Abstract
Achromatopsia is an autosomal recessively inherited visual disorder that is present from birth and that features the absence of color discrimination. We here report the identification of five independent families with achromatopsia that segregate protein-truncation mutations in the GNAT2 gene, located on chromosome 1p13. GNAT2 encodes the cone photoreceptor–specific α-subunit of transducin, a G-protein of the phototransduction cascade, which couples to the visual pigment(s). Our results demonstrate that GNAT2 is the third gene implicated in achromatopsia.
Achromatopsia, also referred to as “rod monochromacy” (ACHM2 [MIM 216900] and ACHM3 [MIM 262300]), is a congenital ocular disorder characterized by total color blindness, low visual acuity, photophobia, and nystagmus. It is inherited as an autosomal recessive trait, with an estimated prevalence of 1/30,000 (François 1961).
Recently, we and others have shown that mutations in CNGA3 and CNGB3, which encode the α- and β-subunits of the cone photoreceptor cGMP-gated channel, cause clinically indistinguishable forms of achromatopsia, including Pingelapese blindness (Kohl et al. 1998, 2000; Sundin et al. 2000).
However, screening for CNGA3 and CNGB3 mutations in a large cohort of patients with achromatopsia revealed further genetic heterogeneity in approximately one-third of the cases. The pursuit of additional genes involved in achromatopsia led us to consider GNAT2 as a prime candidate gene, because of its cone photoreceptor specificity and functional importance in the visual transduction process (Lerea et al. 1986; Blatt et al. 1988; Morris and Fong 1993). GNAT2 encodes the cone-specific α-subunit of transducin (MIM 139340), a heterotrimeric G-protein that couples to the cone visual pigments. Excited pigment molecules induce both the exchange of GDP to GTP at the guanosine-binding site of the transducin α-subunit and its subsequent release from the inhibitory β/γ-subunits. The activated GTP • transducin complex then binds and activates a phosphodiesterase that hydrolyzes cGMP and thereby effectively reduces its intracellular concentration. This results in closure of the cGMP-gated channels and, subsequently, in membrane hyperpolarization (Müller and Kaupp 1998). Transducin thus mediates one of the first steps of the phototransduction cascade, whereas the cGMP-gated channel represents the final component of the very same process.
It has been shown that a missense mutation in GNAT1, which encodes the rod photoreceptor–specific transducin α-subunit, is implicated in autosomal dominant congenital stationary night blindness of Nougaret (Dryja et al. 1996), and a homologous phenotype has been described for the corresponding knockout-mouse model (Calvert et al. 2000). In contrast, no association between the GNAT2 locus and hereditary retinal disorders has been detected in previous studies (Gerber et al. 1995; Magovcevic et al. 1995). The human GNAT2 gene comprises eight exons and covers ∼10 kb of genomic sequence on human chromosome 1p13. GNAT2 encodes a polypeptide of 354 amino acid residues that shows 82% sequence identity with its paralog in rod photoreceptors (Lerea et al. 1989; Kubo et al. 1991; Morris and Fong 1993).
We screened the entire coding sequence of the GNAT2 gene, in a collection of 77 unrelated patients with achromatopsia, in whom mutations in the CNGA3 and CNGB3 genes were excluded. PCR fragments covering all eight exons and flanking intron/UTR sequences of GNAT2 were amplified from genomic DNA (for primer sequences, see table 1) and were subjected to direct DNA sequencing employing Big Dye Terminator chemistry (Applied Biosystems). Reaction products were separated on an ABI 3100 DNA sequencer, and sequences were analyzed by the Lasergene Software package (DNASTAR).
Table 1.
Sequences of PCR Primers Used to Amplify GNAT2 Coding Segments
|
Primer |
|||
| Exon(s) | Forward | Reverse | PCRProduct Size(bp) |
| 1 | GCTTTTAGGGGAGCCAAGTT | CCCCTAGCCAGACTGTGTAGA | 452 |
| 2 and 3 | CCCCCAAGCAAGCAG | CAGCAGGTGGGATTTTAGTT | 691 |
| 4 | TCTGCATCTGGGGAGCATAC | TTGGGTTGGGTGAGGTTT | 434 |
| 5 and 6 | TGGCTTCTAGTCCTGAGGTC | GATTGGTCTGCTGGAGG | 716 |
| 7 and 8 | TTTTTACATTTGGAGCACTG | CCCACTTTGAAAAGAACG | 1,023 |
| IVS 3–5a | TTAAGGTAGACGGTACCTTTC | TAAGGTCTTACTGCGTAGATG | 2,022 |
Amplification was by long-distance PCR.
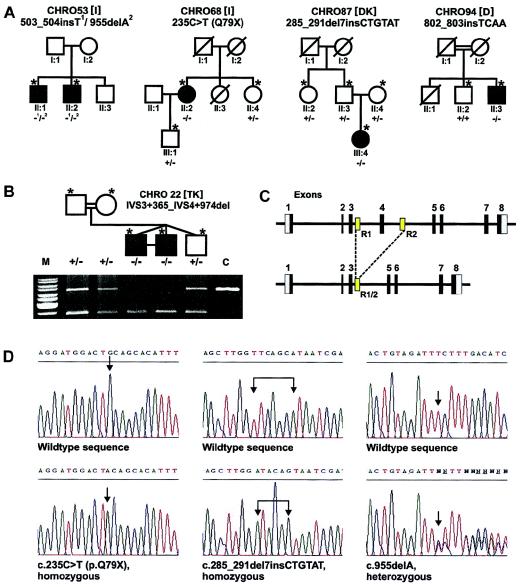
In the 77 patients with achromatopsia whom we analyzed, we were able to identify six different disease-related sequence alterations (table 2) segregating in five independent families of European descent (fig. 1a and b). The mutations comprise one nonsense mutation, c.235C→T (p.Q75X), and four small deletion and/or insertion mutations, which all cause frameshifts and lead to premature translation termination. The sixth mutation represents a large intragenic deletion of 2,019 bp (g.3407_5425del [GenBank accession number Z18859]), which includes exon 4 and flanking intron sequences. This deletion probably results from a recombination between two 229-bp direct-repeat sequences present in intron 3 and intron 4 of GNAT2 (fig. 1c). BLASTN analysis revealed that this repeat is unique to the GNAT2 locus, on chromosome 1p13. Detection of the deletion and analysis of its segregation in family CHRO22 were easily accomplished by long-distance PCR (fig.1b and table 1).
Table 2.
Mutations in GNAT2 in Patients with Achromatopsia
|
Alterationa |
||
| Exon | Nucleotide Sequence | Polypeptide (Consequence) |
| 3 | c.235C→T | p.Q79X |
| 3 | c.285_291del7insCTGTAT | p.Y95fsX61 |
| 4 | IVS3+365_IVS4+974del | Exon 4 deleted (p.A101fsX12) |
| 5 | c.503_504insT | p.L168fsX3 |
| 7 | c.802_803insTCAA | p.L268fsX9 |
| 8 | c.955delA | p.I319fsX5 |
Figure 1.
GNAT2 mutations in families segregating achromatopsia. A and B, Pedigrees of the five achromatopsia families with GNAT2 mutations, including the genotypes of available subjects (denoted by asterisks [*]) as determined by segregation analysis. + = wild type; − = mutation. The geographical origin of the families is indicated by square brackets: I = Italy; DK = Denmark; D = Germany; TK = Turkey. Panel b presents the results of segregation analysis of the exon 4 deletion in family CHRO22. The photograph shows the electrophoretic separation of wild-type alleles (upper band) and mutant alleles (lower band), after amplification by long-distance PCR. M = marker; C = normal control subject. C, Organization of the GNAT2 gene (top) and location of the two repeats (R1 and R2) putatively implicated in the exon 4 deletion (bottom). D, Electropherograms of mutant sequence (bottom) and corresponding wild-type reverse-strand sequence (top), for homozygous mutations c.235C→T (p.Q79X) and c.285_291del7insCTGTAT and for heterozygous mutation c.955delA. Mutation sites are indicated by arrows (↓).
In four of the five families, patients apparently carry homozygous mutations. The presence of heterozygous mutations in both parents could be shown by segregation analysis in families CHRO22 and CHRO87 (fig. 1a and b). Moreover, parental consanguinity is documented in families CHRO22 and CHRO94. We also noticed that, in the remaining two families, CHRO68 and CHRO87, both parents traced their ancestry back to the same geographic region in southern Italy and Denmark, respectively. Therefore, we think that, in the end, consanguinity exists between those parents too, albeit earlier than known family history.
All the observed mutations result in premature translation termination and in mutant polypeptides that lack considerable portions of the genuine carboxy terminus. It has been shown that the conserved carboxy terminus of the rod photoreceptor paralog contains major sites of interaction with the excited rod photopigment (Cai et al. 2001). Considering the high conservation between the rod and cone transducin α-subunits, a similar structural function can be assumed to exist in the cone system. We therefore argue that the mutations represent functional null alleles of GNAT2, which prevent either the formation of the trimeric G-protein complex or its interaction with the excited photopigments.
Notably, all genes now known to be involved in achromatopsia encode crucial components of the cone phototransduction cascade. Because of the lack of cone function in patients with achromatopsia, it is reasonable to argue that, like the cone-specific cGMP-gated channel, all three different types of human cone photoreceptors utilize a common transducin α-subunit and that there is functional conservation of the phototransduction process in cones.
The present study provides convincing evidence that mutations in the GNAT2 gene cause achromatopsia. Patients with mutations in GNAT2 show a phenotype typical of achromatopsia, with low visual acuity, photophobia, nystagmus, and absent or barely detectable cone function in electroretinographic recordings and psychophysical color-vision testing. Genetic classification now allows further detailed clinical studies to investigate whether there are phenotypic differences that distinguish between GNAT2-associated and other forms of achromatopsia.
GNAT2 is the third gene found to be implicated in achromatopsia. Considering that our complete sample of patients consists of nearly 280 unrelated patients with achromatopsia, mutations in the GNAT2 gene are responsible for <2% of all patients affected by this disorder. Compared with the other two genes known to cause achromatopsia—CNGA3 and CNGB3, which account for ∼20%–30% and ∼40%–50% of the cases, respectively (Wissinger et al. 2001; authors' unpublished data)—GNAT2 is only a minor achromatopsia locus. This emphasizes what we call the “orphan disease-gene concept,” which is based on the exclusion of known common disease genes in order to define a subset of patients for subsequent identification of rare disease genes. This will not only improve genetic diagnostics but also provide examples of the effect of mutations in a particular gene and, thereby, increase our knowledge about the pathophysiological spectrum of human genetic disorders.
Acknowledgments
We thank all patients and family members for participating in this study, and we thank Sten Andreasson, Pierre Bitoun, Claudio Castellan, Frans Cremers, Helene Dollfus, Agnes Farkas, Roberto Giorda, Carel Hoyng, Herbert Jägle, Bernhard Jurklies, Gert Matthijs, Francis Munier, Günter Rudolph, Roberto Salati, E. Cumhur Sener, Lindsay T. Sharpe, and Sinan Tatlipinar for collection of patients. This work was supported by grant 01 KS 9602/Q1, jointly funded by the Bundesministerium für Forschung und Technologie and the Interdiziplinäres Zentrum für Klinische Forschung Tübingen, and by Deutsche Forschungsgemeinschaft grants SFB430/A5 and Lo457/1-1,2,3.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human cone photoreceptor transducin α-subunit [GNAT2] [accession number Z18859] and human rod photoreceptor transducin α-subunit [GNAT1] [accession number X15088])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ACHM2 [MIM 216900], ACHM3 [MIM 262300], and GNAT2 [MIM 139340])
References
- Blatt C, Eversole-Cire P, Cohn VH, Zollman S, Fournier RE, Mohandas LT, Nesbitt M, Lugo T, Jones DT, Reed RR, Weiner LP, Sparkes RS, Simon MI (1988) Chromosomal localization of genes encoding guanine nucleotide-binding protein subunits in mouse and human. Proc Natl Acad Sci USA 85:7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Itoh Y, Khorana HG (2001) Mapping of contact sites in complex formation between transducin and light-activated rhodopsin by covalent crosslinking: use of a photoactivatable reagent. Proc Natl Acad Sci USA 98:4877–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Krasnoperova NV, Lyubarsky AL, Sayama T, Nicolo M, Kosaras B, Wong G, Gannon KS, Margolskee RF, Sidman RL, Pugh EN Jr, Makino CL, Lem J (2000) Phototransduction in transgenic mice after targeted deletion of the rod transducin alpha-subunit. Proc Natl Acad Sci USA 97:13913–13918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Dunnen JT, Antonarakis SE (2001) Nomenclature for the description of human sequence variations. Hum Genet 109:121–124 [DOI] [PubMed] [Google Scholar]
- Dryja TP, Hahn LB, Reboul T, Arnaud B (1996) Missense mutation in the gene encoding the alpha subunit of rod transducin in the Nougaret form of congenital stationary night blindness. Nat Genet 13:358–360 [DOI] [PubMed] [Google Scholar]
- François J (1961) Heredity in ophthalmology. Mosby, St Louis [Google Scholar]
- Gerber S, Rozet JM, Bonneau D, Souied E, Weissbach J, Frezal J, Munnich A, Kaplan J (1995) Exclusion of the cone-specific alpha-subunit of the transducin gene in Stargardt's disease. Hum Genet 95:382–384 [DOI] [PubMed] [Google Scholar]
- Kohl S, Baumann B, Broghammer M, Jägle H, Sieving P, Kellner U, Spegal R, Anastasi M, Zrenner E, Sharpe LT, Wissinger B (2000) Mutations in the CNGB3 gene encoding the β-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet 9:2107–2116 [DOI] [PubMed] [Google Scholar]
- Kohl S, Marx T, Giddings I, Jägle H, Jacobson SG, Apfelstedt-Sylla E, Zrenner E, Sharpe LT, Wissinger B (1998) Total colorblindness is caused by mutations in the gene encoding the α-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet 19:257–259 [DOI] [PubMed] [Google Scholar]
- Kubo M, Hirano T, Kakinuma M (1991) Molecular cloning and sequence analysis of cDNA and genomic DNA for the human cone transducin alpha subunit. FEBS Lett 291:245–248 [DOI] [PubMed] [Google Scholar]
- Lerea CL, Bunt-Milam AH, Hurley JB (1989) Alpha-transducin is present in blue-, green-, and red-sensitive cone photoreceptors in the human retina. Neuron 3:367–376 [DOI] [PubMed] [Google Scholar]
- Lerea CL, Somers DE, Hurley JB, Klock IB, Bunt-Milam AH (1986) Identification of specific transducin alpha subunits in retinal rod and cone photoreceptors. Science 234:77–80 [DOI] [PubMed] [Google Scholar]
- Magovcevic I, Weremowicz S, Morton CC, Fong SL, Berson EL, Dryja TP (1995) Mapping of the human cone transducin alpha-subunit (GNAT2) gene to 1p13 and negative mutation analysis in patients with Stargardt disease. Genomics 25:288–290 [DOI] [PubMed] [Google Scholar]
- Morris TA, Fong SL (1993) Characterization of the gene encoding human cone transducin alpha-subunit (GNAT2). Genomics 17:442–448 [DOI] [PubMed] [Google Scholar]
- Müller F, Kaupp UB (1998) Signaltransduktion in Sehzellen. Naturwissenschaften 85:49–61 [DOI] [PubMed] [Google Scholar]
- Sundin OH, Yang JM, Li Y, Zhu D, Hurd JN, Mitchell TN, Silva ED, Hussels-Maumenee I (2000) Genetic basis of total colorblindness among the Pingelapese islanders. Nat Genet 25:289–293 [DOI] [PubMed] [Google Scholar]
- Wissinger B, Gamer D, Jagle H, Giorda R, Marx T, Mayer S, Tippmann S, et al (2001) CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet 69:722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]