Abstract
CHEK2 (previously known as “CHK2”) is a cell-cycle–checkpoint kinase that phosphorylates p53 and BRCA1 in response to DNA damage. A protein-truncating mutation, 1100delC in exon 10, which abolishes the kinase function of CHEK2, has been found in families with Li-Fraumeni syndrome (LFS) and in those with a cancer phenotype that is suggestive of LFS, including breast cancer. In the present study, we found that the frequency of 1100delC was 2.0% among an unselected population-based cohort of 1,035 patients with breast cancer. This was slightly, but not significantly (P=.182), higher than the 1.4% frequency found among 1,885 population control subjects. However, a significantly elevated frequency was found among those 358 patients with a positive family history (11/358 [3.1%]; odds ratio [OR] 2.27; 95% confidence interval [CI] 1.11–4.63; P=.021, compared with population controls). Furthermore, patients with bilateral breast cancer were sixfold more likely to be 1100delC carriers than were patients with unilateral cancer (95% CI 1.87–20.32; P=.007). Analysis of the 1100delC variant in an independent set of 507 patients with familial breast cancer with no BRCA1 and BRCA2 mutations confirmed a significantly elevated frequency of 1100delC (28/507 [5.5%]; OR 4.2; 95% CI 2.4–7.2; P=.0002), compared with controls, with a high frequency also seen in patients with only a single affected first-degree relative (18/291 [6.2%]). Finally, tissue microarray analysis indicated that breast tumors from patients with 1100delC mutations show reduced CHEK2 immunostaining. The results suggest that CHEK2 acts as a low-penetrance tumor-suppressor gene in breast cancer and that it makes a significant contribution to familial clustering of breast cancer—including families with only two affected relatives, which are more common than families that include larger numbers of affected women.
Cell-cycle–checkpoint kinase 2 (CHEK2 [MIM 604373]) (previously known as “CHK2”), a human homologue of budding yeast (Schizosaccharomyces pombe) Cds1 and fission yeast Rad53, is one of the key mediators of cellular responses to DNA damage (Zhou and Elledge 2000; Bartek et al. 2001). CHEK2 is part of the p53 pathway and modulates the function of p53 by phosphorylating it in response to DNA damage, leading to cell-cycle arrest at G1 (Chehab et al. 2000; Shieh et al. 2000). CHEK2 also regulates the function of the BRCA1 protein after DNA damage (Lee et al. 2000). Its upstream regulator in the p53 pathway is the checkpoint protein kinase ATM (Kastan and Lim 2000), which activates CHEK2 by phosphorylation in response to double-strand DNA breaks (Matsuoka et al. 1998, 2000). CHEK2 is thus a component of major functional networks that control cell cycle and DNA repair, as are the most important susceptibility genes in hereditary cancer syndromes. In addition, CHEK2 has been found to undergo somatic mutations in a number of cancer types, including lung, ovarian, and lymphoid tumors and sarcomas, as well as in myelodysplastic syndrome (Bell et al. 1999; Haruki et al. 2000; Hofmann et al. 2001; Tavor et al. 2001; Miller et al. 2002). Recently, rare germline missense variants of CHEK2 were reported in families with multicancer Li-Fraumeni syndrome (LFS [MIM 151623]) (Bell et al. 1999; Lee et al. 2001; Vahteristo et al. 2001b). Interestingly, however, the same recurrent protein-truncating mutation—1100delC in exon 10, which abolishes the kinase function of CHEK2 (Wu et al. 2001)—has been reported both among families with LFS in the United States and among Finnish families with a cancer phenotype that is suggestive of LFS, including breast cancer (Bell et al. 1999; Vahteristo et al. 2001b). This finding prompted us to determine the significance of this CHEK2 genetic variant for breast cancer susceptibility in a large study involving germline-mutation screening of >3,000 individuals. In addition, we compared germline CHEK2 mutation data with analysis of CHEK2 protein expression in breast tumors by use of tissue microarrays.
We first determined the frequency of the 1100delC variant in a population-based cohort of 1,035 unselected patients with breast cancer from the Helsinki (N=627) and Tampere (N=408) University Hospitals and in 1,885 healthy population controls from the Finnish Red Cross Blood Transfusion Service. The breast cancer cohort includes 82% (87% in Helsinki and 75% in Tampere) of all newly diagnosed patients with breast cancer in these hospital districts during 1997–1999. This population-based cohort is described in more detail elsewhere (Syrjakoski et al. 2000), and 4 breast cancer type 1 (BRCA1 [MIM 113705]) and 15 breast cancer type 2, early onset (BRCA2 [MIM 600185]) mutations were discovered in these patients (total frequency 1.8%). For all the cohorts of patients included in the present study, blood samples were collected after obtaining written informed consent and appropriate permissions from the ethics committees of the Departments of Obstetrics and Gynecology and of Oncology at Helsinki University Central Hospital and Tampere University Hospital, as well as from the Ministry of Social Affairs and Health in Finland. Because the DNA sequence containing exon 10 of CHEK2 is present in multiple homologous copies in the genome, specific PCR primers for the CHEK2 exon 10 on chromosome 22 were designed to avoid all other homologous sequences, as described elsewhere (forward primer 5′-TTA ATT TAA GCA AAA TTA AAT GTC-3′; reverse primer 5′-GGC ATG GTG GTG TGC ATC-3′) (Vahteristo et al. 2001b). We applied minisequencing (primer extension) (Syvanen et al. 1993) for the mutation detection, and all positive minisequencing results were confirmed by reamplification from the original genomic DNA sample and by direct sequencing. In the statistical analyses, differences in continuous variables were evaluated by use of the Mann-Whitney test (SPSS, version 8.0), and differences in dichotomous variables were evaluated by χ2 test or by Fisher's exact test. All P values are two sided.
The frequency of the 1100delC variant among population controls was 1.4% (26/1,885) (table 1). Among the unselected patients with breast cancer, a slightly higher frequency (2.0% [21/1,035]) was seen (odds ratio [OR] 1.48; 95% CI 0.83–2.65; P=.182). Of those 358 patients who reported at least one first- or second-degree relative affected with breast or ovarian cancer, 11 (3.1%) carried the 1100delC variant (OR 2.27; 95% CI 1.11–4.63; P=.021, compared with population control subjects). The frequency of 1100delC among 677 patients with no family history of breast cancer (1.5%) was similar to that seen in control individuals. In the same cohort of 1,035 patients with breast cancer, 19 (1.8%) carried BRCA1 or BRCA2 mutations (4.2% of the 358 patients with a positive family history). CHEK2 and BRCA mutations were seen in different kinds of patients with familial breast cancer. Most BRCA1 and BRCA2 mutations (11/19 [58%]) were found in patients with a strong family history (three or more first- or second-degree relatives with breast or ovarian cancer in the family [including the index patient]), whereas only 3/21 (14%) of CHEK2 variants were found among this group. All CHEK2 variants were found in patients who tested negative for BRCA1 and BRCA2 mutations. Taken together, the results suggest that CHEK2 1100delC variant is associated with positive family history of breast cancer, independently of BRCA1 and BRCA2 mutations.
Table 1.
Frequency of the CHEK2 1100delC Mutation in the Cohorts of Unselected Patients and Patients with Familial Breast Cancer, According to Family History
| Cohorts and Subgroups | No. ofSubjects | No. with Mutation (%) | ORa | 95% CI | P |
| Population controls | 1,885 | 26 (1.4) | |||
| Unselected patients with breast cancer: | 1,035 | 21 (2.0) | 1.48 | .83–2.65 | .182 |
| No family history | 677 | 10 (1.5) | |||
| ⩾1 Affected first- or second-degree relative | 358 | 11 (3.1) | 2.27 | 1.11–.63 | .021 |
| Breast cancer only | 318 | 11 (3.5) | 2.56 | 1.25–5.23 | .008 |
| Breast and ovarian cancer | 40 | 0 | |||
| Patients with familial breast cancer: | 507 | 28 (5.5) | 4.18 | 2.4–7.2 | .0002 |
| By type(s) of cancer: | |||||
| Breast cancer only | 448 | 23 (5.1) | 3.87 | 2.19–6.85 | <.00005 |
| Breast and ovarian cancer | 59 | 4 (6.8) | 5.20 | 1.75–15.41 | .012 |
| By no. of affected relatives: | |||||
| Index with 1 affected first-degree relative | 291 | 18 (6.2) | 4.70 | 2.55–8.71 | <.00005 |
| ⩾3 Affected family membersb: | 216 | 10 (4.6) | 3.50 | 1.65–7.3 | .002 |
| Index with affected first-degree relatives | 164 | 7 (4.3) | 3.19 | 1.36–7.46 | .013 |
| Index with affected second-degree relatives only | 52 | 3 (5.8) | 4.38 | 1.28–14.95 | .041 |
Compared with population control subjects.
First- or second-degree relatives with breast or ovarian cancer (including the index patient).
We further examined the significance of CHEK2 in non-BRCA1/2 familial breast cancer in an independent, large cohort of 507 patients, consisting exclusively of those with positive family history. These included 216 BRCA1/2-mutation–negative patients with breast cancer (index cases from the families) who had a strong family history (three or more first- or second-degree relatives with breast or ovarian cancer [including the index patient]), as verified by links to the Finnish Cancer Registry and to hospital records. In addition, we examined 291 unrelated, BRCA1/2 mutation-negative patients with breast cancer who reported only a single affected first-degree relative. The patients with familial breast cancer were identified at the Helsinki University Central Hospital, as described elsewhere (Vehmanen et al. 1997; Eerola et al. 2000; Vahteristo et al. 2001a,), and BRCA1/2 mutations were excluded, as described elsewhere (Vehmanen et al. 1997, Vahteristo et al. 2001a).
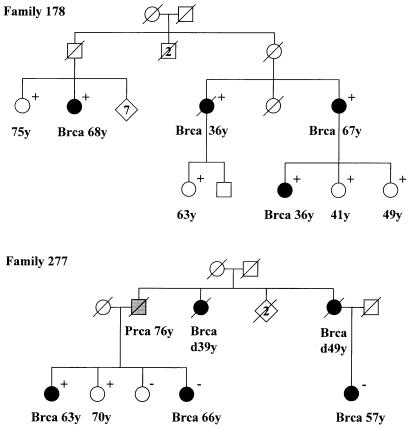
The 507 patients with familial breast cancer included 28 (5.5%) patients who carried the 1100delC variant, a significantly higher frequency than is seen in population control individuals (OR 4.18, 95% CI 2.4–7.2; P=.0002). The frequency of 1100delC was significantly elevated, in comparison with population control subjects, both among patients with only one affected first-degree relative (18/291 [6.2%]; OR 4.7; 95% CI 2.55–8.71; P<.00005), as well as in patients with a strong family history (10/216 [4.6%]; OR 3.5; 95% CI 1.65–7.3; P=.002). Frequencies among index patients with a strong family history, with or without an affected first-degree relative, are shown in table 1. All carriers identified in the different study cohorts were heterozygous for the mutation. Segregation of the CHEK2 variant was tested in the five CHEK2-positive families that had a sufficient number of affected family members available for sampling. Pedigrees of two families are shown in figure 1. Although the mutation was shared among several patients in the five families, segregation of the mutation with the disease was incomplete, in that both unaffected mutation carriers and mutation-negative patients with breast cancer were observed. The observation of unaffected carriers, as well as the strong association of the 1100delC variant with patients who had only a moderate family history, suggests that the CHEK2 1100delC variant has low penetrance. Furthermore, although the CHEK2 1100delC mutation is more frequent in patients with familial breast cancer, it alone may not explain the clustering of the disease in the pedigrees, in which, presumably, other, unknown predisposition alleles may also segregate. According to recent epidemiological evidence from studies of patients with breast cancer and their families, non-BRCA1/2 familial aggregation of breast cancer may be caused by multiplicative effects of low-penetrance genes (Antoniou et al. 2001, 2002). Although several common polymorphisms have been reported to have a slightly higher frequency among patients with breast cancer than among control individuals (Dunning et al. 1999), none of these have been shown to be associated with family history of the disease. The CHEK2 1100delC mutation may therefore be the first example of a low-penetrance genetic alteration that contributes significantly to familial clustering of breast cancer at the population level and also in families with small numbers of affected relatives.
Figure 1.
Pedigrees of two families with a CHEK2 1100delC germline variant. In family 178 (top), four patients with breast cancer share the mutation, but four unaffected women are also carriers. In family 277 (bottom), the mutation is found only in one patient with breast cancer and her unaffected sister. Prca = prostate cancer; Brca = breast cancer; + = positive for mutation; − = negative for mutation. Numbers followed by “y” represent age at diagnosis, except for two women in family 277, whose age is preceded by “d,” representing age at death.
Some phenotypic features of patients with breast cancer who carry the CHEK2 mutation deserve attention. First, we observed a strong association of the 1100delC variant with bilateral breast cancer. Among the 627 unselected patients with breast cancer for whom information on laterality was available, there were 33 patients with bilateral breast cancer, of whom 4 (12.1%) carried the mutation (OR 6.17; 95% CI 1.87–20.32; P=.007), compared with 12 (2.0%) of the 594 patients with unilateral disease. Two of these CHEK2-positive patients had a family history of breast cancer. We have elsewhere reported an elevated risk of a second, contralateral breast cancer in BRCA1/2-mutation–negative families with breast cancer (Eerola et al. 2001). The possibility that this could be related to the CHEK2 1100delC variant warrants further investigation. Second, there was no association of 1100delC mutation with ovarian cancer. In the familial cohort, the frequency of 1100delC was similar in families with and without members who had ovarian cancer (P=.54; table 1), and all 11 mutations among patients with familial disease in the unselected cohort were found in patients whose family history included only breast cancer (table 1). Third, the mean age at onset of the CHEK2 carriers in all of the patient cohorts was 52.7 years, an age only marginally different from the mean age of patients with no mutations (54.8 years; P=.062). A similar trend was observed in the cohort of unselected patients (54.7 vs. 57.4 years; P=.292). These results contrast with the ovarian cancer association and the earlier age at disease onset that are observed among carriers of BRCA1 and BRCA2 mutations, compared with those of noncarriers. In our earlier study of the same cohort (Syrjakoski et al. 2000), this difference was 8 years (49.7 years vs. 57.5 years; P=.026). Therefore, these results further support the role of CHEK2 1100delC mutation as a distinct allele that predisposes carriers to disease and whose effects are not seen as a shift in the age at diagnosis.
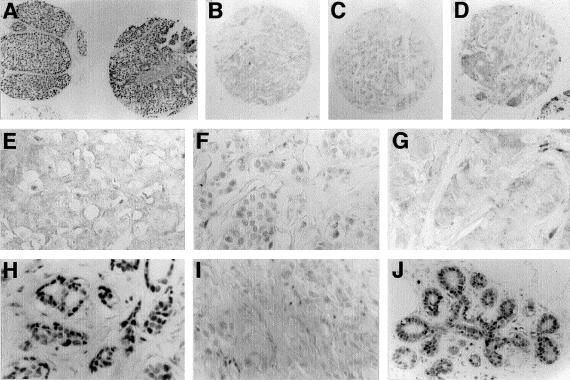
We also explored whether the germline 1100delC mutation is associated with the expression of the CHEK2 protein in tumor tissues. A tissue microarray containing 124 tumors from BRCA1/2-mutation–negative families was constructed and was stained with a mouse monoclonal antibody, DCS-270, against the human CHEK2 protein. The antibody and protocols for sensitive immunoperoxidase staining of human tissues have been described elsewhere (Bartkova et al. 2001; Lukas et al. 2001). The staining results were evaluated without prior knowledge of the mutation status of CHEK2. Specificity and reproducibility controls included lack of staining with DCS-270 preabsorbed with recombinant CHEK2 protein, consistency of staining patterns between two different samples from each of the tumors present on the array, parallel staining of normal breast tissues under identical conditions, and parallel staining of the array with an unrelated monoclonal antibody. Given the reproducible detection of CHEK2 in 70%–90% of luminal epithelial cells in normal breast tissues present on the microarray (as well as cells examined on parallel sections), we considered that the staining of tumors was aberrantly reduced when either a lower CHEK2-staining intensity was seen or when the percentage of CHEK2-positive cells was <50%. Overall, reduced CHEK2 protein expression (fig. 2) was observed in 21/124 (16.9%) breast tumors. The array included four breast tumors from four CHEK2 1100delC germline carriers from different families (including pedigrees from families 178 and 277 shown in fig. 1). All four of these patients showed reduced CHEK2 expression (fig. 2). The 1100delC germline variant was therefore seen in 4/21 (19%) patients with reduced expression of the CHEK2 protein; in contrast, none of the 103 patients with normal CHEK2-staining pattern showed mutations (P<.0005). The fact that all tumors from patients with germline CHEK2 1100delC showed reduced immunostaining for CHEK2 supports the role of the germline variant in influencing CHEK2 protein levels, as well as its possible functional inactivation in tumorigenesis. Three of these four tumors were among the most grossly defective in CHEK2 staining, suggesting that the second allele was probably lost. Because the DCS-270 epitope lies within the N-terminus of CHEK2 (Lukas et al. 2001) (a part of the protein that is preserved in CHEK2 1100delC), this genetic variant may affect the expression and/or stability of CHEK2 in addition to the kinase activity.
Figure 2.
Immunohistochemical detection of CHEK2 in human breast tissues and tumors on a tissue microarray. Examples include breast carcinomas with normal (panels A and H) or aberrantly reduced (panels B, C, D, E, F, G and I) CHEK2 levels, compared with normal breast tissue control (panel J). The samples shown in panels B, C, and D are from three different tumors with 1100delC, and corresponding detailed staining patterns are documented in panels E, F, and G, respectively. Panel I shows reduced CHEK2 immunostaining in a breast carcinoma wild type for exon 10. The incubation with the DCS-270 antibody and biotin-streptavidin-peroxidase detection was not followed by nuclear counterstaining (Bartkova et al. 2001; Lukas et al. 2001), to facilitate evaluation of the CHEK2 staining intensity in the nuclei of tumor cells, normal epithelial cells, and stromal cells. Note the reduced staining for CHEK2 in panels B, C, D, E, F, G, and I, compared with normal levels seen in panels A, H, and J. Magnification 30×, in panels A–D, and 120×, in panels E–J.
In conclusion, our results indicate that the1100delC genetic variant of the CHEK2 tumor-suppressor gene, previously demonstrated to abolish the kinase activity of the CHEK2 protein (Wu et al. 2001), is significantly associated with positive family history of breast cancer in two distinct patient cohorts. A high frequency of this CHEK2 mutation was seen also in patients belonging to families with small numbers of affected relatives. The clustering of this variant in families with small numbers of affected relatives, the incomplete segregation in families with a larger number of affected relatives, and the absence of an association with ovarian cancer and early age at breast cancer onset all support the hypothesis that 1100delC is acting as a low-penetrance predisposition allele, with distinctly different phenotypic consequences, as seen in carriers of BRCA1/2 mutations. The strong association of the CHEK2 1100delC with families that include two affected patients is important, because these are the most common types of breast cancer families. In our recent large epidemiological survey, 16.7% of patients with breast cancer reported only a single affected first-degree relative, whereas 6.9% had two or more affected relatives (Eerola et al. 2000). A genetic alteration such as CHEK2 1100delC is therefore likely to have a significant contribution to familial breast cancer at the population level, although it alone may not be suitable in guiding genetic counseling of individual patients or families. Finally, the 1100delC was sixfold more frequent in patients with bilateral breast cancer than in patients with unilateral disease. If confirmed by additional studies, this observation could, in the future, have a direct impact on patient management and follow-up.
Note added in manuscript.—While this manuscript was under review, another study was published that reported a similar strong association between the CHEK2 1100delC allele and familial breast cancer (CHEK2 Breast Cancer Consortium). Female carriers of the 1100delC allele were estimated to have a twofold increase in breast cancer risk, and the allele was found to have a 1.1% population prevalence among healthy individuals in the United Kingdom, The Netherlands, and North America. The carriers were found to share a common allele at a CHEK2 intragenic microsatellite marker D22S275, which may indicate that this predisposition allele with a wide distribution in different populations may have a very ancient common origin. Taken together with our results, the studies confirm CHEK2 1100delC as a low-penetrance breast cancer–predisposition allele.
Acknowledgments
Dr. Pertti Sistonen and the Finnish Red Cross Blood Transfusion Service are gratefully acknowledged, for the population controls, as is Finnish Cancer Registry, for cancer data. We thank Sirpa Stick and Kati Rouhento, for technical assistance, and Minna Merikivi, for patient contacts. This work was supported by the Helsinki University Science Foundation, the Finnish Cancer Society, the Finnish Cultural Foundation, the Ida Montin Foundation, the Helsinki University Central Hospital Research Fund, the Sigrid Juselius Fund, the Danish Cancer Society, and the Danish Medical Research Council.
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BRCA1 [MIM 113705], BRCA2 [MIM 600185], CHEK2 [MIM 604373], and LFS [MIM 151623])
References
- Antoniou AC, Pharoah PD, McMullan G, Day NE, Ponder BA, Easton DF (2001) Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol 21:1–18 [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J, Ponder BJ, Easton DF (2002) A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer 86:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Falck J, Lukas J (2001) CHK2 kinase: a busy messenger. Nat Rev Mol Cell Biol 2:877–886 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Falck J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J (2001) Chk2 tumour suppressor protein in human spermatogenesis and testicular germ-cell tumours. Oncogene 20:5897–5902 [DOI] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, Garber JE, Haber DA (1999) Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528–2531 [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD (2000) Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 14:278–288 [PMC free article] [PubMed] [Google Scholar]
- CHEK2 Breast Cancer Consortium (2002) Low-penetrance susceptibility to breast cancer due to CHEK2*1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 31:55–59 [DOI] [PubMed] [Google Scholar]
- Dunning AM, Healey CS, Pharoah PD, Teare MD, Ponder BA, Easton DF (1999) A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev 8:843–854 [PubMed] [Google Scholar]
- Eerola H, Blomqvist C, Pukkala E, Pyrhonen S, Nevanlinna H (2000) Familial breast cancer in southern Finland: how prevalent are breast cancer families and can we trust the family history reported by patients? Eur J Cancer 36:1143–1148 [DOI] [PubMed] [Google Scholar]
- Eerola H, Pukkala E, Pyrhonen S, Blomqvist C, Sankila R, Nevanlinna H (2001) Risk of cancer in BRCA1 and BRCA2 mutation-positive and -negative breast cancer families (Finland). Cancer Causes Control 12:739–746 [DOI] [PubMed] [Google Scholar]
- Haruki N, Saito H, Tatematsu Y, Konishi H, Harano T, Masuda A, Osada H, Fujii Y, Takahashi T (2000) Histological type-selective, tumor-predominant expression of a novel CHK1 isoform and infrequent in vivo somatic CHK2 mutation in small cell lung cancer. Cancer Res 60:4689–4692 [PubMed] [Google Scholar]
- Hofmann WK, Miller CW, Tsukasaki K, Tavor S, Ikezoe T, Hoelzer D, Takeuchi S, Koeffler HP (2001) Mutation analysis of the DNA-damage checkpoint gene CHK2 in myelodysplastic syndromes and acute myeloid leukemias. Leuk Res 25:333–338 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Lim DS (2000) The many substrates and functions of ATM. Nat Rev Mol Cell Biol 1:179–186 [DOI] [PubMed] [Google Scholar]
- Lee JS, Collins KM, Brown AL, Lee CH, Chung JH (2000) hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201–204 [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim SH, Bell DW, Wahrer DC, Schiripo TA, Jorczak MM, Sgroi DC, Garber JE, Li FP, Nichols KE, Varley JM, Godwin AK, Shannon KM, Harlow E, Haber DA (2001) Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni syndrome. Cancer Res 61:8062–8067 [PubMed] [Google Scholar]
- Lukas C, Bartkova J, Latella L, Falck J, Mailand N, Schroeder T, Sehested M, Lukas J, Bartek J (2001) DNA damage–activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res 61:4990–4993 [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893–1897 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ (2000) Ataxia telangiectasia–mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA 97:10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CW, Ikezoe T, Krug U, Hofmann WK, Tavor S, Vegesna V, Tsukasaki K, Takeuchi S, Koeffler HP (2002) Mutations of the CHK2 gene are found in some osteosarcomas, but are rare in breast, lung, and ovarian tumors. Genes Chromosomes Cancer 33:17–21 [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 14:289–300 [PMC free article] [PubMed] [Google Scholar]
- Syrjakoski K, Vahteristo P, Eerola H, Tamminen A, Kivinummi K, Sarantaus L, Holli K, Blomqvist C, Kallioniemi OP, Kainu T, Nevanlinna H (2000) Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst 92:1529–1531 [DOI] [PubMed] [Google Scholar]
- Syvanen AC, Sajantila A, Lukka M (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am J Hum Genet 52:46–59 [PMC free article] [PubMed] [Google Scholar]
- Tavor S, Takeuchi S, Tsukasaki K, Miller CW, Hofmann WK, Ikezoe T, Said JW, Koeffler HP (2001) Analysis of the CHK2 gene in lymphoid malignancies. Leuk Lymphoma 42:517–520 [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Eerola H, Tamminen A, Blomqvist C, Nevanlinna H (2001a) A probability model for predicting BRCA1 and BRCA2 mutations in breast and breast-ovarian cancer families. Br J Cancer 84:704–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, Blomqvist C, Aittomaki K, Nevanlinna H (2001b) p53, Chk2, and Chk1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of Chk2 in inherited cancer predisposition. Cancer Res 61:5718–5722 [PubMed] [Google Scholar]
- Vehmanen P, Friedman S, Eerola H, McClure M, Ward B, Sarantaus L, Kainu T, Syrjakoski K, Pyrhönen S, Kallioniemi O, Muhonen T, Luce M, Frank T, Nevanlinna H (1997) Low proportion of BRCA1 and BRCA2 mutations in breast cancer families: evidence for additional susceptibility genes. Hum Mol Genet 6:2309–2315 [DOI] [PubMed] [Google Scholar]
- Wu X, Webster SR, Chen J (2001) Characterization of tumor-associated Chk2 mutations. J Biol Chem 276:2971–2974 [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408:433–439 [DOI] [PubMed] [Google Scholar]