Significance
Cystathionine β-synthase (CBS), the pivotal enzyme of the transsulfuration pathway, regulates the flux through the pathway to yield compounds such as cysteine, glutathione, taurine, and H2S that control the cellular redox status and signaling. Our crystal structures of the full-length wild-type and D444N mutant human CBS enzymes show a unique arrangement of the regulatory CBS motifs, thus making it possible to infer how the enzyme is stimulated by its allosteric activator S-adenosyl-L-methionine and how native tetramers are formed. The structure will allow modeling of numerous mutations causing inherited homocystinuria and the design of compounds modulating CBS activity.
Abstract
Cystathionine β-synthase (CBS) controls the flux of sulfur from methionine to cysteine, a precursor of glutathione, taurine, and H2S. CBS condenses serine and homocysteine to cystathionine with the help of three cofactors, heme, pyridoxal-5′-phosphate, and S-adenosyl-l-methionine. Inherited deficiency of CBS activity causes homocystinuria, the most frequent disorder of sulfur metabolism. We present the structure of the human enzyme, discuss the unique arrangement of the CBS domains in the C-terminal region, and propose how they interact with the catalytic core of the complementary subunit to regulate access to the catalytic site. This arrangement clearly contrasts with other proteins containing the CBS domain including the recent Drosophila melanogaster CBS structure. The absence of large conformational changes and the crystal structure of the partially activated pathogenic D444N mutant suggest that the rotation of CBS motifs and relaxation of loops delineating the entrance to the catalytic site represent the most likely molecular mechanism of CBS activation by S-adenosyl-l-methionine. Moreover, our data suggest how tetramers, the native quaternary structure of the mammalian CBS enzymes, are formed. Because of its central role in transsulfuration, redox status, and H2S biogenesis, CBS represents a very attractive therapeutic target. The availability of the structure will help us understand the pathogenicity of the numerous missense mutations causing inherited homocystinuria and will allow the rational design of compounds modulating CBS activity.
Cystathionine β-synthase (CBS; E.C. 4.2.1.22) is a pyridoxal-5′-phosphate (PLP)–dependent enzyme that plays a pivotal role in sulfur amino acid metabolism. CBS catalyzes a β-replacement reaction in which the hydroxyl group of l-serine (Ser) is replaced by l-homocysteine (Hcy), yielding cystathionine (Cth) (1). Hcy is a nonessential amino acid synthesized from l-methionine (Met) via transmethylation and is a well-recognized multisystem toxic substance (2). An increased plasma level of Hcy, chiefly caused by CBS deficiency, represents an independent modifiable risk factor for thrombosis, atherosclerosis, and vascular disease (3–5). CBS-deficient homocystinuria (CBSDH) is an autosomal, recessive inborn error of metabolism resulting from pathogenic mutations in both CBS alleles (6). In addition to increased Hcy plasma levels, CBSDH is biochemically characterized by very high Met, S-adenosyl-l-methionine (AdoMet), and S-adenosyl-l-homocysteine levels, decreased Cys levels, and very low Cth levels in plasma. Clinical symptoms manifest in the ocular, skeletal, vascular, and central nervous systems. Untreated patients suffer from connective tissue defects such as dislocated optic lenses and multiple skeletal deformities. Vascular thromboses may lead to a fatal stroke early in life, and mental retardation often is diagnosed in the affected patients.
CBS diverts Hcy from its conversion to Met in the methionine cycle and condenses it with Ser to form Cth. The next enzyme in the transsulfuration pathway, cystathionine γ-lyase (CGL), cleaves Cth to α-ketobutyrate and Cys, a limiting substrate for glutathione biosynthesis (7). In addition to their crucial roles in the sulfur amino acid metabolism, CBS and CGL recently have been implicated as major physiological sources of hydrogen sulfide (H2S), the third physiological gasotransmitter joining nitric oxide and carbon monoxide (8). H2S is an important signaling molecule in the cardiovascular and nervous systems, induces smooth muscle relaxation, and has anti-inflammatory and cytoprotective effects on cells. Thus, better understanding of how CBS functions and regulates the sulfur amino acid metabolism, redox cellular status, and H2S biogenesis is essential to address and potentially to modulate the pathophysiological consequences of CBS deficiency and redox stress in general.
Human CBS (hCBS) is a particularly interesting PLP-dependent enzyme with a complex domain structure and regulatory mechanism (Fig. 1A) (1, 9). Each polypeptide of the homotetrameric enzyme consists of three functional domains. The N-terminal domain binds the heme cofactor, which is axially coordinated by C52 and H65. The role of heme in CBS is not fully understood, and both structural and regulatory functions have been attributed to it (10–13). However, CBS from lower eukaryotes, such as yeast, lacks heme entirely, thus clearly indicating that is not essential for catalytic activity (14, 15). The central catalytic domain contains covalently attached PLP via a Schiff bond to the ε-amino group of K119. The C-terminal regulatory domain contains a tandem of CBS motifs known as a “Bateman module” (16). Removal of the regulatory domain yields a highly active catalytic core no longer able to bind AdoMet, an allosteric CBS activator (17, 18). Recently, we identified two sets of AdoMet-binding sites with different structural and energetic features; these sites are capable of accommodating up to six ligands in the regulatory domain of the tetrameric full-length enzyme (19). The C-terminal truncation is accompanied by a change in the oligomeric status from a homotetramer to a homodimer (17), whose crystal structure was solved more than a decade ago (10, 20). In 2010, Koutmos et al. (21) presented the crystal structure of a full-length CBS from Drosophila melanogaster (dCBS). Unfortunately, dCBS is not a suitable model for elucidating how hCBS is regulated by AdoMet, because dCBS has very high basal activity and is not regulated by AdoMet.
Fig. 1.
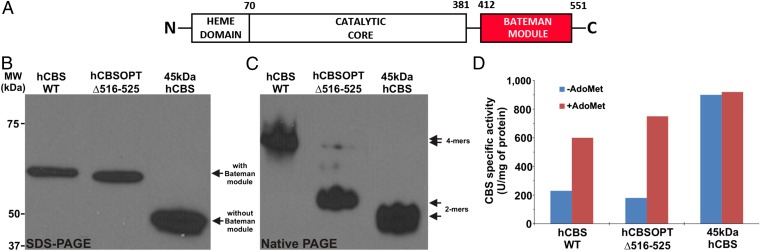
Architecture and biochemical properties of hCBSOPTΔ516–525. (A) In hCBSOPTΔ516–525, unlike the truncated 45-kDa hCBS lacking the Bateman module (red), the distribution of the functional domains is not affected as compared with the native hCBS. (B and C) SDS/PAGE (B) and native PAGE (C) Western blots of the purified enzymes probed with monoclonal anti-CBS antibody show the predominantly dimeric form of hCBSOPTΔ516–525 in contrast to native tetramers of wild-type hCBS and dimers of 45-kDa hCBS. (D) CBS activities ± 300 µM AdoMet show that our hCBSOPTΔ516–525 construct has basal activity and is activated by AdoMet to a similar extent as wild-type hCBS, unlike the constitutively activated 45-kDa hCBS.
Here, we present 3D crystal structures of wild-type and the pathogenic D444N mutant in our modified full-length hCBS construct (hCBSOPTΔ516–525) (22). The relative orientation of the CBS domains pairs in the hCBS dimer is strikingly different from that observed in the dCBS structure, allowing their direct interaction with the functional domain, instead of between each other. Such a unique structural conformation allowed us to explain and discuss the molecular mechanism that is most likely to be behind hCBS allosteric regulation by AdoMet, the effect of pathogenic mutations in the regulatory domain, and the oligomerization of the human enzyme. Because of its central role in sulfur amino acid metabolism and H2S biogenesis, hCBS represents a very attractive therapeutic target. The availability of the structural information will allow the rational design of potent, specific drugs modulating hCBS activity.
Results
Structure of hCBS.
The sequence alignment and superimposition of the dCBS structure (21) and an hCBS model defined a loop protruding from the central β-strand of the human CBS2 domain corresponding to the residues 516–525, which we subsequently removed (22). Like the wild-type hCBS, the modified hCBS construct (hCBSOPTΔ516–525) lacking this loop still contains a tandem of CBS motifs (i.e., a Bateman module) in the C-terminal regulatory region (Fig. 1A) (16, 23). First, we compared our construct with both the wild-type hCBS and the truncated 45-kDa hCBS lacking the regulatory domain (Fig. 1B), whose crystal structure was solved more than a decade ago (10, 20). Under native conditions wild-type hCBS migrates as a tetramer, whereas both the hCBSOPTΔ516–525 and the 45-kDaCBS constructs are predominantly dimeric (Fig. 1C). Additionally, purified hCBSOPTΔ516–525 exhibited basal enzyme activity and could be up-regulated by AdoMet similarly to wild-type hCBS (Fig. 1D). Importantly, the modified hCBSOPTΔ516–525 construct yielded X-ray–diffracting crystals (22).
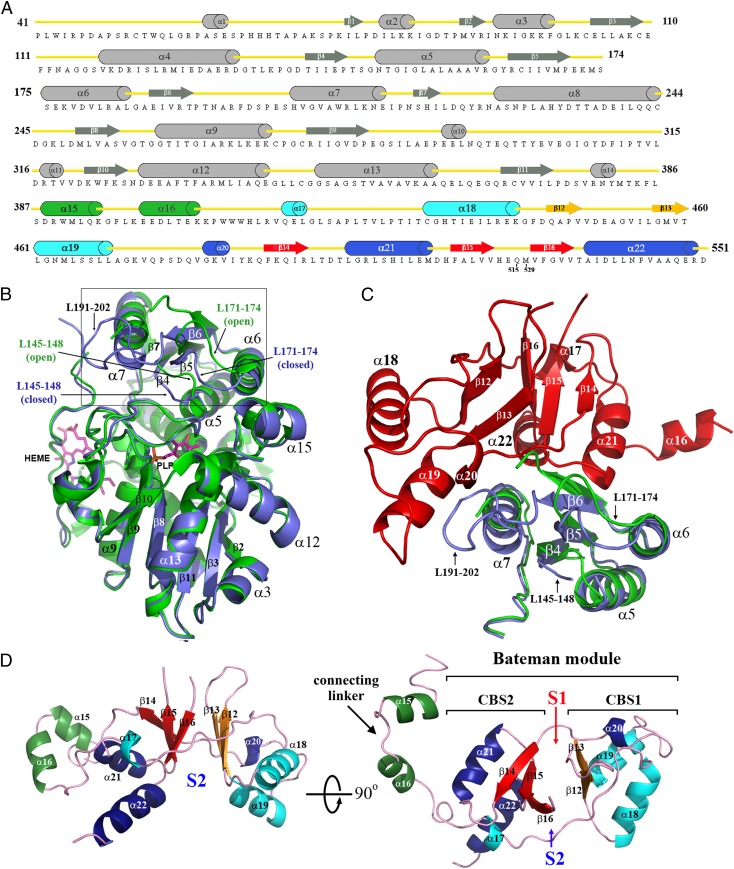
We have determined the 3D structure of the modified hCBS (hCBSOPTΔ516–525) construct from two different types of crystals belonging to space groups I222 and C2221 and diffracting X-rays to 3.0Å and 3.6Å, respectively (Table S1). Structurally, the fold of the highly conserved catalytic core (Fig. 2) belongs to the β-family of PLP-dependent enzymes. It is composed of 13 α-helices and two β-sheets consisting of four (β4–β7) and six (β2, β3, and β8–β11) strands, respectively (Fig. 2A). Interestingly, our structures reveal several regions of the catalytic core that previously were not visible because of their intrinsic flexibility in the absence of the regulatory domain. Among these are the loops L145–148, L171–174, and L191–202, which are sandwiched between the core and the Bateman module at the entrance of the catalytic site, and strands β4, β5, and β6, which precede them in the polypeptide chain, as well as helix α7, which follows the L191–202 loop (Fig. 2 B and C). The catalytic core and the Bateman module are connected via a long linker (382–411) that comprises two α-helices (α15 and α16) (Fig. 2D). The Bateman module includes a pair of interleaved CBS motifs (CBS1, 412–471; CBS2, 477–551) that share 7% sequence identity over 133 residues and show an ααββα and an αβαββα fold, respectively (Fig. 2 C and D). The two CBS motifs interact with each other via their two- or three-stranded β-sheets, and both long edges of this bilayer interface form putative AdoMet-binding sites (designated S1 and S2) (Fig. 2D). Furthermore, each short N-terminal helix (α17 or α20) forms an integral part of the other CBS motif by antiparallel packing between its C-terminal β-strand (β16 or β13) and the α-helix (α22 or α19), so that both CBS motifs form a nested overall structure with pseudo-C2 symmetry.
Fig. 2.
Topology and structure of hCBSOPTΔ516–525. (A) Cartoon presentation of the secondary structures within the C-hCBSOPTΔ516–525 amino acid sequence. (B) Structural superimposition of the truncated 45-kDa hCBS protein (green; PDB ID code 1JBQ) with the catalytic core of the full-length hCBS protein (blue). The loops L145–148, L171–174, and L191–202 are located at the entrance of the PLP cavity and regulate the access of substrates into the catalytic site. As depicted, the loop L191–202 is disordered in the truncated 45-kDa hCBS protein and thus is not visible. The open conformation adopted by the loops L145–148 and L171–174 in the absence of the regulatory domain explain why the truncated enzyme is activated. (C) Interface between the catalytic core (blue) and the Bateman module (red) in the basal form of the full-length enzyme. The truncated 45-kDa hCBS protein (green) is superimposed for comparison. As depicted, the Bateman module pushes the entrance loops toward the PLP cavity, thus hindering the access of substrates. (D) The 3D structure of the Bateman module, which contains two tandem-repeated CBS motifs (CBS1, residues 412–471, and CBS2, residues 477–551). Secondary structure elements have been colored as in A. S1 and S2 designate the proposed AdoMet-binding sites.
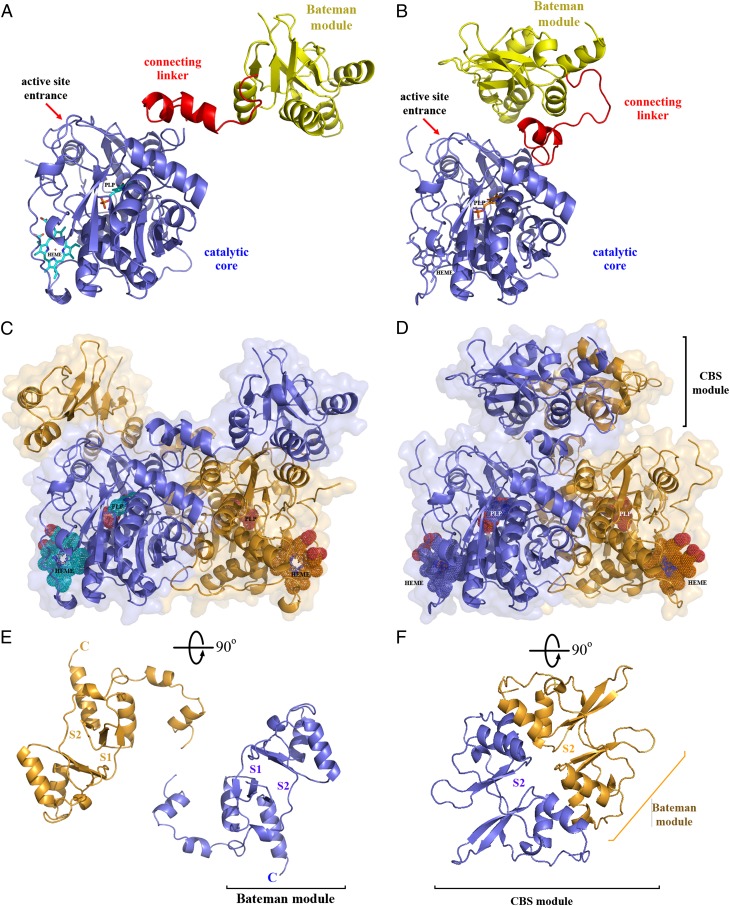
A major contribution of our structure is the unveiling of the relative orientations of the regulatory and catalytic domains in hCBS, which are in striking contrast to the orientation of both the previous in silico models (24, 25) and the dCBS structure (21). The hCBS forms a basket-shaped symmetrical dimer in which the catalytic core of each subunit interacts with both the catalytic core and the regulatory domain of the complementary subunit (Fig. 3). Although the pairing mode and the orientation of the catalytic cores in dCBS and hCBS are similar, the position of their regulatory domains is markedly different (Fig. 3 C and D). In dCBS, the Bateman module does not interact with the catalytic core except via the connecting linker and associates in a tight dimer through its interfacial α-helices, forming a rare head-to-tail–oriented disk-shaped structure referred to as “antiparallel CBS module” (21, 26), which differs from the most commonly found head-to-head–oriented assembly (Fig. 3 D and F) (23, 27). This arrangement allows an unrestricted flow of small molecules into the catalytic cavity, keeping the insect enzyme in a constitutively activated state (21). In contrast to dCBS, the regulatory domains of hCBS subunits are far apart and do not interact with each other, thus demonstrating that, in the dimer, the association between the subunits is dictated by the catalytic cores and not by the Bateman modules (Fig. 3 C and E). Also, the relative arrangement of the catalytic core and regulatory domain places the latter just above the entrance of the catalytic site (Figs. 2C and 3), thus hampering the access of substrates into this cavity (Fig. 4). In hCBS, the interaction of the Bateman module with the core is distributed asymmetrically and involves more secondary structure elements from the CBS2 than from the CBS1 motif. Helices α21 and α22 from the CBS2 motif, α19 and the loop L482–486 (linking helix α20 with strand β14) from CBS1, and helix α7, strand β6, and the loops L171–174 and L192–202 from the core domain all participate in the interface between the regulatory domain and the catalytic core (Fig. 2C).
Fig. 3.
Structural differences between the basal hCBS and the constitutively activated dCBS. (A and B) hCBS (A) and dCBS (B) monomers comprise an N-terminal catalytic core (dark blue) connected via a linker (red) with the C-terminal regulatory domain (yellow). (C and D) The structures of dimeric hCBS (C) and dCBS (D) show the strikingly different orientation of the regulatory domains toward the catalytic cores. The polypeptides within the dimers are shown in orange and blue. (E and F) Top views on the regulatory domains in hCBS (E) and dCBS (F) highlight their distinct arrangement. The Bateman modules of the two subunits in the dCBS dimer form a tight disk-shaped CBS module in contrast to hCBS, in which the regulatory domains are far apart and do not interact with each other.
Fig. 4.
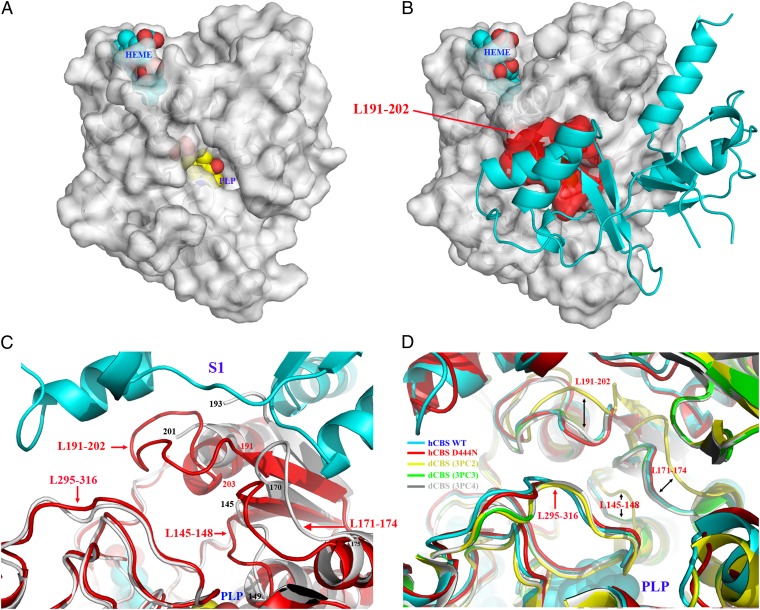
Structural elements determining access to the active site. (A) Surface representation of the 45-kDa hCBS shows that, in the absence of the regulatory domain, the catalytic cavity remains open and the PLP is exposed. (B) In contrast, in the basal form of hCBS, the entrance to the cavity is occluded by the Bateman module by the closed conformation of the loops L145–148, L171–174, and L191–202. (C) A zoom-in view of the interface region between the catalytic core and Bateman module showing the structural elements at the entrance of the catalytic site in hCBS (red) and in the 45-kDa hCBS (gray). (D) Superimposition of the region shown in C for the basal form of hCBS (cyan), for the pathogenic D444N hCBS mutant (red), and for dCBS in the absence (yellow) or the presence (green and gray, respectively) of bound substrates.
Access to the Catalytic Cavity.
In hCBS, the entrance to the PLP-containing cavity is defined by four loops, namely the L145–148, L171–174, L191–202, and L295–316 (Figs. 2 and 4), which adopt a conformation similar to that observed in the substrate-bound dCBS [Protein Data Bank (PDB) ID codes 3PC3 and 3PC4), in which the substrates induce a general collapse of the active site pocket and subsequent closure of the cavity (Fig. 4D and Fig. S1). This scenario differs from both the open (accessible) conformation observed in truncated (activated) hCBS missing the regulatory domain (PDB ID codes 1JBQ and 1M54) and the conformation of these loops in the dCBS with no substrate present in the catalytic cavity (PDB ID code 3PC2) (Fig. 4 A, C, and D and Fig. S1). In the absence of AdoMet, the active site cavity of hCBS adopts a closed conformation because the loop L191–202 is shifted toward the entrance of the cavity by the Bateman module (Figs. 2 and 4 B, C, and D and Fig. S1). A similar effect is observed for the loop L171–174, which is closer to L295–316 (at the opposite side of the cavity) in the presence of the regulatory domain. The closed conformation of L171–174 is stabilized by H-bond interactions involving residues K172–Y301–V303, E304–S174, M173–R190, and R190–P170. The loop L191–202 shows three folds of the main chain that give it a characteristic cloverleaf form. The first turn (residues 191–195) is facilitated by the presence of P192 and allows an H-bond between the ND2 atom of N194 and the carbonyl oxygen of T193. The second turn (residues 196–199) is stabilized by a salt bridge between R196 and D198 and by H-bonds between R196 and S199, which in turn H-bonds N201 favored by the preceding residue P200. This second turn also is stabilized by an H-bond between the carbonyl oxygen of F197 and the hydroxyl group of S202, contributing to the stabilization of the third turn of the loop through another H-bond with S199.
Proposed AdoMet-Binding Sites.
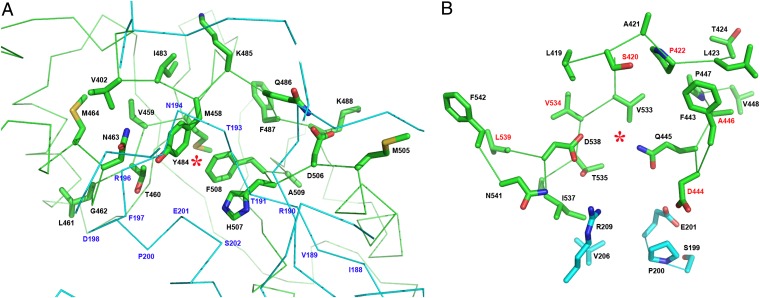
Our structures show that the Bateman module of hCBS contains two major cavities at the β-sheet–lined cleft between the CBS1 and CBS2 motifs that are similar to those that are established as adenosine analog-binding sites in unrelated CBS domain proteins, such as the archeal MJ0100 (Fig. 2D) (27). Accordingly, these cavities represent two possible binding sites for AdoMet in hCBS. Site S1 shows a hydrophobic cage (M458, V459, Y484, F487, F508, A509) that may easily accommodate an adenine ring, a conserved asparagine (N463) along with a threonine (T460) for the interaction with ribose, and a positively charged residue (H507) for stabilizing the carboxylate group of the nucleotide (Fig. 5A). Asparagine residues have been demonstrated, albeit rarely, to play a role equivalent to that of the conserved aspartate in stabilizing the position of the ribose ring of adenosine derivatives in CBS domains (PDB ID codes 3L2B, 3L31, and 3LFZ). On the other hand, site S2 likewise features a hydrophobic environment (P422, L423, F443, A446, P447, V448, V533, V534), conserved aspartate (D538) and threonine (T535) for interaction with ribose, a hydrophobic isoleucine (I537) that may stabilize the methionine alkyl chain of AdoMet, and an acidic cluster (D444, E201) that would compensate the positive charge of the sulphonium ion of AdoMet, thus stabilizing the allosteric regulator in the cavity (Fig. 5B). Our structure of hCBS in the basal form suggests it is unlikely that S1 could accommodate AdoMet, because the access to this cavity is hindered by the presence of structural elements (loop L191–202 and strand β6) from the core domain of the complementary subunit (Figs. 3, 4, and 5A). In contrast, the S2 cavity is exposed and accessible to host AdoMet without steric hindrance (Fig. 5B). Moreover, binding of AdoMet to the S2 site may cause reorientation of the side chains of bulky residues occupying the S1 site, such as Y484, H507 and F508, thus allowing additional binding of AdoMet.
Fig. 5.
Putative AdoMet-binding sites in hCBS. (A) Site S1. The entrance of S1 is sterically hindered by the presence of structural elements from the catalytic core (cyan) of a complementary monomer in the dimer. Additionally, bulky hydrophobic residues (Y484, H507, and F508) occupy the cavity and probably impede the binding of AdoMet at this site in the basal form. Residue N463, instead of the conserved aspartate that would stabilize the nucleotide ribose ring, could play a similar role. (B) Site S2. In contrast, the cavity of S2 is solvent exposed and is not blocked by bulky residues. The site S2 shows features similar to the AdoMet-binding protein MJ0100 (27): a hydrophobic cage to host the adenine ring of AdoMet, conserved aspartate (D538) and threonine (T535) residues to stabilize the ribose ring, and a hydrophobic residue (I537) preceding D538 to accommodate the alkyl chain of AdoMet. Noteworthy are the residues linked with pathogenic mutations (identified in red).
Allosteric Activation of hCBS by AdoMet.
With the aim of unraveling the AdoMet-mediated allosteric regulation of hCBS activity, we further tried to obtain crystals of the hCBS–AdoMet complex. Because of the controversy found in the literature about whether (28–30) or not (19, 31) AdoMet induces a large conformational change upon binding to hCBS and to avoid possible structural artifacts caused by preexisting crystal packing, we first performed cocrystallization experiments of hCBS with AdoMet instead of soaking previously grown crystals in solutions containing the allosteric regulator. To assure complete occupancy of all potential binding sites, an excess of AdoMet (molar ratio 10:1) was added to the protein solution subjected to crystallization. Interestingly, the hCBS–AdoMet solutions yielded small, needle-shaped crystals in similar crystallization conditions as used for crystal growth in the absence of AdoMet Unfortunately, and despite great experimental effort, we could not obtain crystals of sufficient quality (or size) for a crystallographic study.
As an alternative to the hCBS–AdoMet complex, we determined the crystal structure of the D444N pathogenic mutant (30, 32), which shows an approximately twofold increase in basal activity and impaired response to AdoMet stimulation as compared with the wild type (Table S1 and Figs. S1 and S2). Two different protein constructs, referred to as C-hCBSOPTΔ516–525 D444N and C-hCBSOPTΔ1–39Δ516–525 D444N, yielded two distinct types of crystals that belong to space groups P212121 and P65 and that diffracted X-rays to 2.6 Å and 3.40 Å, respectively (Table S1). Interestingly, both structures clearly show that the overall fold of the D444N mutant resembles that of hCBSOPTΔ516–525 protein and clearly differ from the activated conformation of dCBS (Fig. 3 and Fig. S1). The most significant differences between the native and the mutant protein are a slight displacement of the Bateman modules toward the large central cavity of the dimer and a small shift of helices α18 and α22 without significant changes in the fold of the catalytic core (Fig. S2). Unexpectedly, we found that the loops occluding the entrance to the PLP cavity remained in a closed conformation, as observed for the basal state of the wild-type protein (Figs. S1 and S2). These findings are in agreement with the recent biochemical, spectroscopic, and calorimetric studies demonstrating that AdoMet alleviates the intrasteric block imposed by the regulatory domain without significant conformational rearrangements (19, 31). Instead, our data strongly suggest that the active conformation of hCBS differs significantly from that found in dCBS (21). Furthermore, they also indicate that it is highly unlikely that AdoMet would trigger a displacement of the Bateman modules of hCBS to form an antiparallel CBS module, the dimeric association found in dCBS (21), and CBSX2 bound to AMP (26) (Fig. 3 and Movies S1 and S2). Alternatively, our data indicate that the effect of AdoMet could be similar to that identified for MJ0100 (27); i.e., AdoMet induces a relative rotation or a small displacement of the CBS motifs within each Bateman module, thus providing sufficient space to allow free movement of the loops occluding the entrance to the PLP cavity (Fig. 6 and Movie S3). Taking these findings together, we postulate the following mechanism of hCBS activation by AdoMet:
i) A molecule of AdoMet binds to the exposed site S2 of each Bateman module. Interactions with amino acid residues of each monomer would help nest AdoMet within the cavity and would promote a more compact structure of the Bateman module in which the CBS1 and CBS2 motifs rotate with respect to each other (Movie S3). This torsion would take place around the flexible loops connecting helices α17–α18 and helix α20 with strand β14 (Fig. 2D) without affecting the secondary structure elements, as documented in other proteins containing a CBS domain (23, 27, 33).
ii) As a consequence of the relative rotation of the CBS motifs, loops L145–148, L171–174, and L191–202 relax toward an open conformation. The wider entrance of the PLP cavity, similar to dCBS (21), would allow unrestricted flow of substrates to the catalytic center (Fig. 6). Additionally, an intermediate step would take place during which the torsion of the CBS motifs would move the regulatory domain away from the catalytic core, thus alleviating the blockage of the site S1. The exposed site S1 would accommodate an AdoMet molecule, which in turn would favor the opening of the entrance to the PLP cavity and thus activation of the enzyme. This hypothesis could explain why the low-affinity site is responsible of hCBS activation (19).
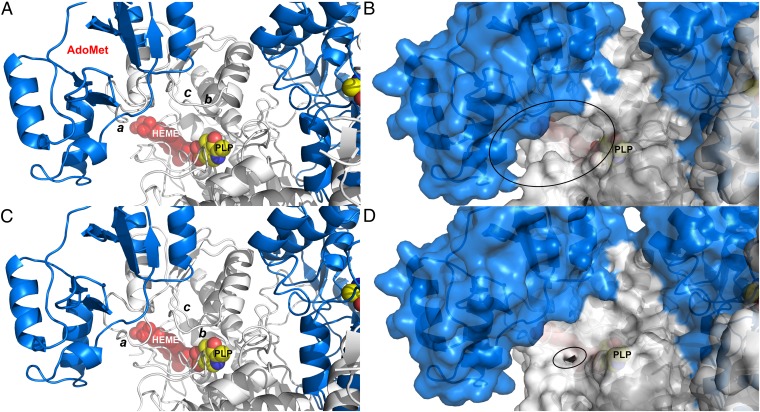
Fig. 6.
The proposed model of hCBS activation. (A and B) Ribbon (A) and surface (B) representations of a model of the activated form of hCBS upon binding of AdoMet at site S2. The loops controlling the access to the PLP cavity (a: L191–202; b: L171–174; c: L145–148) are open, thus allowing the access of substrates. (C and D) Ribbon (C) and surface (D) representations of the basal form of hCBS obtained from the crystals. The loops controlling access to the PLP cavity are closed and occlude the entrance of substrates into the catalytic cavity.
Tetramerization of hCBS and Possible Mechanisms of Aggregation.
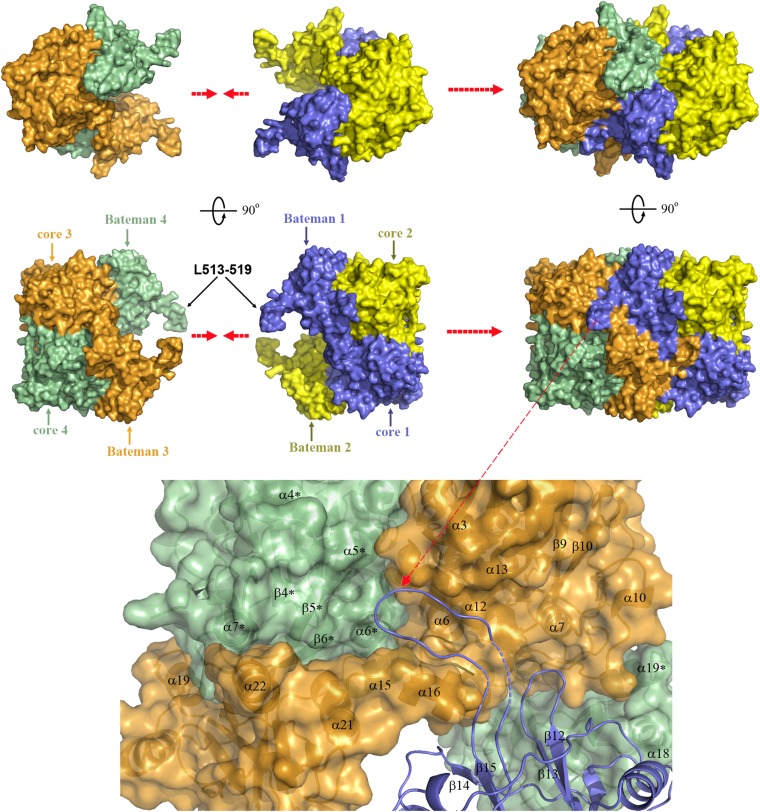
The overall fold of C-hCBSOPTΔ516–525 dimers found in our crystals, the absence of large conformational changes in the D444N hCBS mutant, and the electrostatic potential of the protein surface allow us to suggest how wild-type hCBS likely forms native tetramers. As shown in Fig. 7, we postulate that the native tetramer is an assembly of two dimers, which, like two complementary pieces of a puzzle, are matched together through the main cavity between their Bateman modules without the need of major structural changes. Such a partnership results in a symmetrical dimer of dimers in which the Bateman module of each subunit interacts with the core and Bateman modules of the complementary dimer. An association of this kind places each L513–519 loop on the surface of the complementary dimer, at the level of a relatively large crevice formed by helices α5, α6, α12, α15, and α16 and strands β5 and β6, where it serves as a hook, tightly locking the two dimers together. The stabilizing interactions that may exist between L513–519 and residues from the cavity help explain the fundamental role of L513–519 in stabilizing tetramers and why deletion of residues 516–525 in our constructs inevitably disrupted the oligomeric equilibrium toward the formation of dimers (Fig. 1C).
Fig. 7.
The proposed model of the hCBS tetramer. The hCBS tetramerization is sustained by the interactions of each Bateman module with the Bateman module and with the catalytic cores of the complementary dimer. The tetramer is stabilized by interactions between loop 513–529, which serves as a hook locking the two dimers together, and the residues located at the cavity formed by the helices α6, α12, α15, and α16.
Crystal-packing interactions also may provide clues about the causes that lead to aggregation of the full-length hCBS (Fig. S3). In our crystals, the protein dimer forms polymeric chains that interact through their Bateman modules following two distinct patterns. In the C-hCBSOPTΔ516–525 (space group I222) crystals, the hCBS dimers interact through the α-helices of their CBS1 motifs (Fig. S3A) as well as via salt bridges between the K75, D238, D245, and K247 residues of their core domains (Fig. S3C). On the other hand, the C-hCBSOPTΔ516–525 C2221 crystals and also the C-hCBSOPTΔ516–525 D444N (space group P212121) crystals contain polymeric chains of hCBS dimers interacting through their Bateman modules (Fig. S3B). In this case, the protein contacts include both hydrophobic and polar interactions that affect the flexible loops connecting the central β-strands of the Bateman modules as well as the long loops preceding the first helix of CBS2 motifs.
Discussion
More than a decade ago, the crystal structure of the truncated hCBS lacking the C-terminal regulatory domain demonstrated the overall fold of the catalytic module, the location of residues involved in catalysis, and interactions involved in stabilization of the dimeric enzyme (10, 20). However, it did not explain why removal of the regulatory domain disintegrates the enzyme into hyperactivated dimers or, more importantly, the molecular mechanism by which the allosteric activator, AdoMet, modulates the activity of the enzyme. Moreover, structures of the dimeric catalytic cores could not explain how the most abundant oligomeric species among mammalian CBSs, a tetramer (1), is formed and stabilized by then-unknown interactions involving the regulatory domains of a full-length enzyme.
The recently determined structure of dCBS revealed the relative orientations of the regulatory and catalytic domains and showed that substrate binding induces conformational changes in the protein core that are restricted to a small shift of three loops delineating the entrance of the catalytic cavity and the adjacent β-strands toward the PLP cofactor (Fig. 4D) (21). The size of the entrance is determined exclusively by the absence or presence of substrates within the PLP cavity (PDB codes 3PC2, 3PC3, and 3PC4) (Fig. S1). Thus, in the absence of bound substrates, the loops are open, and the enzyme is ready to perform the catalytic reaction. Conversely, binding of substrates within the cavity induces a collapse of the loops and subsequent closure of the entrance. The facts that the Bateman module remains unaltered throughout this process (21) and that the basal activity of dCBS is much higher than that of hCBS indicate that the solved structure of dCBS represents a constitutively activated conformation of the enzyme. Based on these findings, hCBS might undergo a radical structural change upon binding of AdoMet, which evolves from the basal conformation presented herein to that described for dCBS (21). However, our data indicate that such a transition would be sterically hindered. In fact, despite the high sequence and structural similarity in the catalytic cores of dCBS and hCBS, several lines of evidence already have indicated that these two enzymes differ significantly in their overall architecture (Fig. 3).
Our structures of the basal form of full-length hCBS and of the D444N mutant show that the most significant differences in the CBS enzymes from various organisms lie in the relative orientations of their regulatory domains with respect to the catalytic cores. We have found that, unlike dCBS, the regulatory domain of hCBS in the basal state occludes the entrance to the PLP site. The loops limiting the access to the PLP cavity adopt a closed conformation, being sandwiched between the core and the Bateman module (Figs. 2C, 3C, and 4). These results are concordant with the previously proposed regulatory mechanism in the full-length protein in which the catalytic core is sterically hindered by the C-terminal regulatory domain and the autoinhibition is relaxed upon AdoMet binding (30, 34). Therefore, our findings provide a structural framework for understanding the unique allosteric regulation of hCBS by AdoMet. Unexpectedly, the Bateman module of hCBS does not associate according to the most commonly observed pattern, a disk-like dimeric species known as the “CBS module,” as in dCBS (Fig. 3 D and F). Accordingly, the effect of the bound nucleotide cannot be explained by this structural arrangement. Given the distance between the regulatory domains of both subunits within each dimer, it seems reasonable to rule out the possibility that AdoMet binding plays a role in the degree of enzyme oligomerization. Instead, it becomes obvious that the dimerization is dictated solely by the interaction between the catalytic cores of two subunits. Conversely, our structures help explain why the tetramer stability and the formation of higher-order oligomers are highly dependent on the presence of the regulatory domain (Fig. 7 and Fig. S3).
The D444N mutation belongs to a specific class of CBS missense pathogenic mutations. Patients carrying a mutation such as I435T, D444N, or S466L in the C-terminal regulatory domain are rare, and, although biochemically they are identified as homocystinurics, they lack the usual connective-tissue disorders associated with CBS deficiency (30, 32, 35). The common denominator of these mutants is their normal or increased basal activity and impaired or totally abolished response to AdoMet stimulation compared with the wild-type hCBS. As shown here, introducing an asparagine in position D444 induces a change in the relative orientation of the CBS motifs and a shift of helices α18, α19, and α22. The polar environment in which D444 is immersed suggests that, together with residues E201 and E538, it helps compensate the helix α19 dipole; more importantly, it significantly stabilizes the orientation of the CBS2 motif through long-range columbic interactions with R209. Accordingly, the D444N mutation weakens this electrostatic interaction and most likely is responsible for the displacement suffered by the aforementioned structural elements. Such rearrangement in turn leads to the higher basal activity and impaired AdoMet response seen in our D444N constructs as compared with wild-type hCBS.
In conclusion, the structures of the wild-type and the D444N mutant hCBS enzymes revealed the unique domain organization of this pivotal enzyme in the metabolism of sulfur amino acids. Our structures provide important insights into both the mechanism of allosteric regulation by AdoMet and the effect of pathogenic mutations located in the regulatory domain. Additionally, they shed light on the oligomerization and aggregation tendencies of this enzyme and reveal the distinct structural features in mammalian enzymes, such as hCBS, and those from other organisms, such as dCBS. More importantly, because hCBS represents a highly valuable therapeutic target for homocystinuria itself as well as for other conditions such as vascular disease and cancer (6, 36), the data presented here pave the way for the rational design of inhibitors or activators modulating hCBS activity.
Materials and Methods
Preparation of the recombinant modified hCBSOPTΔ516–525, C-hCBSOPTΔ516–525, C-hCBSOPTΔ516–525 D444N, and C-hCBSOPTΔ1–39Δ516–525 D444N enzymes followed the protocols that we developed for various hCBS constructs either expressed with cleavable GST at their N terminus (37) or carrying a permanent 6xHis tag at their C terminus (38), with a few modifications (22). The deletions and the D444N pathogenic mutation were introduced by using a QuikChangeII XL mutagenesis kit (Agilent) according to the manufacturer’s recommendations. Denaturing and native protein gel electrophoresis, Western blot, and the CBS activity assay in the absence and presence of AdoMet were performed essentially as described previously (37, 38). The crystals were grown by the sitting and/or hanging-drop vapor-diffusion method at 293 K in 96-well and 24-well crystallization plates, respectively, according to the protocol described previously (22). All datasets used in this work were collected at the European Synchrotron Radiation Facility (Grenoble, France) beamlines ID23-1 and ID29 and were processed using HKL2000 (39) or XDS (40) software. The hCBS structures were determined by molecular replacement with the PHENIX program (41) using the crystal structure of the truncated 45-kDa hCBS (PDB ID code 1JBQ) as the initial search model. After several cycles of refinement using PHENIX (41) and REFMAC5 (42), CBS domains were built manually using Coot (43). The crystal characteristics and final refinement statistics are summarized in the Table S1.
Supplementary Material
Acknowledgments
We thank Dr. Alexander Popov at beamline ID23.1 of the European Synchrotron Radiation Facility for valuable support during synchrotron data collection, the staff of beamline ID29 for technical assistance, and Dr. Adriana Rojas for maintenance of the in-house X-ray equipment. This work was supported by Postdoctoral Fellowship 0920079G from the American Heart Association (to T.M.), by National Institutes of Health Grant HL065217, American Heart Association Grant In-Aid 09GRNT2110159, and by a grant from the Jerome Lejeune Foundation (all to J.P.K.), and by Grants from the Department of Education, Universities and Research of the Basque Government (PI2010-17), from the Department of Industry of the Basque Government (ETORTEK Program IE05-147 and IE07-202), from the Bizkaia County (Exp.7/13/08/2006/11 and 7/13/08/2005/14), and from the Spanish Ministry of Economy and Innovation (BFU2010-17857 and SICI-CONSOLIDER Program CSD2008-00005) (all to L.A.M.-C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank database, www.pdb.org (PDB ID codes 4L0D, 4L3V, 4L27, and 4L28).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313683110/-/DCSupplemental.
References
- 1.Miles EW, Kraus JP. Cystathionine beta-synthase: Structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004;279(29):29871–29874. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- 2.Castro R, Rivera I, Blom HJ, Jakobs C, Tavares de Almeida I. Homocysteine metabolism, hyperhomocysteinaemia and vascular disease: An overview. J Inherit Metab Dis. 2006;29(1):3–20. doi: 10.1007/s10545-006-0106-5. [DOI] [PubMed] [Google Scholar]
- 3.Boers GHJ. Hyperhomocysteinemia as a risk factor for arterial and venous disease. A review of evidence and relevance. Thromb Haemost. 1997;78(1):520–522. [PubMed] [Google Scholar]
- 4.Makris M. Hyperhomocysteinemia is a risk factor for venous and arterial thrombosis. Br J Haematol. 1998;101(Suppl 1):18–20. [PubMed] [Google Scholar]
- 5.Welch GN, Loscalzo J. Homocysteine and atherothrombosis. N Engl J Med. 1998;338(15):1042–1050. doi: 10.1056/NEJM199804093381507. [DOI] [PubMed] [Google Scholar]
- 6. Mudd SH, Levy HL, Kraus JP (2001) The Metabolic and Molecular Bases of Inherited Disease, eds Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler K, Vogelstein B (McGraw-Hill, New York), 8th Ed, pp 2007–2056.
- 7.Finkelstein JD. Pathways and regulation of homocysteine metabolism in mammals. Semin Thromb Hemost. 2000;26(3):219–225. doi: 10.1055/s-2000-8466. [DOI] [PubMed] [Google Scholar]
- 8.Szabó C. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 2007;6(11):917–935. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- 9.Singh S, Madzelan P, Banerjee R. Properties of an unusual heme cofactor in PLP-dependent cystathionine beta-synthase. Nat Prod Rep. 2007;24(3):631–639. doi: 10.1039/b604182p. [DOI] [PubMed] [Google Scholar]
- 10.Taoka S, et al. Human cystathionine beta-synthase is a heme sensor protein. Evidence that the redox sensor is heme and not the vicinal cysteines in the CXXC motif seen in the crystal structure of the truncated enzyme. Biochemistry. 2002;41(33):10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- 11.Janosík M, et al. Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet. 2001;68(6):1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majtan T, Singh LR, Wang L, Kruger WD, Kraus JP. Active cystathionine beta-synthase can be expressed in heme-free systems in the presence of metal-substituted porphyrins or a chemical chaperone. J Biol Chem. 2008;283(50):34588–34595. doi: 10.1074/jbc.M805928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weeks CL, Singh S, Madzelan P, Banerjee R, Spiro TG. Heme regulation of human cystathionine beta-synthase activity: Insights from fluorescence and Raman spectroscopy. J Am Chem Soc. 2009;131(35):12809–12816. doi: 10.1021/ja904468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jhee KH, McPhie P, Miles EW. Yeast cystathionine beta-synthase is a pyridoxal phosphate enzyme but, unlike the human enzyme, is not a heme protein. J Biol Chem. 2000;275(16):11541–11544. doi: 10.1074/jbc.c000056200. [DOI] [PubMed] [Google Scholar]
- 15.Maclean KN, Janosík M, Oliveriusová J, Kery V, Kraus JP. Transsulfuration in Saccharomyces cerevisiae is not dependent on heme: Purification and characterization of recombinant yeast cystathionine beta-synthase. J Inorg Biochem. 2000;81(3):161–171. doi: 10.1016/s0162-0134(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22(1):12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 17.Kery V, Poneleit L, Kraus JP. Trypsin cleavage of human cystathionine beta-synthase into an evolutionarily conserved active core: Structural and functional consequences. Arch Biochem Biophys. 1998;355(2):222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 18.Shan X, Kruger WD. Correction of disease-causing CBS mutations in yeast. Nat Genet. 1998;19(1):91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- 19.Pey AL, Majtan T, Sanchez-Ruiz JM, Kraus JP. Human cystathionine β-synthase (CBS) contains two classes of binding sites for S-adenosylmethionine (SAM): Complex regulation of CBS activity and stability by SAM. Biochem J. 2013;449(1):109–121. doi: 10.1042/BJ20120731. [DOI] [PubMed] [Google Scholar]
- 20.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: A unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 2001;20(15):3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutmos M, Kabil O, Smith JL, Banerjee R. Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine beta-synthase. Proc Natl Acad Sci USA. 2010;107(49):20958–20963. doi: 10.1073/pnas.1011448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyenarte I, et al. Purification, crystallization and preliminary crystallographic analysis of human cystathionine β-synthase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 11):1318–1322. doi: 10.1107/S1744309112037219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baykov AA, Tuominen HK, Lahti R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem Biol. 2011;6(11):1156–1163. doi: 10.1021/cb200231c. [DOI] [PubMed] [Google Scholar]
- 24.Sen S, Banerjee R. A pathogenic linked mutation in the catalytic core of human cystathionine beta-synthase disrupts allosteric regulation and allows kinetic characterization of a full-length dimer. Biochemistry. 2007;46(13):4110–4116. doi: 10.1021/bi602617f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnízda A, et al. Cross-talk between the catalytic core and the regulatory domain in cystathionine β-synthase: Study by differential covalent labeling and computational modeling. Biochemistry. 2010;49(49):10526–10534. doi: 10.1021/bi101384m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong BC, Park SH, Yoo KS, Shin JS, Song HK. Change in single cystathionine β-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding. J Struct Biol. 2013;183(1):40–46. doi: 10.1016/j.jsb.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Lucas M, et al. Binding of S-methyl-5′-thioadenosine and S-adenosyl-L-methionine to protein MJ0100 triggers an open-to-closed conformational change in its CBS motif pair. J Mol Biol. 2010;396(3):800–820. doi: 10.1016/j.jmb.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Taoka S, Widjaja L, Banerjee R. Assignment of enzymatic functions to specific regions of the PLP-dependent heme protein cystathionine beta-synthase. Biochemistry. 1999;38(40):13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- 29.Shan X, Dunbrack RL, Jr, Christopher SA, Kruger WD. Mutations in the regulatory domain of cystathionine beta synthase can functionally suppress patient-derived mutations in cis. Hum Mol Genet. 2001;10(6):635–643. doi: 10.1093/hmg/10.6.635. [DOI] [PubMed] [Google Scholar]
- 30.Evande R, Blom H, Boers GH, Banerjee R. Alleviation of intrasteric inhibition by the pathogenic activation domain mutation, D444N, in human cystathionine beta-synthase. Biochemistry. 2002;41(39):11832–11837. doi: 10.1021/bi026248d. [DOI] [PubMed] [Google Scholar]
- 31.Hnízda A, et al. Conformational properties of nine purified cystathionine β-synthase mutants. Biochemistry. 2012;51(23):4755–4763. doi: 10.1021/bi300435e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kluijtmans LA, et al. Defective cystathionine beta-synthase regulation by S-adenosylmethionine in a partially pyridoxine responsive homocystinuria patient. J Clin Invest. 1996;98(2):285–289. doi: 10.1172/JCI118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gómez-García I, Oyenarte I, Martínez-Cruz LA. The crystal structure of protein MJ1225 from Methanocaldococcus jannaschii shows strong conservation of key structural features seen in the eukaryal gamma-AMPK. J Mol Biol. 2010;399(1):53–70. doi: 10.1016/j.jmb.2010.03.045. [DOI] [PubMed] [Google Scholar]
- 34.Janosík M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: Evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40(35):10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 35.Maclean KN, et al. High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum Mutat. 2002;19(6):641–655. doi: 10.1002/humu.10089. [DOI] [PubMed] [Google Scholar]
- 36.Szabo C, et al. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci USA. 2013;110(30):12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majtan T, Liu L, Carpenter JF, Kraus JP. Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: Purification and characterization of eight CBS mutant enzymes. J Biol Chem. 2010;285(21):15866–15873. doi: 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majtan T, Kraus JP. Folding and activity of mutant cystathionine β-synthase depends on the position and nature of the purification tag: Characterization of the R266K CBS mutant. Protein Expr Purif. 2012;82(2):317–324. doi: 10.1016/j.pep.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.