immediately following β6 hydrogen bonding to the adenine N1 and N6 positions (15, 16). BtrN does not have a β6 strand; instead, the backbone amide nitrogen and carbonyl oxygen of S150, the last residue in the protein’s abridged AdoMet radical fold, hydrogen bond to the adenine (2.9 and 3.1 Å from the N1 and N6 positions, respectively) (Fig. 1D). These interactions are like those provided by traditional β6 motif positions but lie in a loop following β5. This loop is able to replace two helices and a strand usually found in the AdoMet radical fold, while exhibiting an AdoMet primary coordination sphere very similar to those seen previously.
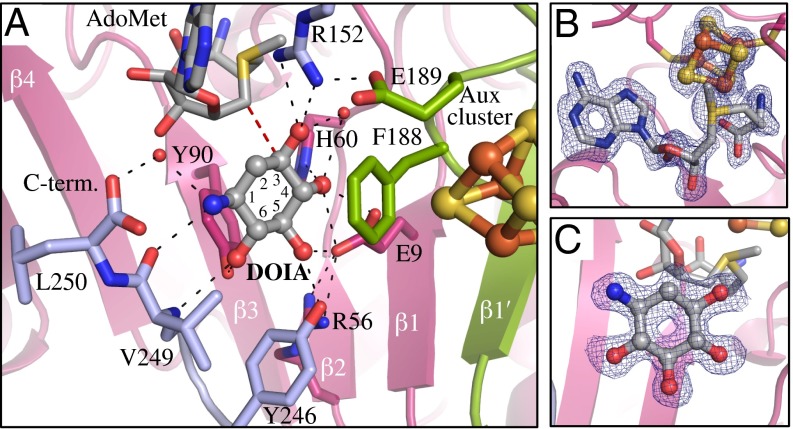
Following this modified “β6 motif” in BtrN, the linker region between the AdoMet radical and C-terminal domains contributes R152. This residue, which is disordered in the OPEN structure, makes an interaction with AdoMet in the CLOSED structure that has not been previously observed. In particular, the guanidinium group of R152 stacks with the adenine base and interacts with the ring oxygen of the cofactor’s ribose moiety (Fig. 2A and Fig. S3).
Fig. 2.
Substrate binding in BtrN. (A) Residues contacting DOIA (gray) (colored as in Fig. 1). Red dashes indicate the separation of the 5′ position of AdoMet and the DOIA hydrogen abstraction site, C3. (B) 2Fo − Fc composite omit electron density countered at 1.0 σ for AdoMet and the AdoMet cluster and (C) substrate.
The C-Terminal Auxiliary Cluster Domain.
The auxiliary cluster domain initiates with C169, the first Aux cluster ligand, and is composed of a β-hairpin and an α-helix (Fig. 1). The β-hairpin lies in between the first (C169) and second (C187) Aux cluster ligands, and β1′ of the hairpin interacts with β1 of the AdoMet domain, continuing the β-sheet. These secondary structural elements and cysteine ligand positions have high structural similarity to the SPASM domain from anSMEcpe (20); residues C169–E223 of BtrN align to residues C261–F311 of anSMEcpe with an rmsd of 1.1 Å. The two proteins diverge after the α helical region. BtrN terminates in a series of loops that contain the final two ligands to its Aux cluster (C232 and C235) whereas anSMEcpe continues, binding an additional [4Fe-4S] cluster (Fig. 3 and Fig. S4). Although this region does not align as well to MoaA (MoaA residues C264–I311; rsmd 4.4 Å), the iron-ligating cysteine positions and secondary structural elements are conserved (Fig. 3 and Fig. S4). Additionally, all three proteins share a conserved glycine residue in the β-hairpin motif between two auxiliary cluster ligands, in a CX8–12GX4C motif (Fig. 3B). The effect of these instances of structural homology is that the AdoMet and Aux cluster positions are nearly identical; AdoMet cluster to Aux cluster distances in the three proteins are 15.8, 16.9, and 16.5 Å for BtrN, anSMEcpe, and MoaA, respectively.
Fig. 3.
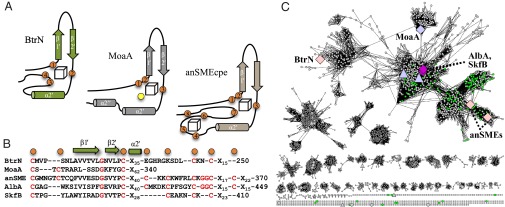
SPASMs and twitches in the AdoMet radical superfamily. (A) Topologies of the Auxiliary cluster domains in BtrN, MoaA, and anSMEcpe (iron-ligating cysteines in orange; MoaA substrate in yellow). (B) C-terminal domains in five auxiliary FeS cluster-containing AdoMet radical proteins. Orange circles represent characterized (BtrN, MoaA, anSMEcpe) or possible (Alba, SkfB) iron-ligating cysteine positions. In green are structural elements found in BtrN, MoaA, and anSMEcpe. In red are conserved elements of the SPASM and twitch domains (20). (C) The AdoMet radical superfamily (nodes are enzymes with 40% sequence identity) using a threshold cutoff of an E-Value of 1 × 10−22. The SPASM/twitch subfamily is enlarged with twitch members (BtrN, MoaA, SkfB) and SPASM members (anSMEs, AlbA) labeled. Figure generated using Cytoscape (44). See Fig. S9 for a more detailed diagram of the superfamily.
DOIA Binding.
Although we were unable to solve a cocrystal structure with DOIA using a C-terminally His6-tagged BtrN construct, the substrate was readily apparent in the electron density from N-terminally His6-tagged BtrN crystals (Fig. 2C). DOIA binds in a hydrophilic pocket between the AdoMet and Aux clusters. The aminoglycoside is in a low energy chair conformation, with all five functional groups in equatorial positions stabilized by numerous hydrogen bonds to amino acid positions spanning the BtrN sequence, from E9 to V249 (Fig. 2A and Fig. S2). The latter interaction is made between the amine group of DOIA and the amide nitrogen and carbonyl of the V249 peptide backbone, one residue from the C terminus of the protein. The C terminus, which itself provides a water-mediated hydrogen bond to DOIA (Fig. 2A), forms a loop that caps the underside of the BtrN active site. This capping function suggests that DOIA binds after AdoMet, consistent with previous substrate inhibition studies (5). Numerous van der Waals and hydrogen-bonding interactions to the AdoMet radical domain stabilize the conformation of this cap (Fig. S5). Although the C-terminally His6-tagged BtrN (used to obtain the OPEN structure) is active (18), we believe that the tag impedes these capping interactions, thereby preventing the formation of a CLOSED complex stable enough for structural characterization.
The C3 position of DOIA (the hydrogen abstraction site) is 3.7 Å away from the 5′ carbon of AdoMet (Fig. 2A), consistent with prior structural characterizations of AdoMet radical enzymes (3.8–4.1 Å) (16). Following hydrogen-atom abstraction, oxidation of the C3 and deprotonation of the C3-OH are required to complete turnover. Possible electron-transfer partners for oxidation are both the AdoMet and Aux clusters. These clusters are 8.6 and 9.6 Å away, respectively, from the C3 position (Fig. S5). For deprotonation, only R152 and a water molecule (hydrogen bonding to another water and the F61 backbone carbonyl) are within hydrogen bonding distance of the C3-OH. Although H60 is within 5 Å, it is hydrogen bonding to the C4-OH, not the C3-OH. R152, however, makes two 3.1 Å hydrogen bonds to the C3-OH position via both η nitrogens (Fig. 2A). R152 also represents the only major difference between the binding pockets of BtrN and anSMEcpe. Although the active sites of these enzymes show little sequence conservation (Fig. S2), a number of residues important for substrate binding are found in similar positions in 3D space (SI Results and Fig. S6).
Discussion
With structural information for BtrN (this work) and anSMEcpe (20), we have completed the structural characterization of the currently identified members of the AdoMet radical dehydrogenase subfamily. Using AdoMet radical chemistry, these enzymes catalyze nearly identical two-electron oxidations (Scheme 1, A and B) and exhibit different variants of the AdoMet radical fold. One fold, the partial TIM barrel fold found in anSMEcpe and common to nearly all AdoMet radical enzymes (15, 16), provides a highly conserved primary coordination sphere around AdoMet. In BtrN, however, this fold lacks 25% of the expected (β/α)6 architecture secondary structure, adopting an abridged, (β5/α4) partial barrel. This interesting deviation contrasts with the striking level of structural similarity between the auxiliary cluster folds of BtrN and anSMEcpe. The two enzymes share an rmsd of 1.1 Å in this region even though BtrN binds one auxiliary (Aux) [4Fe-4S] cluster proximal to its active site whereas anSMEcpe binds two, one near the active site and another near the protein surface. Despite this difference in FeS cluster content, the two proteins were hypothesized to use an active site auxiliary cluster for direct substrate ligation to catalyze similar reactions (9, 18, 21). We have previously indicated that this hypothesis is incorrect in the case of anSMEcpe (20), and we similarly show in this work that the BtrN Aux cluster is fully protein-ligated in the presence and absence of substrate. The DOIA substrate is instead largely stabilized by interactions provided by three of the five BtrN C-terminal residues, potentially explaining the difficulty in crystallizing a substrate-bound structure of a C-terminally His6-tagged BtrN construct.
Direct ligation of DOIA to its Aux cluster was proposed to aid in deprotonating the DOIA C3 hydroxyl group and oxidizing the substrate radical intermediate (18). Without direct ligation to an auxiliary cluster to activate the hydroxyl group, deprotonation before AdoMet-mediated hydrogen abstraction is more difficult due to the ∼16 unit pKa of the C3-OH. Formation of the α-hydroxyalkyl radical by hydrogen abstraction at the C3 position is expected to activate the hydroxyl functional group by decreasing its pKa by approximately five units (32). For subsequent deprotonation, we find only one titratable residue, R152, within 4 Å of the C3-OH group of DOIA. This residue makes contacts, 3.1 Å away, using both η nitrogens of the guanidinium group (Fig. 2A). Whereas arginine sidechains have high pKas (∼12.5) that are usually unsuitable for accepting protons during catalysis, nearby carboxyl side chains have been known to adjust arginine pKas for general base function (33). In BtrN, E189 forms a hydrogen bond/salt bridge with R152 (Fig. 2A), possibly activating it in this manner. Taken together, these results are consistent with R152 accepting a proton from the •C3-OH intermediate during catalysis.
After or concerted with deprotonation, collapse of the •C3-O– radical to the C3=O product necessitates loss of an electron, most likely to one of BtrN’s [4Fe-4S] clusters (5, 17, 18, 34). The AdoMet and Aux clusters are 8.6 and 9.6 Å away, respectively, from the C3 position of DOIA, making them both within range for a suitable electron transfer partner (35) (Fig. S5). A similar situation exists in anSMEcpe, where an oxidation event is required from a cysteinyl Cβ positioned nearly equidistant from its AdoMet and Aux clusters (8.9 and 8.6 Å, respectively) (20). Like in anSMEcpe, we suggest that the role of the Aux cluster in BtrN is to accept an electron from substrate during turnover. In anSMEcpe, the reduced active site Aux cluster is positioned to reduce a second Aux cluster at the protein surface for transfer to a terminal electron acceptor. This pathway appears to be required in anSMEcpe because the large peptidyl substrate in the reaction buries the anSMEcpe active site Aux cluster, necessitating transfer to a more solvent exposed cluster before transfer to a partner protein (20). The small molecule substrate of BtrN, in contrast, does not change the solvent accessibility of the Aux cluster, removing the need for a second Aux cluster. To reoxidize the Aux cluster in BtrN, electron transfer to the AdoMet cluster has been proposed, which would ready this cluster for the next round of catalysis (18). At 15.9 Å, however, the distance between the two clusters is on the longer side for facile electron transfer (35), suggesting that, like in anSMEcpe, electron transfer between the Aux and AdoMet clusters does not occur (20). We therefore propose that, following turnover in BtrN, a partner protein reoxidizes the reduced Aux cluster directly, without an internal electron transfer event.
Another carbohydrate modifying AdoMet radical protein, DesII, also has interesting redox requirements. In its native deaminase reaction (Scheme 1, D), the removal of an amine group from TDP-4-amino-4,6-dideoxy-d-glucose is coupled to the oxidation of an alcohol to a ketone. Whereas this overall reaction is net redox neutral, it requires the reduction of a substrate radical intermediate following hydrogen abstraction (36). Interestingly, DesII can also catalyze a dehydrogenase reaction when given the unnatural substrate, TDP-d-quinovose (Scheme 1, C). This reaction, like BtrN, requires the oxidation of the substrate radical intermediate, which was found by kinetic isotope effects to occur either following or concerted with deprotonation (34). In both cases, the AdoMet cluster is hypothesized to be the electron transfer partner because DesII does not have any auxiliary clusters (11, 37). How DesII accomplishes the same chemistry as BtrN (and anSMEcpe) in the absence of an auxiliary cluster could be explained by the DesII AdoMet cluster’s innate ability to react with substrate, as required in its native reaction. Supplying an unnatural substrate with a poor leaving group presumably prolongs the lifetime of the potent •C3-O– species (E°′ < −1.6 V), reversing the flow of electrons relative to the native reaction (32). In the BtrN and anSMEcpe reactions, nature has made available an auxiliary cluster. In BtrN, this cluster cannot be reduced by titanium citrate (E°′ ∼ −480 mV), implying that it has a low redox potential tuned to match that of the substrate radical intermediate (18). These hypotheses await further electrochemical characterization of the clusters.
As mentioned above, the native DesII reaction requires the elimination of an amine (Scheme 1, D). Although the mechanism for this elimination is unknown, one proposal is that the C4-NH2 group is eliminated in an E1cb-type mechanism, creating an enol radical species that, upon collapse of the ketone, can protonate and reduce to the product (38) (Fig. S7A). This mechanism relies on the ability of the enzyme to activate the leaving group, stabilizing a C4-NH3+ species. A second hypothesis, reminiscent of B12-dependent radical mechanisms, suggests that the amine group migrates to a •C3 intermediate, forming a carbinolamine radical species that readily deaminates forming a ketone (38) (Fig. S7A). This pathway also requires protonation/reduction and relies on a high degree of overlap between the radical and C4-NH3+ orbitals for efficient migration (39). Although it is unknown what configuration substrate assumes in the native DesII reaction, when given the unnatural TDP-d-quinovose substrate, EPR studies have indicated little orbital overlap between these two positions (37). The environment surrounding the C4 functional group is also unknown.
In contrast, the structure of BtrN shows that the enzyme is well designed to avoid an elimination reaction by either mechanism. First, an E1cb-type mechanism is disfavored, as the C4-OH group is not activated for elimination. The functional group is involved in two tight (2.7 Å) hydrogen bonds with the E9 sidechain, which itself is being activated as a hydrogen bond acceptor through interactions with a nearby arginine (R56, Fig. 2A). Second, elimination via a migration-assisted mechanism is not supported as DOIA binds in an equatorial chair conformation with little overlap between the hydrogen abstraction site and the C4-OH bonding orbital (Fig. S8A). The substrate is held in this conformation by multiple hydrogen bonds and van der Waals interactions. Modeling a partial boat DOIA conformation to increase orbital overlap distorts the active-site hydrogen-bonding network and introduces steric clashes with Y90 and F188, two residues positioned above and below the DOIA cyclohexane ring (Fig. S8B). Thus, BtrN disfavors a DesII-like elimination, given either mechanism. The existence of the Aux cluster in BtrN, which we suggest drives oxidation of the radical intermediate, further distinguishes it from DesII.
The Aux cluster binding domains of BtrN, anSMEcpe, and MoaA place them in a large subfamily within the AdoMet radical superfamily. AnSMEcpe is a member of the recently described SPASM subfamily (20, 21), whose members contain two [4Fe-4S] clusters in a C-terminal seven-cysteine motif. BtrN only binds one cluster using a cysteine motif similar to the first half of the SPASM motif, which we have termed a twitch domain. We now conclude that this twitch domain comprises the β1′/β2′ hairpin and the α2′ helix (Fig. 3). In addition to BtrN, MoaA contains a twitch domain with the same C-terminal secondary structural elements as BtrN and anSMEcpe (25) (Fig. 3 and Fig. S4). Mapping these three proteins onto a representative diagram of the AdoMet radical superfamily (Fig. 3C and Fig. S9) suggests that this twitch/SPASM domain will be a common feature to ∼6,400 uncharacterized AdoMet radical enzymes, comprising nearly 16% of the superfamily (40). Although it is difficult to predict iron sulfur cluster content in these twitch/SPASM subfamily members, all currently characterized members contain at least one Aux [4Fe-4S] cluster (8, 9, 18, 20, 22–24, 41).
Contained in this subfamily is the recently described thioether formation class of AdoMet radical enzymes, represented by AlbA (22) and SkfB (41) (Fig. 3C). SkfB has a C-terminal motif with five cysteines, which we suggest forms a twitch domain much like that of BtrN (Fig. 3B). AlbA, a founding member of the SPASM subfamily, contains seven cysteines in its C-terminal domain, in an arrangement much like anSMEcpe (Fig. 3B). Upon removal of the AdoMet cluster, AlbA still has a [4Fe-4S] cluster EPR signal, which was attributed to one auxiliary cluster (22). Based on sequence analysis, we suggest that AlbA binds two auxiliary [4Fe-4S] clusters in a SPASM fold similar to that of anSMEs. Like BtrN and anSMEs, AlbA and SkfB catalyze very similar reactions, namely, the linkage of a peptidyl cysteine thiol to the Cα position of an up or downstream residue on a peptide substrate (Fig. S7B). We propose that this system will be another example of a SPASM containing protein (AlbA) and a twitch domain containing protein (SkfB) catalyzing similar chemistry on different substrates.
The positioning of enzymes in the SPASM/twitch sequence diagram in Fig. 3C is consistent with our Aux cluster architecture designations. The SPASM proteins (anSMEs and green nodes) are located on the opposite end of the SPASM/twitch subfamily sequence space from twitch proteins BtrN and MoaA. AlbA and SkfB, however, map nearly on top of each other, right where we believe the junction exists between SPASM and twitch-containing nodes (Fig. 3C). Functionally, whether the direct ligation feature of MoaA is conserved in SkfB, AlbA, or any other SPASM/twitch subclass member is an interesting issue and awaits further biochemical and structural studies. Regardless, the structures of BtrN provide the third example of a highly conserved Aux cluster binding architecture, suggesting that it will be present in the thioether bond forming enzymes and across this abundant and interesting SPASM/twitch AdoMet radical subclass.
Materials and Methods
The codon-optimized btrN gene was amplified by PCR from the plasmid pBtrNWt, which was previously derived from pET26b (18), and transferred to a pET28a vector. Both the C-terminal and N-terminal constructs of BtrN Wt were produced and purified as previously described (9, 18, 42). The selenomethionine (SeMet) substituted BtrN was produced by BL-21(DE3) with expression in M9 media supplemented with SeMet as previously described (43). Both were crystallized anaerobically using the vapor-diffusion technique. The structure of anSMEcpe was solved using Se anomalous data collected at 0.9792 Å. This dataset and high resolution datasets were collected on beamlines 24-ID-E and 24-ID-C at the Advanced Photon Source. More detailed experimental protocols can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
For helpful discussions, we thank Jennifer Bridwell-Rabb. We thank Dr. Gemma L. Holliday (University of California, San Francisco) for the similarity networks and accompanying information, which are also available from the Structure–Function Linkage Database (http://sfld.rbvi.ucsf.edu/django/superfamily/29/). This work was supported by National Institutes of Health Grants GM-63847 (to S.J.B.) and GM-103268 (to S.J.B.) and by National Science Foundation Grant MCB-0543833 (to C.L.D.). C.L.D. is a Howard Hughes Medical Institute Investigator. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by Award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4M7S and 4M7T).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312228110/-/DCSupplemental.
References
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16(3):430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehmeier UF, Piepersberg W. Enzymology of aminoglycoside biosynthesis-deduction from gene clusters. Methods Enzymol. 2009;459:459–491. doi: 10.1016/S0076-6879(09)04619-9. [DOI] [PubMed] [Google Scholar]
- 4.Kudo F, Yamamoto Y, Yokoyama K, Eguchi T, Kakinuma K. Biosynthesis of 2-deoxystreptamine by three crucial enzymes in Streptomyces fradiae NBRC 12773. J Antibiot (Tokyo) 2005;58(12):766–774. doi: 10.1038/ja.2005.104. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama K, Numakura M, Kudo F, Ohmori D, Eguchi T. Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J Am Chem Soc. 2007;129(49):15147–15155. doi: 10.1021/ja072481t. [DOI] [PubMed] [Google Scholar]
- 6.Kudo F, Eguchi T. Biosynthetic genes for aminoglycoside antibiotics. J Antibiot (Tokyo) 2009;62(9):471–481. doi: 10.1038/ja.2009.76. [DOI] [PubMed] [Google Scholar]
- 7.Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Crit Rev Biochem Mol Biol. 2008;43(1):63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 8.Benjdia A, et al. Anaerobic sulfatase-maturating enzymes, first dual substrate radical S-adenosylmethionine enzymes. J Biol Chem. 2008;283(26):17815–17826. doi: 10.1074/jbc.M710074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grove TL, Lee KH, St Clair J, Krebs C, Booker SJ. In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry. 2008;47(28):7523–7538. doi: 10.1021/bi8004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanz ND, Booker SJ. Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes. Biochim Biophys Acta. 2012;1824(11):1196–1212. doi: 10.1016/j.bbapap.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Szu PH, Ruszczycky MW, Choi SH, Yan F, Liu HW. Characterization and mechanistic studies of DesII: A radical S-adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-desosamine. J Am Chem Soc. 2009;131(39):14030–14042. doi: 10.1021/ja903354k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29(5):1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiscox MJ, Driesener RC, Roach PL. Enzyme catalyzed formation of radicals from S-adenosylmethionine and inhibition of enzyme activity by the cleavage products. Biochim Biophys Acta. 2012;1824(11):1165–1177. doi: 10.1016/j.bbapap.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An anchoring role for FeS clusters: Chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase activating enzyme. J Am Chem Soc. 2002;124(38):11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 15.Vey JL, Drennan CL. Structural insights into radical generation by the radical SAM superfamily. Chem Rev. 2011;111(4):2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowling DP, Vey JL, Croft AK, Drennan CL. Structural diversity in the AdoMet radical enzyme superfamily. Biochim Biophys Acta. 2012;1824(11):1178–1195. doi: 10.1016/j.bbapap.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama K, Ohmori D, Kudo F, Eguchi T. Mechanistic study on the reaction of a radical SAM dehydrogenase BtrN by electron paramagnetic resonance spectroscopy. Biochemistry. 2008;47(34):8950–8960. doi: 10.1021/bi800509x. [DOI] [PubMed] [Google Scholar]
- 18.Grove TL, Ahlum JH, Sharma P, Krebs C, Booker SJ. A consensus mechanism for Radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters. Biochemistry. 2010;49(18):3783–3785. doi: 10.1021/bi9022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjdia A, et al. Anaerobic sulfatase-maturating enzyme—a mechanistic link with glycyl radical-activating enzymes? FEBS J. 2010;277(8):1906–1920. doi: 10.1111/j.1742-4658.2010.07613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman PJ, et al. X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc Natl Acad Sci USA. 2013;110(21):8519–8524. doi: 10.1073/pnas.1302417110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haft DH, Basu MK. Biological systems discovery in silico: Radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J Bacteriol. 2011;193(11):2745–2755. doi: 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flühe L, et al. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat Chem Biol. 2012;8(4):350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 23.Wecksler SR, et al. Pyrroloquinoline quinone biogenesis: Demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry. 2009;48(42):10151–10161. doi: 10.1021/bi900918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haft DH. Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics. 2011;12:21. doi: 10.1186/1471-2164-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci USA. 2004;101(35):12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hover BM, Loksztejn A, Ribeiro AA, Yokoyama K. Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J Am Chem Soc. 2013;135(18):7019–7032. doi: 10.1021/ja401781t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hänzelmann P, Schindelin H. Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci USA. 2006;103(18):6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lees NS, et al. ENDOR spectroscopy shows that guanine N1 binds to [4Fe-4S] cluster II of the S-adenosylmethionine-dependent enzyme MoaA: Mechanistic implications. J Am Chem Soc. 2009;131(26):9184–9185. doi: 10.1021/ja903978u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. EMBO J. 2003;22(23):6214–6224. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolet Y, Amara P, Mouesca JM, Fontecilla-Camps JC. Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins. Proc Natl Acad Sci USA. 2009;106(35):14867–14871. doi: 10.1073/pnas.0904385106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quitterer F, List A, Eisenreich W, Bacher A, Groll M. Crystal structure of methylornithine synthase (PylB): Insights into the pyrrolysine biosynthesis. Angew Chem Int Ed Engl. 2012;51(6):1339–1342. doi: 10.1002/anie.201106765. [DOI] [PubMed] [Google Scholar]
- 32.Hayon E, Simic M. Acid-base properties of free-radicals in solution. Acc Chem Res. 1974;7(4):114–121. [Google Scholar]
- 33.Guillén Schlippe YV, Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch Biochem Biophys. 2005;433(1):266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Ruszczycky MW, Choi SH, Liu HW. EPR-kinetic isotope effect study of the mechanism of radical-mediated dehydrogenation of an alcohol by the radical SAM enzyme DesII. Proc Natl Acad Sci USA. 2013;110(6):2088–2093. doi: 10.1073/pnas.1209446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser CC, Anderson JL, Dutton PL. Guidelines for tunneling in enzymes. Biochim Biophys Acta. 2010;1797(9):1573–1586. doi: 10.1016/j.bbabio.2010.04.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruszczycky MW, Choi SH, Liu HW. Stoichiometry of the redox neutral deamination and oxidative dehydrogenation reactions catalyzed by the radical SAM enzyme DesII. J Am Chem Soc. 2010;132(7):2359–2369. doi: 10.1021/ja909451a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruszczycky MW, Choi SH, Mansoorabadi SO, Liu HW. Mechanistic studies of the radical S-adenosyl-L-methionine enzyme DesII: EPR characterization of a radical intermediate generated during its catalyzed dehydrogenation of TDP-D-quinovose. J Am Chem Soc. 2011;133(19):7292–7295. doi: 10.1021/ja201212f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szu PH, He X, Zhao L, Liu HW. Biosynthesis of TDP-D-desosamine: Identification of a strategy for C4 deoxygenation. Angew Chem Int Ed Engl. 2005;44(41):6742–6746. doi: 10.1002/anie.200501998. [DOI] [PubMed] [Google Scholar]
- 39.Ruszczycky MW, Ogasawara Y, Liu HW. Radical SAM enzymes in the biosynthesis of sugar-containing natural products. Biochim Biophys Acta. 2012;1824(11):1231–1244. doi: 10.1016/j.bbapap.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pegg SC, et al. Leveraging enzyme structure-function relationships for functional inference and experimental design: The structure-function linkage database. Biochemistry. 2006;45(8):2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 41.Flühe L, et al. Two [4Fe-4S] clusters containing radical SAM enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor. J Am Chem Soc. 2013;135(3):959–962. doi: 10.1021/ja310542g. [DOI] [PubMed] [Google Scholar]
- 42.Grove TL, et al. Further characterization of Cys-type and Ser-type anaerobic sulfatase maturating enzymes suggests a commonality in the mechanism of catalysis. Biochemistry. 2013;52(17):2874–2887. doi: 10.1021/bi400136u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long F, et al. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.