Significance
Circadian clocks are endogenous molecular oscillators that drive daily rhythms of physiology and behavior. The mammalian clock, intrinsic to most cells and tissues, is built on a conserved negative feedback loop in which a heterodimeric transcription factor, circadian locomotor output cycles kaput (CLOCK)-brain, muscle Arnt-like 1 (BMAL1), drives the transcription of its specific inhibitor proteins. This study shows that CLOCK-BMAL1 is associated with a clock-driven, rhythmically expressed coactivator protein, TRAP150 (thyroid hormone receptor-associated protein-150), that promotes the binding of CLOCK-BMAL1 to DNA and links it to the basic transcriptional machinery. The activity and oscillation of TRAP150 define a positive feedback loop within the core clock and provide a potential mechanism for the precise timing of circadian transcription cycles.
Abstract
Circadian clocks in mammals are built on a negative feedback loop in which the heterodimeric transcription factor circadian locomotor output cycles kaput (CLOCK)-brain, muscle Arnt-like 1 (BMAL1) drives the expression of its own inhibitors, the PERIOD and CRYPTOCHROME proteins. Reactivation of CLOCK-BMAL1 occurs at a specific time several hours after PERIOD and CRYPTOCHROME protein turnover, but the mechanism underlying this process is unknown. We found that mouse BMAL1 complexes include TRAP150 (thyroid hormone receptor-associated protein-150; also known as THRAP3). TRAP150 is a selective coactivator for CLOCK-BMAL1, which oscillates under CLOCK-BMAL1 transcriptional control. TRAP150 promotes CLOCK-BMAL1 binding to target genes and links CLOCK-BMAL1 to the transcriptional machinery at target-gene promoters. Depletion of TRAP150 caused low-amplitude, long-period rhythms, identifying it as a positive clock element. The activity of TRAP150 defines a positive feedback loop within the clock and provides a potential mechanism for timing the reactivation of circadian transcription.
Circadian clocks are endogenous oscillators that drive daily rhythms of physiology and behavior. The mammalian clock, intrinsic to most cells and tissues (1, 2), is built on a conserved negative feedback loop that generates circadian rhythms at the molecular level (3). The core positive element of the clock is the heterodimeric transcription factor circadian locomotor output cycles kaput (CLOCK)-brain, muscle Arnt-like 1 (BMAL1), which drives transcription of Period (Per) and Cryptochrome (Cry) genes from E-box sites (4). PER and CRY proteins, acting as negative elements of the clock, enter the nucleus, associate with CLOCK-BMAL1 (5) at E-box sites (6), and suppress the transcriptional activity of CLOCK-BMAL1 in part by recruiting the SIN3-HDAC histone deacetylase complex (6) and inhibiting transcriptional termination (7). Turnover of PERs and CRYs ends the negative-feedback phase of the cycle (8–11). An interlocked feedback loop involving REV-ERBα and -β (nuclear receptor subfamily 1, group D, members 1 and 2, respectively) contributes to clock function (12–14).
Reactivation of CLOCK-BMAL1 transcription of circadian target genes occurs several hours after the end of negative feedback (15, 16), suggesting that the onset of circadian transcription in each cycle by CLOCK-BMAL1 is not simply a passive consequence of the turnover of negative-feedback proteins, but is positively regulated and timed by unknown clock-controlled factors. Evidence that there is active positive regulation of CLOCK-BMAL1 comes from reports showing enhancement of CLOCK-BMAL1 transcriptional activity by chromatin-modifying proteins by CBP/p300 (17), MLL1 (18), and JARID1a (19). Although not previously described, a clock-controlled, rhythmic positive factor for CLOCK-BMAL1 would provide a potential mechanism for precisely setting the onset of transcription each circadian cycle.
Results
To identify factors associated with CLOCK-BMAL1, we used FLAG antibodies to affinity-purify BMAL1 complexes from a mouse fibroblast cell line expressing a FLAG-Hemagglutinin-BMAL1 fusion protein (FH-BMAL1) (20). After purification and gel electrophoresis, mass spectrometry revealed that proteins specifically copurifying with FH-BMAL1 included CLOCK, as expected, and TRAP150 (thyroid hormone receptor-associated protein-150), a protein not previously linked to circadian clock function. TRAP150 is a transcriptional coactivator for certain nuclear receptors (21, 22), and it has additional functions in RNA splicing (23) and DNA repair (24). TRAP150 has occasionally been confused with Mediator (MED) subunit 23; the two are unrelated proteins encoded by different genes (SI Text). TRAP150 associates with a small subset of Mediator complexes in its function as a transcriptional coactivator, but it is not considered to be a structural component of Mediator (25).
Antibodies to TRAP150 coimmunoprecipitated BMAL1 and CLOCK from nuclear extracts of wild-type mouse fibroblasts but did not coimmunoprecipitate the ubiquitous transcription factor SP1 or nuclear protein LAMIN B1 (Fig. 1A). A similar result was obtained from nuclear extracts of mouse liver (Fig. 1B). In addition, antibodies to CLOCK specifically coimmunoprecipitated BMAL1 and TRAP150 from mouse fibroblast nuclear extracts (Fig. 1C). TRAP150 is thus a constituent of endogenous BMAL1 complexes.
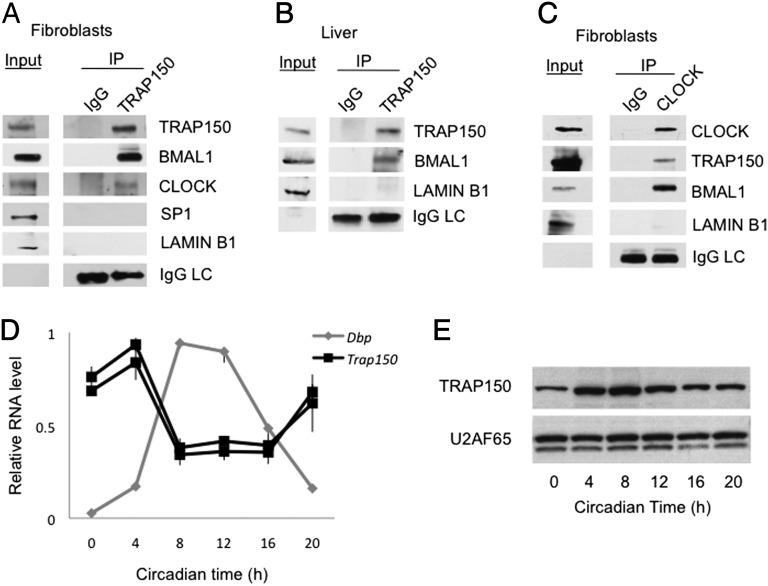
Fig. 1.
TRAP150 is a clock-regulated constituent of endogenous BMAL1 complexes. (A) Coimmunoprecipitation of BMAL1 and CLOCK with TRAP150 from nuclear extracts of wild-type mouse fibroblasts (unsynchronized). Nuclear proteins SP1 and LAMIN B1 are negative controls and IgG light chain (IgG LC) a positive control for immunoprecipitation (IP). IgG, nonspecific IgG. (B) Coimmunoprecipitation of BMAL1 with TRAP150 from nuclear extracts of wild-type mouse liver (CT8). (C) Coimmunoprecipitation of TRAP150 and BMAL1 with CLOCK from mouse fibroblast nuclear extracts (unsynchronized). Input lanes in A–C represent 0.5% of total extract; IP lanes represent 5–12.5% of total extract (vol/vol). (D) Circadian oscillation of Trap150 mRNA in liver. High-amplitude circadian profile of Dbp mRNA is shown for reference. The two Trap150 mRNA profiles were generated with different primer pairs for quantitative RT-PCR. Data show mean ± SEM (n = 3). No products were detected when reverse transcriptase was omitted. (E) Circadian oscillation of TRAP150 protein in liver. U2AF65, loading control.
Trap150 mRNA exhibited a circadian oscillation of abundance in mouse liver with a peak at approximately circadian time (CT) 4 h (Fig. 1D), and TRAP150 protein showed a circadian rhythm (Fig. 1E) that lagged slightly behind the mRNA oscillation, with the peak at approximately CT8, a time when CLOCK-BMAL1 DNA-binding and transcriptional activity are maximal (6, 15, 26, 27). The Trap150 gene contains E-box sequences in its promoter (Fig. S1) and was ranked among significant targets of rhythmic BMAL1 binding in two circadian genome-wide ChIP studies in mouse liver (15, 26). TRAP150 is thus a clock-regulated, rhythmic component of BMAL1 complexes.
To determine if TRAP150 is associated with CLOCK-BMAL1 on DNA we performed ChIP studies of mouse tissues. In the liver (CT8), TRAP150, BMAL1, and CLOCK cooccupied the proximal E-box site of the Per1 gene, but TRAP150 was not found at arbitrary sites in the Per1 gene that lacked detectable CLOCK-BMAL1 binding (Fig. 2A). Circadian analysis of mouse liver and lung revealed that BMAL1, CLOCK, and TRAP150 exhibit a synchronous circadian oscillation at the Per1 E-box site (Fig. 2 B and C). TRAP150 showed a similar circadian pattern of occupancy at the E-box sites of multiple additional CLOCK-BMAL1 circadian target genes (Fig. S2A), and its E-box association was dependent on BMAL1 (Fig. S2B). Thus, TRAP150, CLOCK, and BMAL1 rhythmically colocalize at circadian E-box sites, peaking at a time of the day when CLOCK-BMAL1 is transcriptionally active.
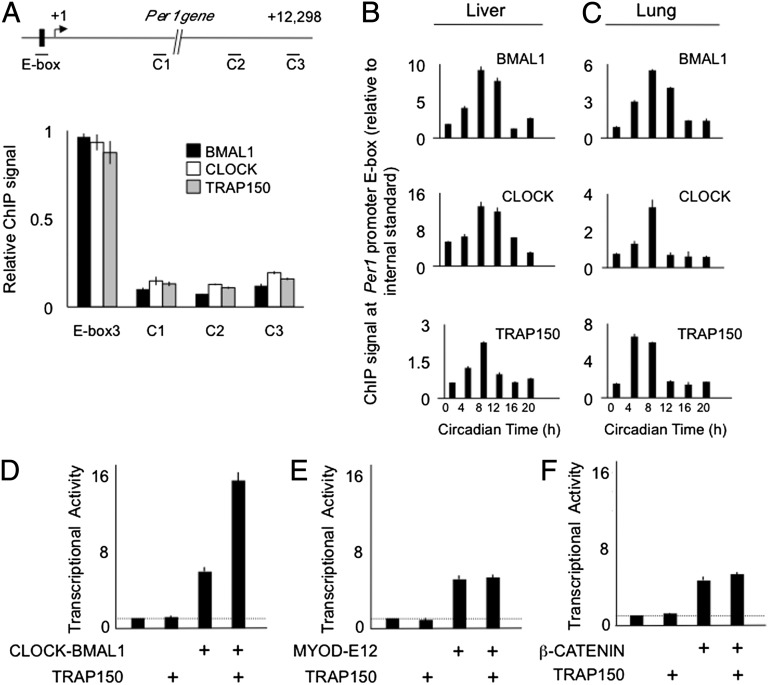
Fig. 2.
Coordinate circadian binding of BMAL1, CLOCK, and TRAP150 to the Per1 promoter. (A, Upper) Mouse Per1 gene showing proximal E-box and arbitrary control sites C1, C2, and C3. Small lines at each site indicate PCR product amplified in ChIP assays. (Lower) ChIP assays from mouse liver harvested at CT8, performed with antibodies against the indicated proteins. ChIP data: mean ± SEM of triplicate experiment; representative of three experiments. Signal was normalized to IgG control ChIP, and within each experiment the signal at the Per1 E-box was set to 1. (B and C) Synchronous circadian cycle of BMAL1, CLOCK, and TRAP150 at Per1 E-box site. Shown are ChIP assays from mouse liver (B) and lung (C) obtained across a circadian cycle performed with antibodies indicated at the upper right corner of each panel. Background was determined from an arbitrary internal control gene and was set to a value of 1. Results are representative of four independent experiments. (D–F) TRAP150 selectively promotes CLOCK-BMAL1 transcriptional activity. Luciferase transactivation assays in mouse fibroblasts showing the effect of TRAP150 on the transcriptional activity of (D) CLOCK-BMAL1, (E) MYOD-E12, or (F) β-CATENIN. Transfection of expression plasmids for the transcription factors was set such that the activity of each transcription factor in the absence of transfected TRAP150 was four- to five-times background (dashed line), well below saturation of the assay.
Because TRAP150 acts as a coactivator for certain nuclear receptors, we tested whether it can similarly promote transcriptional activity of CLOCK-BMAL1. Overexpression of TRAP150 in mammalian cells strongly stimulated CLOCK-BMAL1 transcription from a Per1 E-box (Fig. 2D), but it had no effect on basal transcription (Fig. 2 D–F) or on the activity of other transcription factors (Fig. 2 E and F). Because the reporter gene in this transactivation assay lacks introns, the selective action of TRAP150 likely reflects its transcriptional coactivator properties rather than an effect on pre-mRNA splicing.
To determine if endogenous TRAP150 functions as a coactivator for CLOCK-BMAL1, we depleted TRAP150 from mouse fibroblasts (20) or human U2OS circadian reporter cells (28, 29) and monitored the mean transcription of CLOCK-BMAL1 target genes or real-time circadian oscillations, respectively. Depletion of a coactivator should result in reduced CLOCK-BMAL1 transcriptional activity, thereby selectively decreasing the transcription of CLOCK-BMAL1 target genes and producing circadian oscillations with a long period and low amplitude, as observed with the Clock mutation (30, 31) or after depletion of CLOCK or BMAL1 from mammalian cells (28).
Depletion of TRAP150 (Fig. 3A) from unsynchronized fibroblasts caused varying reductions (10–40%) in the mean transcription of CLOCK-BMAL1 circadian target genes but had little or no effect on the mean transcription of control genes (Fig. 3B). The range of effects suggests that TRAP150 plays a greater role in the transcription of some target genes, such as Per1 and Dec1, than others. A likely reason for this variability is that some clock genes, such as Per1, are driven predominantly by CLOCK-BMAL1 (and even then not exclusively), whereas others appear to be driven substantially by additional transcription factors (32, 33), which would not likely be affected by the TRAP150 depletion. Because the reduction of TRAP150 was only partial, these results likely underestimate the true dependence of CLOCK-BMAL1 transcriptional activity on TRAP150; nonetheless, and despite the variability, the difference between CLOCK-BMAL1 target genes as a class and control genes as a class was highly significant (P < 0.001; t test, one-tailed). Thus, TRAP150 promotes CLOCK-BMAL1 transcriptional activity at multiple circadian target genes.
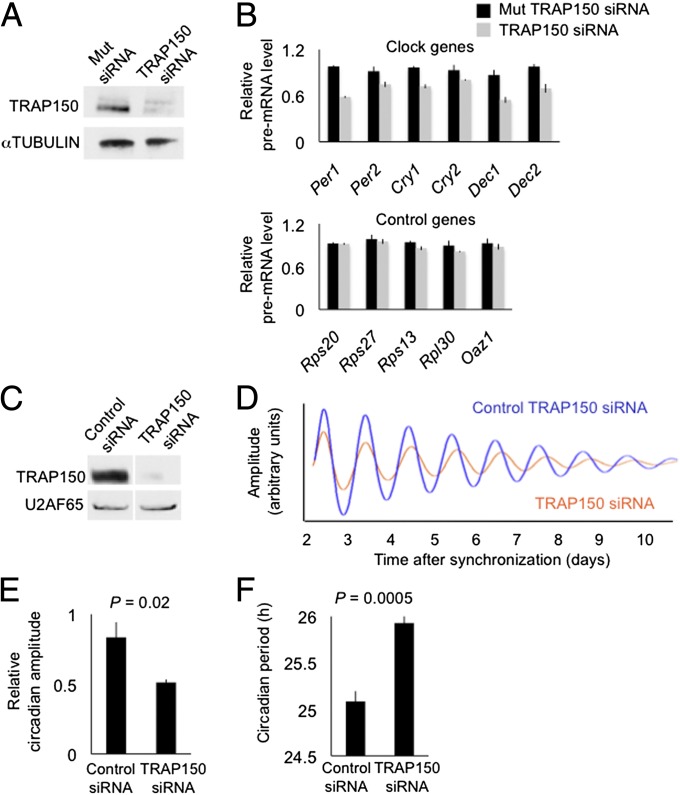
Fig. 3.
Depletion of TRAP150 from mammalian cells impairs transcription of CLOCK-BMAL1 circadian target genes and alters circadian clock function. (A) Western blot showing the effect of control siRNA (Mut TRAP150, three point-mutations) or TRAP150 siRNA on steady-state level of endogenous TRAP150 in mouse fibroblasts. α-TUBULIN, loading control. (B) Effect of TRAP150 depletion on transcription of endogenous CLOCK-BMAL1 circadian target genes and noncircadian control genes, as assessed by quantitative RT-PCR of the respective pre-mRNAs (mean ± SEM of triplicate experiment). For each gene, signals were normalized to the highest value among the six measurements (triplicate control and triplicate TRAP150 depletion). Data are representative of three independent experiments. No products were detected when reverse transcriptase was omitted. (C–F) Human Bmal1-Luc U2OS circadian reporter cells. (C) Western blot showing the effect of control siRNA or TRAP150 siRNA on steady-state level of endogenous TRAP150. U2AF65, loading control. (D) Real-time circadian oscillations of bioluminescence in synchronized cells after electroporation of TRAP150 siRNA (red) or control siRNA (blue). Representative of seven independent experiments. (E and F) Group data for circadian amplitudes and period lengths, respectively, after depletion of TRAP150 (mean ± SEM; n = 7; t test, amplitude one-tailed; period length two-tailed).
Depletion of TRAP150 (Fig. 3C) from synchronized U2OS cells caused a low-amplitude, long-period circadian phenotype that was readily apparent in individual bioluminescence traces (Fig. 3D) and significant in the population data (Fig. 3 E and F). The change in period length is comparable to that observed after depletion of PER1, PER2, REV-ERBα, or REV-ERBβ in similar RNA interference experiments (34). Essentially identical phenotypes were obtained when TRAP150 was depleted with a different, nonoverlapping siRNA (Fig. S3 A–D) or when TRAP150 was depleted from a different circadian reporter cell line (Fig. S3 E–H). Mathematical simulation of circadian oscillations using a model of the core mammalian circadian clock (35) suggests that a reduction in Per and Cry transcription comparable to what we observed after depletion of TRAP150 is sufficient to account for the long-period, low-amplitude phenotype (Fig. S4). These results identify TRAP150 as a positive element of the clock.
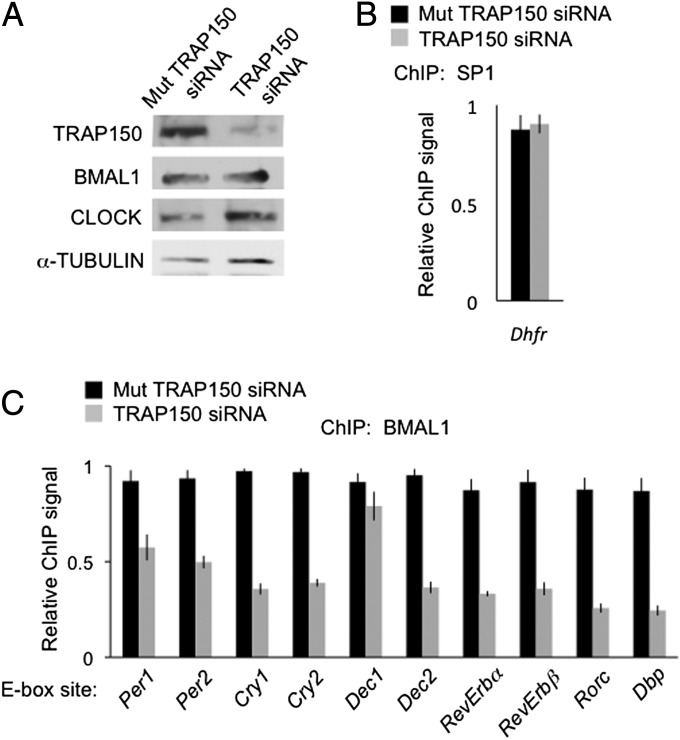
Because TRAP150 has actions in addition (23, 24) to transcriptional coactivation, we tested whether TRAP150 performs several specific functions expected of a CLOCK-BMAL1 coactivator. Depletion of TRAP150 (Fig. 4A) had no effect on the occupancy of the ubiquitous transcription factor SP1 on the Dhfr promoter (Fig. 4B), but it caused a substantial reduction in the occupancy of BMAL1 at various circadian target genes (Fig. 4C). A similar result was found for CLOCK (Fig. S5). The effect on BMAL1 occupancy was stronger and more general than the effect on target gene transcription, a finding consistent with multiple activators contributing to circadian gene expression (32, 33). TRAP150 thus potently and selectively promotes the binding of CLOCK-BMAL1 to circadian target genes.
Fig. 4.
TRAP150 promotes CLOCK-BMAL1 binding to circadian target genes. (A) Depletion of TRAP150 from mouse fibroblasts does not detectably affect BMAL1 and CLOCK steady-state levels. Shown are Western blots of mouse fibroblasts probed with antibodies against the indicated proteins after introduction of ineffective mutant TRAP150 siRNA or effective TRAP150 siRNA. (B) Control ChIP assay showing little or no effect of TRAP150 depletion on the occupancy of the ubiquitous transcription factor SP1 on the Dhfr gene promoter. (C) ChIP assays showing the effect of TRAP150 depletion on the occupancy of BMAL1 on E-box sites of the indicated genes. Data show the mean ± SEM of triplicate experiment. For each gene, signals were normalized to the highest value among the six measurements (triplicate control and triplicate TRAP150 depletion). Data are representative of three independent experiments.
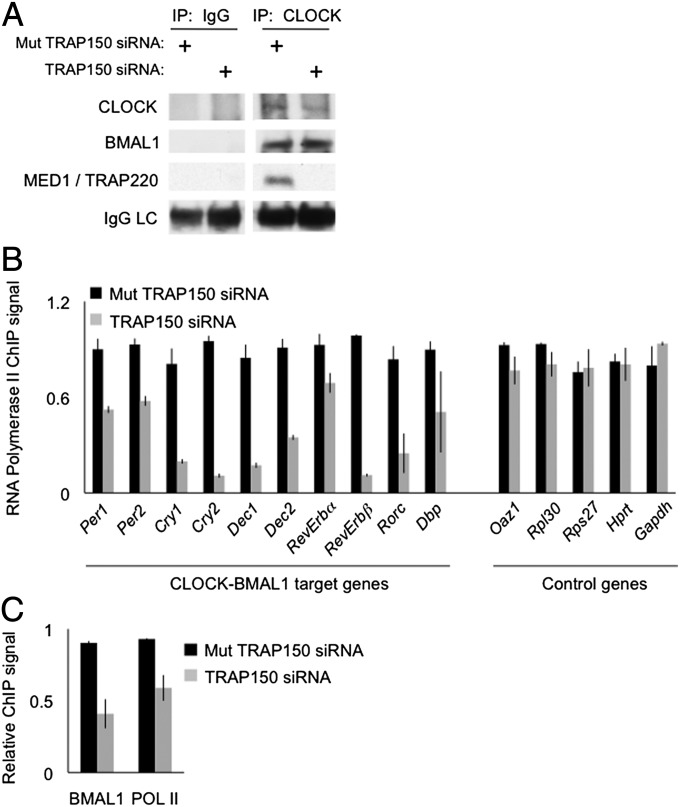
TRAP150 acts as a nuclear receptor coactivator by linking its target nuclear receptors to the MED1/TRAP220 subunit of Mediator (21, 22, 25), a large protein complex that modulates the RNA polymerase II preinitiation complex (25). We examined whether TRAP150 also uses MED1 to bridge CLOCK-BMAL1 to the transcriptional machinery. Depletion of TRAP150 from mouse fibroblasts had no detectable effect on the association of CLOCK with BMAL1, but it markedly reduced or abolished the association of CLOCK with MED1/TRAP220 (Fig. 5A).
Fig. 5.
TRAP150 links CLOCK-BMAL1 to basic transcriptional machinery. (A) IgG control or CLOCK immunoprecipitations showing the effect of TRAP150 depletion on the association of CLOCK (or BMAL1) with the MED1/TRAP220 subunit of Mediator. Note that depletion of TRAP150 had no appreciable effect on the CLOCK-BMAL1 interaction. (B) ChIP assays showing the effect of TRAP150 depletion on the association of RNA polymerase II with E-box sites of CLOCK-BMAL1 circadian target genes or comparable sites of noncircadian control genes, as indicated. Association of RNA polymerase II with E-box sites could reflect direct binding nearby or cross-linking of CLOCK-BMAL1 at E-box to RNA polymerase II at or near transcriptional start site (with looping of intervening DNA). Shown are mean ± SEM of triplicate experiments. For each gene, signals were normalized to the highest value among the six measurements (triplicate control and triplicate TRAP150 depletion). Data are representative of three independent experiments. (C) ChIP assays showing the effect of TRAP150 depletion on BMAL1 binding or RNA polymerase II binding to the Trap150 gene E-box site, as indicated. Data analyzed as for panel B above.
As expected for a coactivator (22), depletion of TRAP150 dramatically reduced the association of RNA polymerase II with the E-box sites of multiple CLOCK-BMAL1 circadian target genes, but it had little or no effect on RNA polymerase II on the promoters of control genes (Fig. 5B). The reduced association of RNA polymerase II with E-box sites could reflect decreased direct binding of the polymerase to chromatin locally or, perhaps more likely, decreased cross-linking of CLOCK-BMAL1 at the E-box to RNA polymerase II at or near the transcriptional start site. Depletion of TRAP150 also reduced the association of BMAL1 and RNA polymerase II with the E-box site just upstream of the Trap150 transcriptional start site, providing further evidence that TRAP150 is a CLOCK-BMAL1-regulated factor (Fig. 5C). This selective and substantial decrease of RNA polymerase II association with circadian E-box sequences indicates a major role for TRAP150 in bridging bound CLOCK-BMAL1 to RNA polymerase II on circadian target genes. The significant disruption of the association between CLOCK-BMAL1 at E-box sites and RNA polymerase II observed for virtually all of the circadian target genes (Fig. 5B) would be expected to impair transcription substantially at those target genes driven predominantly by CLOCK-BMAL1, such as Per1, and less so at those driven by a combination of activators, which have factors in addition to CLOCK-BMAL1 stabilizing the polymerase at transcriptional start sites.
Discussion
Our results show that TRAP150 is a clock-regulated component of CLOCK-BMAL1 complexes that promotes CLOCK-BMAL1 transcriptional activity and is important for circadian clock function. TRAP150 promotes the DNA-binding of CLOCK-BMAL1, associates with CLOCK-BMAL1 at E-box sites of circadian target genes, and it links CLOCK-BMAL1 to Mediator, thereby recruiting the general transcriptional machinery. Taken together, the circadian oscillation of TRAP150 and its intrinsic properties as a CLOCK-BMAL1 coactivator provide a potential mechanism for setting the onset of circadian transcription each cycle. According to this model, a few hours after the end of negative feedback TRAP150 accumulates to its circadian peak concentration, associates with CLOCK-BMAL1 complexes, and incorporates CLOCK-BMAL1 into a cooperative network of protein–protein and protein–DNA interactions, thereby stabilizing the CLOCK-BMAL1-TRAP150 at the E-box site at a defined circadian phase. In this regard, TRAP150 might work synergistically with other CLOCK-BMAL1 coactivators (17–19).
Because TRAP150 is itself clock-regulated, the coactivator function of TRAP150 creates a positive feedback loop nested within the core circadian negative feedback loop (Fig. S6). A similar topology appears in other rhythmic systems, and a recent analysis indicates that an embedded positive feedback loop has advantages for the precision and robustness of negative feedback oscillators (36). It is likely that circadian rhythms of physiology and behavior rely on fine temporal control of transcription within and beyond the circadian clock feedback loop (37). By linking CLOCK-BMAL1 to the basic transcriptional machinery, TRAP150 is critically placed to modulate circadian transcription.
Materials and Methods
Cell Lines.
Cell lines expressing FH-BMAL1 were generated by stably transfecting BLi cells (20) with an expression plasmid in which a proximal 1-kb fragment of the Bmal1 promoter is linked to a cDNA encoding FH-BMAL (pBS/Neo-endo/FH-Bmal1). G418 (Gibco; 500 µg/mL) selection was initiated 48 h after transfection. Following 2–3 wk of selection, the remaining clones were pooled and used for experiments.
Cell Fractionation for Preparative Purification.
Cells were lysed [15 mM NaCl, 60 mM KCl, 12% (wt/vol) sucrose, 2 mM EDTA, 0.5 mM EGTA, 0.65 mM spermidine, 1 mM DTT, 0.05% Triton X-100, protease inhibitor mixture EDTA-free (Roche), phosphatase inhibitor mixtures I and II (Sigma)]. Nuclei were pelleted (13,000 × g, 15 min), and the supernatant (cytoplasmic fraction) was stored at −80 °C. Nuclei were washed three times in lysis buffer and resuspended in 100 mM Hepes pH 7.4, 200 mM NaCl, 0.5% Nonidet P-40, with protease and phosphatase inhibitor mixtures as above. The suspension was sonicated and then centrifuged at 10,000 × g, and the supernatant (nuclear fraction) was collected.
Isolation of BMAL1 Complexes.
EFH nuclear fraction obtained by cell fractionation was either applied to anti-FLAG columns (FLAG M2; Sigma) or to the same column preincubated with flag peptide (Sigma; 0.1 mg/mL). After three washes in 100 mM Hepes pH 7.4, 200 mM NaCl, 0.5% Nonidet P-40 supplemented with protease and phosphatase inhibitors (Sigma), BMAL1-containing complexes were eluted in Laemmli buffer and resolved by SDS/PAGE.
Transactivation Assay.
293T cells were transfected with Lipofectamine-Plus (GibcoBRL), and grown in DMEM supplemented with 10% (vol/vol) FBS (GibcoBRL) in 24-well plates. Cells in each well were transfected with 125 ng to 400 ng of the expression plasmids with the indicated inserts, 15 ng of the indicated firefly luciferase reporter with the indicated inserts, and 7.5 ng of pRL-CMV (Renilla) luciferase control plasmid. Expression vectors were transfected using the following amounts: β-catenin (12.5 ng per well), BMAL1 (20 ng per well), CLOCK (20 ng per well), MYOD (12.5 g per well), and TRAP150 (100 ng per well). After 48 h, cells in each well were lysed with 200 µL of Promega passive lysis buffer, and measured for for firefly and Renilla luciferase emission. The MYOD, BMAL1, and CLOCK expression vectors were previously described (20).
Circadian Profile of Trap150 mRNA.
Total RNA from livers was purified (Ambion RiboPure). Residual genomic DNA was digested using DNaseI (Invitrogen), and RNA was transcribed into cDNA using random hexamers and MultiScribe reverse transcriptase (Applied Biosystems). cDNA derived from 1 µg total RNA was PCR-amplified in a PTC-200 thermocycler with a Chromo4 module (MJ Research) using TaqMan primer-probe assays and Universal Master Mix (Life Technologies) according to the manufacturer’s instructions. For quantification, threshold cycle number difference was calculated based on the sample with the lowest expression, and the values obtained were 2n transformed. The expression level of Gapdh was used to normalize expression values. Primers (Integrated DNA Technologies) were designed such that products would span an intron-exon boundary (Table S1).
Circadian Profile of TRAP150 Protein.
Tissues were homogenized by douncing in cellular fractionation buffer [15 mM NaCl, 60 mM KCl, 12% (wt/vol) sucrose, 2 mM EDTA, 0.5 mM EGTA, 0.65 mM spermidine, 1 mM DTT, 0.05% Triton X-100, protease inhibitor mixture EDTA-free (Roche), phosphatase inhibitor mixtures I and II (Sigma)]. Nuclei were then pelleted by centrifugation (13,000 × g, 15 min), washed three times in lysis buffer, and resuspended in 100 mM Hepes pH 7.4, 200 mM NaCl, 0.5% Nonidet P-40 with protease and phosphatase inhibitor mixtures as above. The suspension was sonicated and then centrifuged at 10,000 × g, and the supernatant (nuclear fraction) was collected. For SDS/PAGE, nuclear extracts were diluted in Guanidine HCl (final Guanidine HCl concentration of 4.5 M) and denatured in Laemmli buffer.
Mass Spectrometry.
Mass Spectrometry was performed as previously described (20).
Mice and Tissue Collection.
Male C57BL/6J mice were entrained to a 12:12-h light-dark cycle (> 10 d) and then transferred to darkness. Mice were killed under infrared light, and tissues were dissected under light. For ChIP and RNA analysis, livers and lungs were flash-frozen in liquid nitrogen. For coimmunoprecipitation, tissues were immersed in lysis buffer and processed immediately. Studies were performed in accordance with a protocol approved by the Harvard Medical School Standing Committee on Animals.
Circadian Reporter Fibroblast Cell Lines.
Circadian reporter cell lines were previously described: human Bmal1-Luc and Per2-Luc reporter U2OS cell line (28, 29).
Real-Time Monitoring of Circadian Oscillations.
U2OS cells (>90% confluence) were synchronized by addition of DMEM containing 10 μM Forskolin. After 2 h, the medium was changed to DMEM with 10% FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, and 250 mM d-Luciferin. Bioluminescence was continuously recorded (LumiCycle, Actimetrics) and analyzed as previously described (38). For amplitude calculation, each trace was analyzed individually to determine the precise times of peak and trough at day 3 (U2OS Bmal1-Luc) or day 5 (U2OS Per2-Luc). Peak and trough luminescence emission counts were obtained for each trace from the raw (baseline-subtracted) data. Amplitude was calculated as the difference in emission between peak and trough.
Coommunoprecipitation.
Cells and tissues were processed as follows. For CLOCK, 4 μg of anti-CLOCK IgG (Thermo-Fisher) or control IgG (Cell Signaling Technologies) were incubated with 500 μg of cell extract overnight at 4 °C. For TRAP150, 4 µg of anti-TR150 (Abcam) or control IgG (Cell Signaling Technologies) were used. Antibody–protein complexes were then incubated with Rabbit-IgG conjugated magnetic beads (Invitrogen). Beads were washed three times (0.5 mL, 0.5% Nonidet P-40, 250 mM NaCl), and protein complexes were eluted in Laemmli buffer supplemented with β-mercaptoethanol and resolved on SDS/PAGE.
Immunoblots.
Anti-BMAL1 antiserum (39) was made by Covance (1/2,500). Antibodies against the following proteins were obtained from the indicated suppliers: CLOCK (Bethyl; 1/200); TRAP150 (Santa Cruz; 1/500); MED1 (Bethyl; 1/2,000); U2AF65 (Abcam, 1/2,000); SP1 (Millipore, 1/500); α-Tubulin (Sigma; 1/5,000); Lamin B1 (Abcam; 1/2,500); HRP-conjugated secondary antibodies (Amersham; 1:5,000). Extracts (20–100 μg of protein) were resolved by SDS/PAGE, blotted onto PVDF (Millipore), and analyzed by enhanced chemiluminescence (GE Healthcare).
RNA Interference Against TRAP150 (THRAP3).
For Bli cells, siRNA was electroporated with Nucleofector system (D-023, Amaxa, Solution V, Lonza). Three independent sets of oligomers (Integrated DNA Technologies) were designed to be highly specific for TRAP150 (Table S2). RNA or Chromatin was isolated 36 or 48 h after electroporation. For U2OS cells, siRNA against human Thrap3 (Qiagen) was electroporated with Nucleofector System (X-001, Amaxa, Solution V, Lonza).
Pre-mRNA Analysis.
Total RNA from Bli cells was purified (RNAqueous-4PCR; Life Technologies). Residual DNA was digested (DNaseI; Life Technologies), and RNA was transcribed into cDNA using random hexamers (MultiScribe reverse transcriptase; Applied Biosystems). Parallel control reactions omitted the reverse transcriptase. cDNA derived from 400 ng to 1 µg of total RNA was PCR-amplified in a PTC-200 thermocycler with a Chromo4 module (MJ Research) with TaqMan primer-probe assays and Universal Master Mix (Life Technologies) according to the manufacturers’ instructions. Primer efficiency was assayed by amplification of serial dilutions of cDNA, and amplification efficiency was calculated using standard curves (Table S3). Only primers with correlation coefficients (R2) > 0.99 and with slopes of −3.3 ± 0.2 were used. For quantification, threshold cycle number difference was calculated based on the sample with the lowest expression, and the values obtained were 2n-transformed. The expression level of Gapdh was used to normalize expression values. PCR products were either intronic or spanned an intron–exon junction.
ChIP.
For chromatin preparation from liver and lungs, frozen tissues were lysed on ice in Pre-Crosslinking Buffer (100 mM NaCl, 1 mM EDTA pH 8, 0.5 mM EGTA pH 8, and 50 mM Hepes pH 8) and homogenized by douncing. Homogenates were cross-linked in 1% Formaldehyde (10 min, room temperature). Formaldehyde was quenched (0.125 M Glycine), and lysates were washed once with ice-cold PBS supplemented with protease inhibitor mixture, EDTA-free (Roche). Nuclei were extracted in Cell Lysis Buffer (5 mM Pipes pH 7.5, 85 mM KCl, 0.5% Nonidet P-40; 20 min on ice), pelleted by centrifugation (12,500 × g, 10 min, 4 °C), and resuspended in Nuclear Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris⋅HCl pH 8). The suspension was sonicated, centrifuged at 10,000 × g, and the supernatant was collected and used for ChIP. The equivalent of 30 mg of tissue was used for each reaction.
For chromatin preparation from cultured cells, cells were trypsinized, resuspended in DMEM supplemented with 10% FBS and penicillin/streptomycin, and cross-linked in 1% formaldehyde (10 min, room temperature). The cross-linking reaction was quenched (0.125 M glycine), and cells were washed once with ice-cold PBS, supplemented with protease inhibitor mixture, EDTA-free (Roche). Nuclei were extracted in Cell Lysis Buffer (5 mM Pipes pH 7.5, 85 mM KCl, 0.5% Nonidet P-40; 20 min on ice), pelleted by centrifugation (12,500 × g, 10 min, 4 °C), and resuspended in Nuclear Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris⋅HCl pH 8). The suspension was sonicated, centrifuged at 10,000 × g, and the supernatant was collected and used for ChIP. Before immunoprecipitation, DNA was purified from a chromatin aliquot, and shearing efficiency was assessed by gel electrophoresis. Concentration was measured by spectrometry, and the chromatin equivalent of 5 µg of genomic DNA was used for each reaction. Anti-BMAL1 antiserum was as above, 1 µL per reaction. Antibodies against the following proteins were obtained from the indicated suppliers: CLOCK (Bethyl; 1 µg), TR150 (Abcam; 5 µg), SP1 (Millipore; 2 µg), polymerase II (Cell Signaling; 1 µg), PER2 (Alpha Diagnostic International; 5 µg) or IgG (Millipore and Cell Signaling; 1–5 µg per reaction).
Chromatin–antibody complexes were incubated overnight at 4 °C in the presence of IgG-conjugated magnetic beads (Invitrogen and Millipore) for CLOCK, TR150, SP1, and polymerase II reactions, and 2 h at 4 °C for BMAL1 reactions. Beads were washed in 0.5 mL of low-salt, high-salt, lithium chloride, and TE buffers (Millipore), and DNA-chromatin complexes were eluted in Elution buffer (Millipore) supplemented with Proteinase K. Cross-links were reversed at 62 °C for 2 h, DNA was purified (Qiagen QIAQUICK), and the eluate was analyzed by quantitative PCR, in a PTC-200 thermocycler with a Chromo4 module (MJ Research) using SYBR green (IQ SYBR Green Supermix; Bio-Rad) according to the manufacturer’s instructions. Templates were amplified with the annealing temperature initially lowered from 70 °C to 60 °C in two-cycle increments followed by 30 cycles of the following three steps: 20 s at 94 °C, 20 s at 60 °C, and 30 s at 72 °C. For quantification, threshold cycle number difference was calculated based on the sample with the lowest expression, and the values obtained were 2n-transformed as described above. For normalization, binding of target genes to the immunoprecipitating protein was compared with irrelevant IgG-bound DNA, or internally to the binding level of an irrelevant gene (Secreted frizzled-related protein 1, Sfrp1). Primer efficiency was assayed by amplification of serial dilutions of genomic DNA, and amplification efficiency was calculated using standard curves (Table S4). Only primer pairs with correlation coefficients (R2) > 0.99 and with slopes of −3.3 ± 0.2 were used.
Supplementary Material
Acknowledgments
We thank P. Nakatani for the FLAG-Hemagglutinin epitope tag cassette; R. Roeder for a TRAP150 expression plasmid; J. DeBruyne and J. Hogenesch for U2OS cells; M. Greenberg for β-catenin plasmid and reporter; Adrián Granada for generous help with the clock mathematical model; and the Harvard Medical School Critical Discussion Group for helpful advice. This work was supported by an award from the G. Harold and Leila Y. Mathers Charitable Foundation (to C.J.W.); a grant from the National Institute of General Medical Sciences (to C.J.W.); a European Molecular Biology Organization postdoctoral fellowship (to C.B.); and an Alice and Joseph E. Brooks postdoctoral fellowship of Harvard Medical School (to L.L.-D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305980110/-/DCSupplemental.
References
- 1.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS, Hong H-K, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 5.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107(7):855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 6.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332(6036):1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337(6094):599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 8.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316(5826):900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 9.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129(5):1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godinho SIH, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316(5826):897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 11.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 12.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 13.Solt LA, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Martelot G, et al. CycliX Consortium Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10(11):e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosoda H, et al. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain. 2009;2:34. doi: 10.1186/1756-6606-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17(12):1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333(6051):1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327(5964):463–466. doi: 10.1126/science.1180067. [DOI] [PubMed] [Google Scholar]
- 21.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93(16):8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang YK, Guermah M, Yuan C-X, Roeder RG. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc Natl Acad Sci USA. 2002;99(5):2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell. 2010;40(1):126–137. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beli P, et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46(2):212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taatjes DJ. The human Mediator complex: A versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35(6):315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 28.Baggs JE, et al. Network features of the mammalian circadian clock. PLoS Biol. 2009;7(3):e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier B, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23(6):708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitaterna MH, et al. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci USA. 2006;103(24):9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10(10):1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 34.Wallach T, et al. Dynamic circadian protein-protein interaction networks predict temporal organization of cellular functions. PLoS Genet. 2013;9(3):e1003398. doi: 10.1371/journal.pgen.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Relógio A, et al. Tuning the mammalian circadian clock: Robust synergy of two loops. PLOS Comput Biol. 2011;7(12):e1002309. doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akman OE, Rand DA, Brown PE, Millar AJ. Robustness from flexibility in the fungal circadian clock. BMC Syst Biol. 2010;4:88. doi: 10.1186/1752-0509-4-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6(5):e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao W-N, et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat Cell Biol. 2007;9(3):268–275. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 39.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.