Significance
Human health is generally maintained even when important genes are mutated because the human genome is diploid, with two sets of genomic DNA. Thus, various genes are heterozygous, with one functional and one defective allele at their loci. Somatic cells often incur DNA double-strand breaks by the direct effects of DNA-damaging agents, including oxidative stress, and indirectly, such as through DNA replication-fork collapse at damaged sites. Double-strand breaks are repaired through recombination processes, including homologous recombination and nonhomologous end-joining. These processes include the risk of losing a functional allele, called loss of heterozygosity (LOH). This study revealed that a putative antirecombinase, Srs2 DNA helicase, is a key enzyme to promote LOH-less recombination repair of double-strand breaks.
Keywords: SDSA, bubble migration, NHEJ
Abstract
DNA damage alone or DNA replication fork arrest at damaged sites may induce DNA double-strand breaks and initiate homologous recombination. This event can result in a crossover with a homologous chromosome, causing loss of heterozygosity along the chromosome. It is known that Srs2 acts as an antirecombinase at the replication fork: it is recruited by the SUMO (a small ubiquitin-related modifier)-conjugated DNA-polymerase sliding clamp (PCNA) and interferes with Rad51/Rad52-mediated homologous recombination. Here, we report that Srs2 promotes another type of homologous recombination that produces noncrossover products only, in collaboration with PCNA and Rad51. Srs2 proteins lacking the Rad51-binding domain, PCNA-SUMO–binding motifs, or ATP hydrolysis-dependent DNA helicase activity reduce this noncrossover recombination. However, the removal of either the Rad51-binding domain or the PCNA-binding motif strongly increases crossovers. Srs2 gene mutations are epistatic to mutations in the PCNA modification-related genes encoding PCNA, Siz1 (a SUMO ligase) and Rad6 (a ubiquitin-conjugating protein). Knocking out RAD51 blocked this recombination but enhanced nonhomologous end-joining. We hypothesize that, during DNA double-strand break repair, Srs2 mediates collaboration between the Rad51 nucleofilament and PCNA-SUMO and directs the heteroduplex intermediate to DNA synthesis in a moving bubble. This Rad51/Rad52/Srs2/PCNA-mediated noncrossover pathway avoids both interchromosomal crossover and imprecise end-joining, two potential paths leading to loss of heterozygosity, and contributes to genome maintenance and human health.
Mutations in the DNA helicase Srs2 gene cause a hyperrecombination phenotype (1) and increase mitotic crossovers (2). These findings suggest that Srs2 negatively regulates somatic homologous recombination, and thus Srs2 is regarded as an antirecombinase. The Srs2 DNA helicase has a recombinase Rad51-binding motif in its C-terminal region (3, 4), and in vitro analyses have demonstrated that it disrupts Rad51 nucleofilaments formed on single-stranded DNA and inhibits heteroduplex formation mediated by Rad51 recombinase (4, 5). In addition, synthetic heteroduplexes with a D-loop with Rad51 nucleofilaments are efficiently dissociated by Srs2 (6). These biochemical results appear to explain the negative regulation of heteroduplex formation by Srs2 during homologous recombination. Other motifs around the C-terminal tip allow the Srs2 helicase to bind to SUMO-conjugated DNA-polymerase sliding clamp (PCNA-SUMO) (3, 7). The interaction between Srs2 and PCNA-SUMO is essential to prevent the Rad51/Rad52-mediated sister-chromatid exchanges that occur in DNA replication forks stalled at DNA lesions, to channel to the translesion DNA synthesis initiated by Rad6/Rad18-ubiquitinated PCNA (3, 8). This postreplication repair is induced by base-modification types of DNA lesions, caused by DNA scission reagents such as UV, 4NQO, and MMS.
Radiation and reactive oxygen species cause DNA double-strand breaks, which are repaired through various recombination-dependent pathways (Fig. S1 A–C). Double Holliday junction-mediated homologous recombination (Fig. S1A) produces either the crossover (A6) or noncrossover (A5) product, upon resolution of the double Holliday junction (A4) (9, 10). Synthesis-dependent strand-annealing (SDSA)-mediated homologous recombination (Fig. S1B), involving bubble migration (B2–B4), produces only the noncrossover type (B5) product (11, 12). Nonhomologous end-joining (NHEJ) (Fig. S1C) rejoins the double-strand break termini without extensive homologous sequences and generates either imprecise (C4) or precise end-joining products. Srs2 is reportedly involved in crossover regulation (2), SDSA-mediated noncrossover promotion (13), and NHEJ promotion (14). However, the roles and the detailed mechanisms of Srs2 in repair of DNA double-strand breaks are still unclear, in contrast to those in postreplication repair.
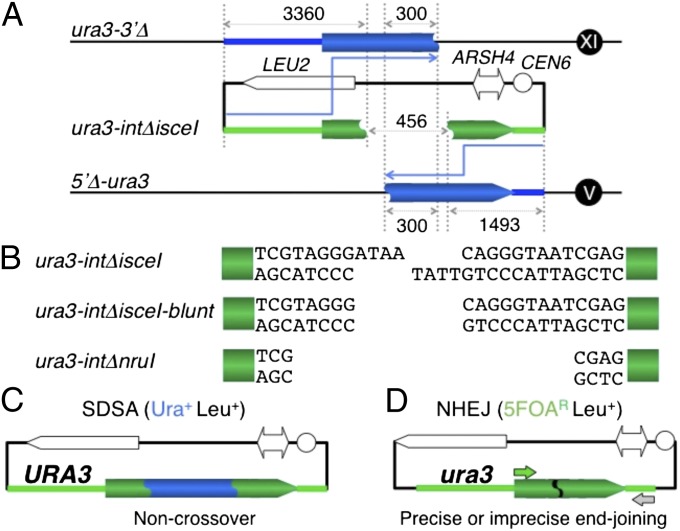
A double-strand break-induced interchromosomal crossover can occur anywhere between the centromere and loci with heterozygous status in G2 somatic cells, and this reaction allows all of the distal loci with heterozygous status to change simultaneously to homozygous status upon cell division (Fig. S1D, Top). Double-strand break-induced imprecise end-joining often generates a deletion on the chromosome, and this reaction changes a locus with heterozygous status, which is in the deletion area, to hemizygous status upon cell division (Fig. S1D, Bottom). Thus, the SDSA-mediated noncrossover and precise end-joining pathways are crucial for maintaining heterozygosity of loci such as tumor suppressor genes in humans (Fig. S1D, Middle). However, the detailed mechanisms of the pathways are still unclear. To elucidate their mechanisms, we developed a unique yeast SDSA/NHEJ assay (Fig. 1) (13) and analyzed the genetic requirements of both pathways. This assay enables quantification of the noncrossover products formed solely by SDSA-mediated homologous recombination (Fig. 1 A and C). As the cleaved gene (ura3-intΔ) does not induce formation of a double Holliday-junction structure from the two discontinuous templates on different chromosomes, ura3-3′Δ (XI) and 5′Δ-ura3 (V) (Fig. 1A), and is repaired by copying both templates and annealing the complementary copies, the noncrossover products are generated solely by SDSA (Fig. 1C). This assay also allows the quantification of the NHEJ products and distinguishes between precise and imprecise end-joining products (Fig. 1 A and D).
Fig. 1.
SDSA/NHEJ assay. (A) For this assay, two different donor-alleles, 5′Δ-ura3 [with a deletion from the promoter (the 221-nt upstream of the initiation codon) to the first base of the 39th codon, residing at the ura3 locus on Chr. V] and ura3-3′Δ [the ura3 ectopic allele bearing only the region from the promoter (the 221-nt upstream of the initiation codon) to the first base of the 139th codon of the URA3 gene, integrated into the AUR1 locus on Chr. XI], were constructed (13). The plasmids with the recipient alleles, ura3-intΔnruI and ura3-intΔisceI (the internal 458-bp deletion of the URA3 gene, sealed with an NruI site and an I-SceI site, respectively, residing within the plasmid with the LEU2 marker), were introduced into the double-template strains with the ura3-3′Δ allele and 5′Δ-ura3 allele, which share 300-bp internal homology. The homologous regions between ura3-intΔ and ura3-3′Δ or 5′Δ-ura3 are 3,360 bp and 1,493 bp in length, respectively. (B) ura3-intΔisceI, ura3-intΔisceI-blunt and ura3-intΔnruI generate 4-nucleotide 3′ tails by I-SceI cleavage for pSDSA/NHEJ (with the I-SceI site) plasmid DNA, blunt ends by T4 DNA polymerase treatment following the I-SceI cleavage, and blunt ends by NruI cleavage for pSDSA/NHEJ (with the NruI site) plasmid DNA, respectively. (C) SDSA products as Ura+ Leu+ transformants bearing the URA3 plasmids (13). The 458-bp gap is repaired via SDSA. (D) NHEJ products as Ura− (5FOAR) Leu+ transformants bearing the ura3 plasmids (13). The retention or deletion of the I-SceI sequence was detected by PCR with the primers (arrows) (13), I-SceI nuclease treatment, and sequence determination as precise or imprecise end-joining, respectively (Table 1).
In this study, we show that the antirecombinase Srs2 helicase promotes SDSA-mediated noncrossover recombination, which requires ATP hydrolysis activity and interactions of Srs2 with Rad51 and PCNA-SUMO. The PCNA modification-related genes POL30 (PCNA), SIZ1 and RAD6, as the SRS2 epistasis group, were required for SDSA-mediated noncrossovers. The Rad51 gene knockout blocked the SDSA-mediated noncrossovers but enhanced NHEJ, which was error-prone. In light of all these findings, we propose that Srs2 collaborates with Rad51 and PCNA to ensure accurate double-strand DNA break repair and thus avoid loss of heterozygosity.
Results
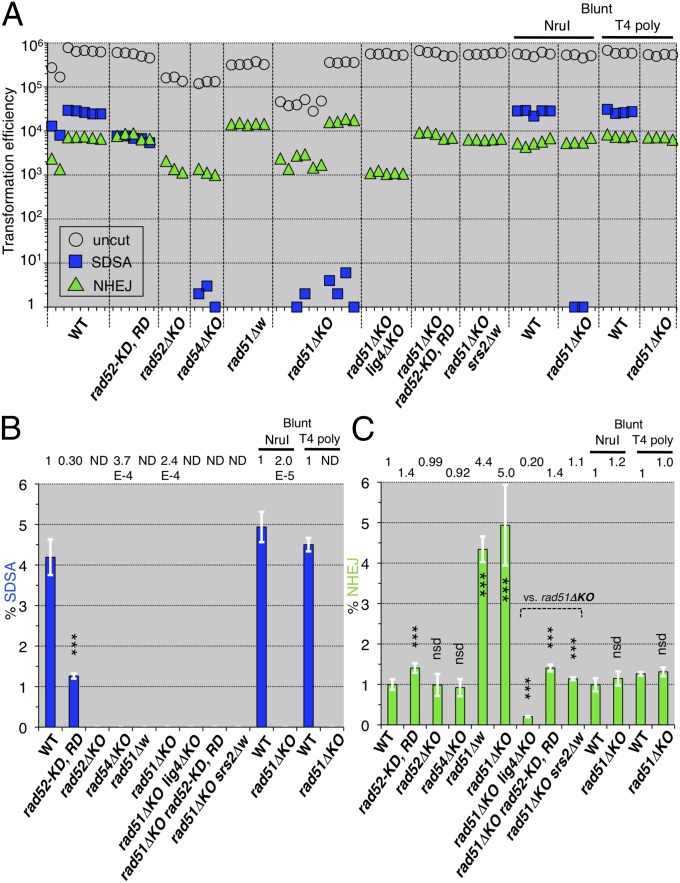
Rad52, Rad51, and Rad54 recombinases are required for heteroduplex formation and processing (15) and must be situated at the branching point to enable double Holliday junction-mediated homologous recombination (either crossover or noncrossover) (9, 10) (Fig. S1A) or SDSA-mediated homologous recombination (noncrossover only) (11, 12) (Fig. S1B). We tested whether these proteins are involved in SDSA-mediated noncrossover recombination (Fig. 1 A and C). The SDSA assay revealed that inactivations of Rad51 and Rad54 caused large reductions (4,200-fold and 2,700-fold vs. wild-type) in SDSA-mediated noncrossover recombination, similar to that of the RAD52 knockout (13) (Fig. 2 A and B), indicating that these heteroduplex forming and processing enzymes are essential to SDSA-mediated noncrossovers. The mutations of two DNA-binding sites in the N-terminal domain of Rad52, rad52-K117D, R148D (SI Materials and Methods) (16), caused a 3.3-fold reduction (vs. wild-type) in SDSA (Fig. 2 A and B). This result indicates that SDSA-mediated noncrossovers require the DNA-binding domain of Rad52.
Fig. 2.
SDSA and NHEJ of homologous recombination-deficient mutants. (A) Leu+ transformation efficiencies with the uncut plasmid bearing ura3-intΔisceI are plotted as the transformation competencies of the cell suspensions (open circles). Ura+ Leu+ and 5-FOAR Leu+ transformation efficiencies with the I-SceI–cut plasmid bearing ura3-intΔisceI are plotted as numbers of SDSA progeny (blue squares) and as numbers of NHEJ progeny (green triangles), respectively (Fig. 1). (B) The normalized frequencies (%) of SDSA events were calculated from the transformation efficiencies (Materials and Methods) and plotted (blue bars). “ND” indicates that no SDSA progeny were detected. (C) The normalized frequencies (%) of the NHEJ events were calculated from the transformation efficiencies (Materials and Methods) and plotted (green bars). rad51ΔKO lacks most of the coding region, and rad51Δw lacks the entire coding region, from the initiation codon to the last sense codon (Tables S1 and S2). rad52-KD, RD shows rad52-K117D, R148D. “NruI” and “T4 poly” indicate the use of the blunt-ended ura3-intΔnruI and ura3-intδΔisceI-blunt DNAs, respectively. The statistical analysis (Table S3): nsd, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed Student’s t test (vs. wild-type except for vs. rad51ΔKO); error bars = SD.
NHEJ analyses with the same linearized plasmid (Fig. 1 A and D) unexpectedly revealed that inactivation of Rad51 caused a four- to fivefold enhancement (vs. wild-type) (Fig. 2 A and C). As the RAD52 and RAD54 knockouts did not generate this enhancement, these results indicate that this NHEJ enhancement specifically appears with the loss of Rad51. Unlike the results with the wild-type strain (13) and the RAD52 knockout, 10–11% of the Rad51 inactivation-induced NHEJ products were resistant to the I-SceI endonuclease and were associated with small or 665-base pair deletions (Table 1), indicating that this type of NHEJ is error-prone. Lig4 (DNA ligase 4) displayed the major NHEJ activities in the same assay (13), and Rad52 promoted NHEJ in linearized vectors (14, 17). Therefore, we determined whether the Rad51 inactivation-induced NHEJ depends on Lig4 and Rad52. The LIG4 knockout reduced this type of NHEJ (Fig. 2 A and C) to the same extent as the single LIG4 knockout (0.18 relative to WT) reported previously (13). The rad52-K117D, R148D mutation also decreased this type of NHEJ to the same extent as the single rad52-K117D, R148D mutation (Fig. 2 A and C). These results indicate the requirement of Lig4 and the Rad52 DNA-binding domain for Rad51 inactivation-induced NHEJ. Furthermore, we tested whether Rad51 inactivation-induced NHEJ requires staggered ends at the I-SceI–created double-strand break. When we used blunt-ended DNA, generated by T4 DNA polymerase treatment following I-SceI cleavage or NruI digestion of another assay plasmid (Fig. 1B), no enhancement was detected (Fig. 2 A and C). Rad52 possesses single-strand DNA annealing activity (18). Taken together, these results suggest that Rad52 promotes single-strand annealing between the I-SceI–created 3′-staggered ends or the microhomologies as shown in Table 1.
Table 1.
Rad51 inactivation-induced NHEJ products
| Relevant genotype | WT* | rad52ΔKO | rad51ΔKO | rad51Δw |
| 5FOAR Leu+ transformants analyzed | 144 | 120 | 132 | 120 |
| I-SceI–resistant products (%) | 0 (0) | 1 (0.83) | 14 (11) | 12 (10) |
| TCGTAGGG ATAa|CAGGGTAATCGAG | 0 | 1 | 3 | 4 |
| AGCATCCC|TATt GTCCCATTAGCTC | ||||
| TCGTAGGG AtAA|CAGGGTAATCGAG | 0 | 0 | 1 | 2 |
| AGCATCCC|TaTT GTCCCATTAGCTC | ||||
| TCGTAGGG AtaA|CAGGGTAATCGAG | 0 | 0 | 4 | 2 |
| AGCATCCC|TatT GTCCCATTAGCTC | ||||
| TCGTAGGG Ataa|CAGGGTAATCGAG | 0 | 0 | 2 | 1 |
| AGCATCCC|Tatt GTCCCATTAGCTC | ||||
| TCGTAGGG ataa|CAGGGTAATCGAG | 0 | 0 | 3 | 1 |
| AGCATCCC|tatt GTCCCATTAGCTC | ||||
| TCGTAGggataa|cagggTAATCGAG | 0 | 0 | 0 | 1 |
| AGCATCcc|tattgtcccATTAGCTC | ||||
| AGAACAAaaacctcgtagggataa|c | 0 | 0 | 1 | 1 |
| TCTTGTTtttggagcatccc|tattg | ||||
| agggtaat(625)actccaaagaacaa | ||||
| tcccatta(625)tgaggtttcttgtt |
Seven types of I-SceI–resistant sequences are shown, as the inserted 25-bp sequence (except for the last sequence) including the I-SceI sequence (italicized letters) and the portion lost (lowercase letters with strikethrough) due to imprecise NHEJ. The vertical lines indicate the I-SceI cleavage. Underlined sequences are putatively base pair annealed during imprecise NHEJ (Fig. S1C). The numbers of each type of imprecise NHEJ are for each strain. ΔKO shows alleles lacking most of the coding region, and Δw shows those lacking the entire coding region, from the initiation codon to the last sense codon.
The data for the wild type are obtained from a previous report (13).
The Srs2 helicase promotes NHEJ in linearized vectors (14). We tested whether Srs2 is involved in the Rad51 inactivation-induced NHEJ. The entire deletion of SRS2 completely inhibited this type of NHEJ (Fig. 2 A and C). This result suggests that Rad51 prohibits the NHEJ-promoting activity of Srs2 and directs Srs2 toward SDSA-mediated noncrossovers. This suggestion is supported by the previous finding that Srs2 promoted an SDSA pathway in the presence of RAD51 (13), but the roles of Srs2 in the Rad51-directed SDSA pathway are unknown.
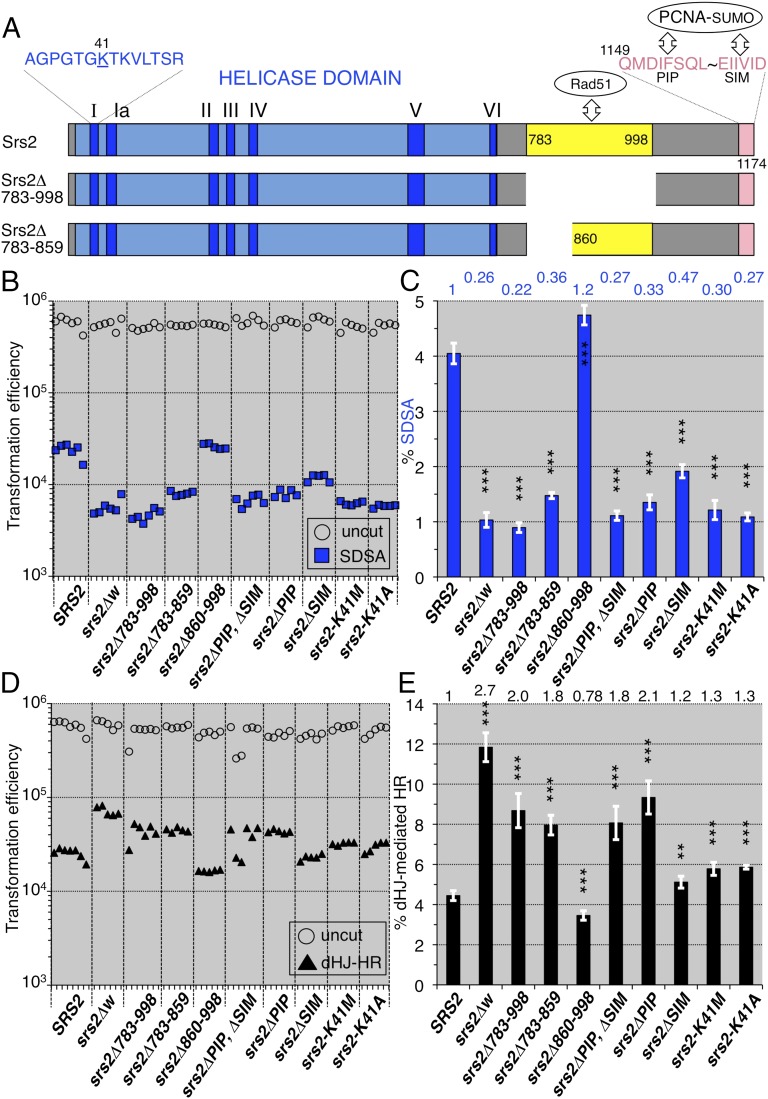
We analyzed the requirement of each functional domain of Srs2 (Fig. 3A) for SDSA-mediated noncrossovers. The removal of the Rad51-binding domain (residues 783–998), identified previously (3, 4), srs2Δ783–998, and a smaller deletion, srs2Δ783–859, generated 4.5-fold and 2.7-fold reductions [vs. wild-type (SRS2)] in SDSA, which are of a similar extent to the deletion of the entire SRS2 coding region, srs2Δw (Fig. 3 B and C). In contrast, in targeted integration via double Holliday junction-mediated homologous recombination, a process that mostly produces crossovers (Fig. S2), srs2Δ783–998 and srs2Δ783–859 showed 2.0-fold and 1.8-fold enhancements (vs. SRS2), respectively, which are of a similar extent to the enhancement caused by srs2Δw (Fig. 3 D and E). The deletion of the distal region of the residues 783–998, srs2Δ860–998, had no strong effect in either assay, indicating that residues 783–859 are important for double-strand break repairs (Fig. 3 C and E). These results suggest that Srs2 acts on the Rad51 nucleofilament through the region that includes residues 783–859, resulting in SDSA-mediated noncrossovers and preventing crossovers.
Fig. 3.
SDSA and double Holliday junction-mediated homologous recombination (dHJ-mediated HR) of various srs2 mutants. (A) The Srs2 helicase domain (blue box), including seven consensus motifs (dark blue), and the locations of the Rad51-binding domain (783-998), the N-terminal half of the Rad51-binding domain (783-859), PIP motif (1149-1156), and SIM motif (1169-1174). The 41st residue (lysine) is substituted with methionine and alanine in the srs2-K41M and srs2-K41A mutants, respectively. (B) Leu+ and Ura+ Leu+ (SDSA progeny) transformation efficiencies with uncut and I-SceI–cut plasmids possessing ura3-intΔisceI, respectively. (C) The normalized frequencies (%) of the SDSA events were calculated from the transformation efficiencies (Materials and Methods) and plotted (blue bars). (D) Targeted integration completed by double Holliday junction-mediated homologous recombination (Fig. S2). The plot shows the AurR transformation efficiencies with the uncut pRS315-AurR plasmid as the transformation competencies of the cell suspensions (open circles), and the AurR transformation efficiencies with the StuI-cut pAUR101 plasmid as the numbers of double Holliday junction-mediated homologous recombination progeny (closed triangles). (E) The normalized frequencies (%) of the double Holliday junction-mediated homologous recombination were calculated from the transformation efficiencies (Materials and Methods) and plotted (black bars). Statistical analysis (Tables S4 and S5): nsd, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed Student’s t test (vs. wild-type); error bars = SD.
The removal of the PCNA-binding motif (PIP-like sequence; 1149–1156) (Fig. 3A), srs2ΔPIP, generated a 3.0-fold reduction (vs. SRS2) in SDSA-mediated noncrossovers (Fig. 3 B and C). As this mutant protein still possesses the Rad51-binding domain, it is expected to inhibit crossovers in a similar manner to the wild-type. However, the PIP deletion unexpectedly showed a 2.1-fold enhancement (vs. SRS2) in crossovers, similar to srs2Δ783–998 (Fig. 3 D and E). These results suggest that the interaction between Srs2 and PCNA is essential in promoting the SDSA pathway and preventing the crossover pathway. Similar results were obtained from double-strand break repair assays with srs2ΔPIP, ΔSIM, which lacks both the PIP and SUMO-interacting motifs (SIM) (1169–1174) (Fig. 3 A, C, and E). Deletion of the SIM sequence, srs2ΔSIM, alone caused a 2.1-fold reduction (vs. SRS2) in SDSA, which is a smaller change than that induced by srs2ΔPIP (Fig. 3 B and C), but no strong enhancement in crossovers, unlike srs2ΔPIP (Fig. 3 D and E). These results indicate that the fully active interaction between Srs2 and PCNA-SUMO is necessary for the promotion of SDSA.
The Srs2 protein has DNA helicase activity with 3′→5′ polarity, which is fueled by ATP hydrolysis (19). We tested whether the ATP hydrolysis-dependent DNA helicase activity of Srs2 is necessary to promote SDSA and prevent crossovers. The mutations of the ATP-binding motif, srs2-K41M and srs2-K41A (SI Materials and Methods) that abolished DNA helicase activity of Srs2 without affecting its DNA- and Rad51-binding activities (20) caused a three- to fourfold reduction (vs. SRS2) in SDSA (Fig. 3 B and C) but did not strongly enhance crossovers (Fig. 3 D and E). These results indicate that the DNA-helicase activity of Srs2 is necessary for SDSA-mediated noncrossover recombination.
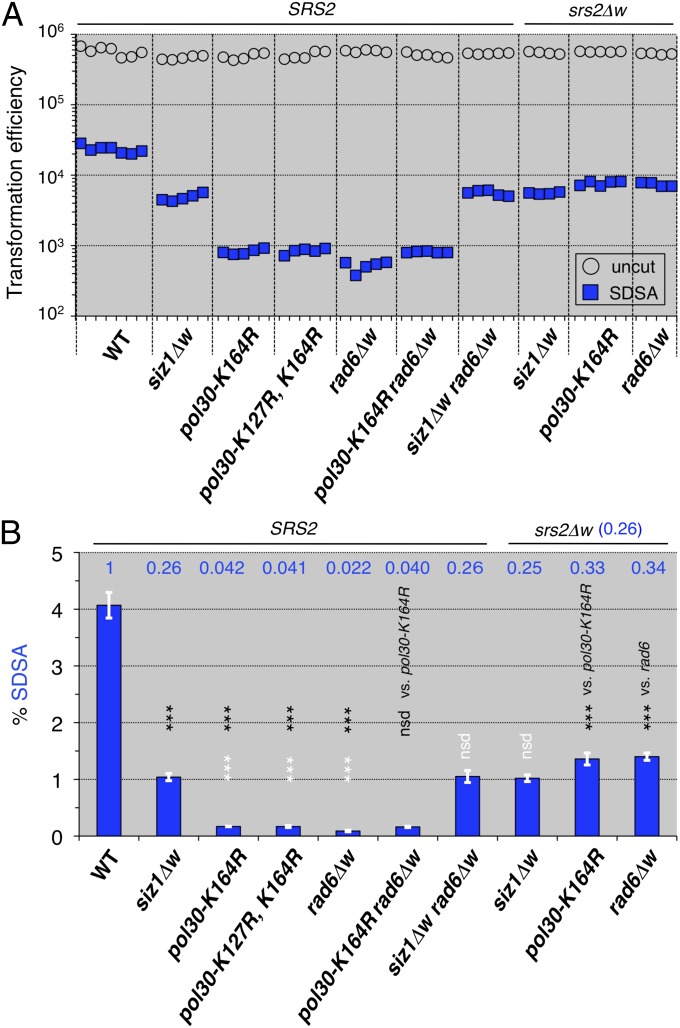
We attempted to identify the role of the interactions between PCNA and Srs2, by evaluating the effects of the PCNA modification-deficient mutations on SDSA-mediated noncrossovers. The SIZ1 deletion (a PCNA-SUMOylation–deficient mutant) (21) decreased SDSA, but the pol30 (pcna)-K127R, K164R and pol30 (pcna)-K164R mutants (the PCNA-modification site mutants) (22) (SI Materials and Methods) and the RAD6 deletion (a PCNA ubiquitination-deficient mutant) (22) decreased SDSA-mediated noncrossovers more effectively than the SIZ1 deletion (Fig. 4 A and B). The results of the pol30 (pcna) rad6 double mutant are similar to that of each single mutant, suggesting the requirement for PCNA ubiquitination for SDSA-mediated noncrossovers (Fig. 4 A and B). The results of the siz1 rad6 double mutant are similar to that of the siz1 single mutant, indicating that siz1 completely suppresses the negative effect of the rad6 mutation on SDSA-mediated noncrossovers. The results of the double mutants of siz1, pol30 (pcna), or rad6 with srs2Δw are similar to that of the srs2 single mutant (0.26 relative to SRS2 in Fig. 3C), indicating that srs2 completely suppresses the negative effect of each of the siz1, pol30 (pcna), and rad6 mutations on SDSA-mediated noncrossover recombination (Fig. 4 A and B). Taken together, these results suggest that SIZ1, POL30 (PCNA), and RAD6 are included in the SRS2 epistasis group, with respect to the Rad51/Rad52-mediated SDSA pathway.
Fig. 4.
SRS2 is epistatic to SIZ1, POL30 (PCNA), and RAD6 (PCNA modification-related genes), with respect to SDSA. (A) Leu+ and Ura+ Leu+ (SDSA progeny) transformation efficiencies with uncut and I-SceI–cut plasmids possessing ura3-intΔisceI, respectively. (B) The normalized frequencies (%) of the SDSA events were calculated from the transformation efficiencies (Materials and Methods) and plotted (blue bars). Statistical analysis (Table S6): nsd, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed Student’s t test [black letters and asterisks, vs. wild-type (except for vs. indicated genotypes); white letters and asterisks, vs. siz1 mutant]; error bars = SD.
Discussion
Crossover was strongly enhanced by the defective physical interactions of Srs2 with Rad51 and PCNA, in contrast to the milder effects, on crossover, of defects in the interaction between SUMO conjugated with PCNA and SIM of Srs2 and in ATP hydrolysis-dependent DNA helicase activity (Fig. 3E). These results indicate that prevention of crossovers by Srs2 is more dependent on the interactions of Srs2 with Rad51 and PCNA than on the SUMO-SIM interaction and the DNA helicase activity of Srs2. The requirement of the interactions of Srs2 with both Rad51 and PCNA suggests that Srs2 mediates the collaboration between PCNA and the Rad51 nucleofilament on heteroduplex DNA to facilitate loading of Srs2 and PCNA on the Rad51–heteroduplex complex to unwind the invading strand from the donor strand at the heteroduplex joint and prevent crossover (6). Although the unwinding of the invading strand at the heteroduplex joint by Srs2 requires DNA helicase activity in an in vitro assay (6), the requirement of DNA helicase activity for the prevention of crossover in the in vivo assay was mild. Another possible explanation for the prevention of crossover therefore arises: that Srs2 and PCNA interact with Rad51 recombinase, forming the heteroduplex joint DNA, and prevent polymerization of Rad51 along the duplex DNA ahead of the heteroduplex joint. The extended Rad51 nucleofilament ahead of the heteroduplex joint provokes the repair DNA synthesis-free capture of the processed single-strand DNA tail from the second end of the same double-strand break, a possibility that has been demonstrated by bacterial RecA recombinase, the prototype of Rad51 recombinase (23).
The promotion of SDSA by Srs2 requires its DNA helicase activity and physical interaction with the SUMO protein conjugated with PCNA, as well as interactions with Rad51 and PCNA (Fig. 3C). The interaction of Srs2 with the PCNA-SUMO form and the ATP hydrolysis-fueled DNA helicase activity of Srs2 may be required for the initiation of repair DNA synthesis at the leading end of a heteroduplex DNA with the Rad51 nucleofilament. The DNA unwinding activity of Srs2 may also be required for dissociation of the heteroduplex intermediate at its trailing end and the subsequent dissociation of the newly synthesized strand for bubble migration. SRS2 and SIZ1 were epistatic to RAD6 with respect to the SDSA pathway (Fig. 4B), suggesting that Rad6 acts during later steps of the Rad51/Srs2/PCNA-SUMO-mediated SDSA pathway. The Rad6-formed PCNA–Ubiquitin complex may be required for DNA-strand elongation associated with bubble migration.
The RAD51 knockout inhibited SDSA-mediated noncrossovers in the repair of double-stranded breaks (Fig. 2B); however, the RAD51 knockout enhanced the error-prone NHEJ, which required Rad52 and Srs2 as well as Lig4, only when the double-strand breaks to be repaired were staggered (Fig. 2C). In the wild-type, Rad51 may inhibit the intrinsic NHEJ activity of Rad52 by controlling Rad52 annealing activity (24) and also inhibit the intrinsic NHEJ activity of Srs2. We propose that, even if SDSA-mediated noncrossovers do not repair some double-stranded breaks, Rad51 ensures the precise rejoining of staggered double-strand break ends, as a secured type of DNA double-strand break repair. This function is crucial for genome stability against radiation- and reactive oxygen species-induced double-strand DNA breaks because the ends of such double-strand breaks are mostly staggered.
Even when a double-strand break is induced between the centromere and loci with heterozygous status at the 4n-stage, the Rad51/Rad52/Srs2/PCNA-SUMO–mediated SDSA pathway produces only the noncrossover type, and thus, SDSA as well as the precise rejoining avoid the double-strand break-induced loss of heterozygosity in somatic cells (Fig. S1D, Middle). Humans have an ortholog of Srs2, PARI (PCNA-Associated Recombination Inhibitor), which possesses the Rad51-binding domain, PIP and SIM motifs (25). Thus, elucidation of the mechanisms of secure double-strand break repair via SDSA-mediated noncrossover recombination and the precise end-joining pathways in yeast and humans will provide crucial insights into the prevention of carcinogenesis.
Materials and Methods
Yeast Strains, Media, and Transformation.
The Saccharomyces cerevisiae strains used in this study are listed in Table S1. YPD [1% yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) glucose] and SD [2% (wt/vol) glucose or 2% (wt/vol) galactose, and 0.67% (wt/vol) Bacto-yeast nitrogen base] media with appropriate supplements were used to select transformed cells bearing selective markers. Yeast transformation was performed using a lithium acetate-based protocol (26) for the strain construction, the SDSA and NHEJ assays, and the targeted integration assay.
Strain Construction.
All of the knockout (disruption) mutants and the whole-deletion mutants, lacking the entire coding regions, used in this study were constructed by the PCR-based one-step gene disruption method (27) (Table S2) (13). The primers used for the knockouts and the whole deletions are listed in Table S2. The structures of the knockouts and the whole deletions were confirmed by PCR on genomic DNA, with primers situated outside of the 5′ and 3′ homology arms. Methods for the generation of in-frame deletion and point mutants are provided in SI Materials and Methods.
SDSA/NHEJ Assay.
For this assay, leucine-free SD plates, leucine, uracil-free SD plates and leucine-free SD plates containing 1 μg/mL 5-fluoroorotic acid monohydrate (5FOA) (Wako) were used to select the uncleaved plasmid-bearing transformants, the SDSA progeny (Fig. 1C), and the NHEJ progeny (Fig. 1D), respectively. The details of the preparation of competent cells for the lithium acetate-based transformation method have been described previously (13). The SDSA frequency was normalized by dividing the Ura+ Leu+ transformation efficiency with 1 μg of the I-SceI–cleaved or NruI-cleaved plasmid with ura3-intΔisceI or ura3-intΔnruI allele, respectively, by the Leu+ transformation competency with 1 μg of the uncleaved plasmid with the same recipient allele as the cleaved plasmid DNA. The NHEJ frequency was normalized by dividing the 5FOAR Leu+ transformation efficiency with the cleaved plasmid by the Leu+ transformation competency with the uncleaved plasmid. To produce cleaved DNA with ura3-intΔisceI-blunt, the plasmid DNA with ura3-intΔisceI was cleaved by I-SceI nuclease and treated with T4 DNA polymerase (Invitrogen) with dNTPs.
Targeted Integration Assay.
The plasmid pAUR101 (TAKARA), bearing the dominant AUR1-C allele conferring resistance to Aureobasidin A (AurR), lacks a centromere (CEN) and an autonomous replication site (ARS) (Fig. S2) (28). SD plates with 0.4 μg/mL Aureobasidin A (TAKARA) were used for selection. The targeted integration frequency (%) via double Holliday junction-mediated homologous recombination was normalized by dividing the AurR transformation efficiency with 1 μg of the StuI-cut pAUR101 DNA by the AurR transformation competency with the same molar amount of the uncut pRS315-AurR plasmid, which possesses CEN, ARS, and the AUR1-C allele (13).
Supplementary Material
Acknowledgments
We thank Dr. Naoto Arai (Nihon University College of Bioresource Sciences) for kindly providing the pNS188 plasmid bearing the rad52-K117D, R148D allele. This research was supported by Grants-in-Aid for Scientific Research on Innovative Areas (Grant 3102-21113004) and for Scientific Research (A) (Grant 22247002) from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303111110/-/DCSupplemental.
References
- 1.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119(4):779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115(4):401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436(7049):428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 4.Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423(6937):305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 5.Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423(6937):309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 6.Dupaigne P, et al. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29(2):243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483(7387):59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19(1):123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Resnick MA, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 10.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 11.McGill C, Shafer B, Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57(3):459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- 12.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14(3):1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura T, et al. Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases. Genetics. 2012;191(1):65–78. doi: 10.1534/genetics.112.139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde V, Klein H. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 2000;28(14):2779–2783. doi: 10.1093/nar/28.14.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66(4):630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arai N, et al. Vital roles of the second DNA-binding site of Rad52 protein in yeast homologous recombination. J Biol Chem. 2011;286(20):17607–17617. doi: 10.1074/jbc.M110.216739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15(18):5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 18.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93(20):10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268(2):1252–1259. [PubMed] [Google Scholar]
- 20.Krejci L, et al. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem. 2004;279(22):23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- 21.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106(6):735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 22.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 23.Zaitsev EN, Kowalczykowski SC. A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 2000;14(6):740–749. [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283(21):14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moldovan GL, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45(1):75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11(4):355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 28.Hashida-Okado T, Ogawa A, Kato I, Takesako K. Transformation system for prototrophic industrial yeasts using the AUR1 gene as a dominant selection marker. FEBS Lett. 1998;425(1):117–122. doi: 10.1016/s0014-5793(98)00211-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.