Significance
In neurons, many mRNAs are transported to synapses in a translationally repressed state, allowing for the spatial and temporal regulation of protein synthesis required for synaptic plasticity. It has been assumed that these mRNAs are repressed at the initiation step of translation. Here we provide evidence for a second mechanism whereby these mRNAs are instead repressed at elongation/termination awaiting translational reactivation upon appropriate synaptic signals. Our results establish that a form of translation-dependent synaptic plasticity, which is dysregulated in neurodevelopmental and psychiatric pathologies, occurs independently of translation initiation. Elucidating the upstream pathways that lead to repression and reactivation of elongation/termination on these mRNAs may provide new avenues for the design of therapies targeting neurodevelopmental disorders.
Keywords: RNA granule, mGluR-LTD, translation elongation, microtubule-associated protein 1b
Abstract
Some forms of synaptic plasticity require rapid, local activation of protein synthesis. Although this is thought to reflect recruitment of mRNAs to free ribosomes, this would limit the speed and magnitude of translational activation. Here we provide compelling in situ evidence supporting an alternative model in which synaptic mRNAs are transported as stably paused polyribosomes. Remarkably, we show that metabotropic glutamate receptor activation allows the synthesis of proteins that lead to a functional long-term depression phenotype even when translation initiation has been greatly reduced. Thus, neurons evolved a unique mechanism to swiftly translate synaptic mRNAs into functional protein upon synaptic signaling using stalled polyribosomes to bypass the rate-limiting step of translation initiation. Because dysregulated plasticity is implicated in neurodevelopmental and psychiatric disorders such as fragile X syndrome, this work uncovers a unique translational target for therapies.
Most studies of translational control focus on initiation, the process where mRNAs recruit ribosomes and catalyze the first step of translation (1). This highly regulated and normally rate-limiting step of translation is followed by elongation and termination, resulting in completed proteins. Although multiple ribosomes on a given mRNA (a polyribosome) imply active peptide synthesis, we and others identified neuronal RNA granules—motile aggregates of nontranslating ribosomes (2, 3). These electron-dense bodies contain single copies of synaptic mRNAs that are translationally silenced during their transport from soma to synapse (1, 4).
Many models assume that neuronally transported mRNAs are translationally paused before completion of the initiation step of translation during transport. An appropriate synaptic signal would then activate translation (initiation/elongation/termination) of the granule mRNA. However, it is not clear how many free ribosomal subunits are present at synapses to support translation initiation. Further, at a typical translation elongation rate of six amino acids per s (5, 6), synthesis of larger synaptic proteins (e.g., microtubule-associated protein 1b; MAP1b) would take over 5 min even if initiation were immediate. These two factors constrain the speed and magnitude of synaptic translation and, thus, plasticity. As some forms of synaptic plasticity require rapid (<10 min) and localized activation of protein synthesis, an alternative model is wanting (7–9).
We have previously proposed the concept of a neuronal RNA granule as a stalled polyribosome (10, 11). Ribosomal stalling has been shown to occur in lysates from a mouse neuroblastoma cell line and in an in vitro rabbit reticulocyte lysate translation assay programmed with brain homogenate (12). Whether neuronal ribosome stalling occurs in vivo is uncertain. We hypothesized that neuronal RNA granules contain paused ribosomes with incomplete proteins initiated in the soma before their packaging and transport to dendrites, where translation can be rapidly and locally completed on demand. Here we show that reactivation of translation on stalled polyribosomes is a unique feature of the neuronal landscape that functions in metabotropic glutamate receptor (mGluR) long-term depression (LTD), providing the neuron with the ability to rapidly and specifically respond to stimuli independently of translation initiation.
Results
An appropriate assay is required to determine whether stalled polyribosomes are present in neurons and, if so, whether they are dynamically regulated by neuronal activity. We reasoned that if polyribosomes are stalled at the level of elongation and/or termination, they should be unaffected by inhibitors that act at initiation or during the first round of elongation at the initiation codon, whereas those actively synthesizing protein would dissociate due to ribosome runoff and could not be replaced. To perform this type of runoff assay, we required a robust method to visualize polyribosome complexes within intact neurons. Here we use the recently described ribopuromycylation (RPM) method (13–15) to visualize ribosomes associated with nascent peptide chains within formaldehyde-fixed primary hippocampal neurons. In this technique, live cells are coincubated with the irreversible translation elongation inhibitor emetine, together with puromycin, a tyrosyl-tRNA analog that covalently attaches to the carboxyl terminus of nascent chains. Although puromycylation is a peptide chain-terminating event, emetine added in this context (through an imperfectly understood mechanism) prevents puromycylated nascent chain release from ribosomes (14). This enables standard immunofluorescence visualization of ribosome-bound nascent chains in fixed and permeabilized cells using a puromycin-specific monoclonal antibody.
Puromycylated Puncta in Rat Hippocampal Neurons Represent Neuronal RNA Granules.
As shown in Fig. 1A, RPM staining in dissociated rat hippocampal neuron cultures revealed puromycin-specific staining indicative of polyribosomes throughout the soma and neurites (“−puro” and “+puro” conditions demonstrate the strict puromycin dependence on staining). Closer inspection revealed intensely stained puncta throughout the neurites (Fig. 1B, arrowheads), consistent with electron micrographs of polyribosomes in neurites (2). Further characterization of neuronal RPM staining revealed that puncta colocalize with the RNA-specific dye Syto14 (Fig. 1C; quantified in Fig. 1 H and I), consistent with previous detection of RNA granules in live neurons using this dye (16). Syto14 also stains mitochondria; however, the Syto14-mitochondria puncta (imaged with an antibody targeting translocase of outer mitochondrial membrane 20 (Tom20), a mitochondrial outer-membrane protein) do not colocalize with RPM staining (Fig. S1, arrowheads). Colocalization of RPM signal with ribosomal protein S6 has been previously shown in a fibroblast cell line (15), and we also observed significant colocalization of S6 with RPM puncta in primary neurons (Fig. 1D; quantified in Fig. 1 H and I). Further, we found that RPM puncta colocalize with punctate fragile X mental retardation protein (FMRP), a component of RNA granules, where it is thought to negatively regulate translation by stalling ribosome processivity (12, 17–19) (Fig. 1E; quantified in Fig. 1 H and I). We found similar colocalization of transiently expressed GFP fused to the RNA-binding protein Staufen 2 (GFP-Stau2), a component of an RNA granule that regulates transport and translation of MAP1b mRNA (3, 11) (Fig. 1F; quantified in Fig. 1 H and I). RPM staining is significantly more colocalized with all of the RNA granule markers than it is with the mitochondrial marker Tom20 (Fig. 1G; quantified in Fig. 1 H and I). However, not all puncta stained with the RNA granule markers colocalized with RPM (Fig. S2), consistent with the presence of multiple types of RNA transport particles that do not contain polyribosomes (10, 19). We also ruled out the possibility that RPM puncta reflect puromycylation on mitochondrial or contaminating bacterial ribosomes by inhibiting prokaryotic/mitochondrial translation or by assessing colocalization of RPM puncta with any bacterial DNA that might be present (Figs. S3 and S4). Together, these data demonstrate that the puromycin-stained puncta show characteristics of neuronal RNA granules.
Fig. 1.
Puromycylated puncta in rat hippocampal neurons represent neuronal RNA granules. (A) Confocal images of polyribosome labeling by RPM in rat hippocampal neurons. Arrows denote high-intensity puncta in neurites. (B) Magnified and straightened neurites from A with arrowheads denoting high-intensity puromycin puncta in distal locales. (C–F) Representative confocal images of neurites illustrating colocalization of RPM puncta (circles) with markers of either ribosomal RNA (Syto14) or RNA granules including ribosomal protein S6, FMRP, or GFP-Stau2. (G) Representative confocal images of neurites illustrating weak colocalization of RPM puncta with mitochondria (Tom20). (H) The fraction of RPM signal that colocalizes with RNA granule and mitochondrial markers as quantified by calculating the Manders colocalization split coefficient; n = 20 neurites from three independent experiments. (I) The correlation of RPM puncta signal with RNA granule and mitochondrial marker signals as quantified using the Pearson correlation function; n = 50 puncta from three independent experiments. All bars represent mean ± SEM, and P values were calculated using a one-way ANOVA with Dunnett’s post hoc tests.
We next performed experiments using the human embryonic kidney (HEK) 293T cell line to verify that we could induce runoff of polyribosomes in intact cells (measured by RPM) as previously described (13–15). Because RNA granules (including putative stalled polyribosomes) have so far been observed only in oligodendrocytes (20) and neurons (16), we did not expect stalled polyribosomes to be abundant in 293T cells. To induce runoff, we used pateamine A (PatA), which binds the RNA helicase eukaryotic initiation factor (eIF)4A, blocking its essential function in eIF4F-mediated translation initiation (21), and homoharringtonine (HHT), which uniquely inhibits the first round of translation elongation by impeding translocation of the ribosome from the initiation codon while leaving downstream ribosomes unaffected, thus depleting elongating ribosomes and preventing any further initiation (Fig. S5) (22, 23). To assess the ability of PatA and HHT to inhibit peptide synthesis, we metabolically labeled neurons with the methionine analog l-azidohomoalanine (AHA) for 60 min, followed by fixation and coupling of AHA-labeled proteins to a fluorophore via click chemistry (24). As expected, preincubation with either PatA or HHT severely repressed active protein synthesis in 293T cells by ∼85% (Fig. 2A; quantified in Fig. 2B). Next, we measured protein synthesis at the level of the polyribosome using RPM in 293T cells. As expected, preincubation with PatA or HHT blocked a significant proportion (∼70%) of cytoplasmic RPM staining, consistent with the majority of polyribosomes in nonneuronal cells being actively engaged in protein synthesis (Fig. 2C; quantified in Fig. 2D). Critically, we were able to create an artificially stalled polyribosome phenotype in 293T cells by irreversibly blocking elongation with emetine before treatment with HHT (Fig. 2 C and D). Under these conditions, HHT was unable to reduce RPM staining, confirming that this technique reliably detects stalled polyribosomes.
Fig. 2.
Inhibitors of initiation and the first round of elongation induce polyribosome runoff that can be detected by ribopuromycylation in a nonneuronal cell line. (A) Confocal images demonstrating that nascent polypeptide chain labeling with the methionine analog AHA in HEK293T cells is prevented by preincubation with 100 nM PatA or 5 mM HHT. Cells were incubated in the presence of AHA for 60 min before fixation. Omitting the copper catalyst (−Cu) prevented fluorophore coupling to AHA peptides, thus serving as a nonspecific staining control. (B) Quantitation of cytoplasmic AHA signal from images in A. (C) RPM labeling of polyribosomes in HEK293T cells in the presence of PatA, HHT, and emetine. Cells were pretreated with inhibitors before RPM as detailed in Materials and Methods. For “Emetine+HHT” treatment, cells were incubated with emetine for 5 min before supplementing with HHT for an additional 10 min. (D) Quantitation of cytoplasmic RPM signal from images represented in C. (E) Confocal images demonstrating that PatA and HHT reduce active translation (measured by metabolic AHA labeling) in rat hippocampal neurons to the same extent as seen in HEK293T cells. (F) Magnified soma and straightened neurites from images in E. (G) Quantification of AHA signal in the soma and neurites from images represented in F. All bars represent mean ± SEM and n = 9 cells from three independent experiments. ***P < 0.001. ns, P > 0.05 calculated using a one-way ANOVA with Dunnett’s post hoc tests.
Neuronal Polyribosomes Are Stalled Downstream of the First Round of Translation Elongation.
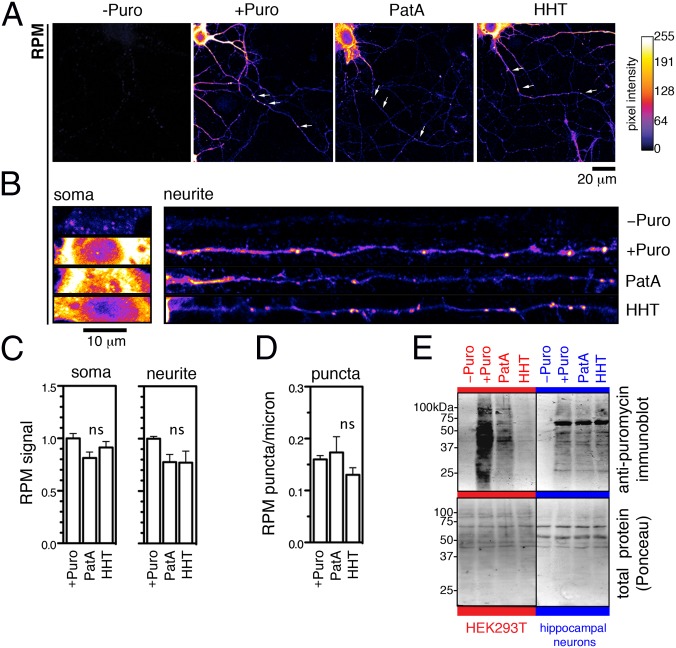
With a reliable assay to detect stalled polyribosomes (preincubation with PatA or HHT followed by RPM), we next asked whether they are present in primary neurons. First, we confirmed that both PatA and HHT could reduce active peptide synthesis in both the soma and neurites as measured by AHA labeling (Fig. 2 E and F; quantified in Fig. 2G). To detect stalled polyribosomes, we examined whether RPM staining was resistant to runoff by preincubating with the inhibitors for 10 min. Despite the ability of either PatA or HHT to block most (∼90%) ongoing translation throughout the neuron (Fig. 2G) and the ability of PatA and HHT to induce polyribosome runoff in 293T cells (Fig. 2D), virtually all RPM staining in neuronal soma and neurites remained (Fig. 3 A and B; quantified in Fig. 3C). We next objectively measured the high-intensity RPM puncta by setting an intensity threshold and found that their number in neurites was significantly affected by neither PatA nor HHT (Fig. 3D). Although we observed a small (∼10–20%) decrease in both diffuse and punctate RPM staining (Fig. 3 C and D), the difference was not statistically significant when corrected for multiple comparisons. Thus, a majority of neuronal polyribosomes are stalled and resistant to runoff. Importantly, we were able to recapitulate the properties of RPM immunostaining by immunoblotting RPM-treated cell extracts with anti-puromycin antibody (Fig. 3E). The resulting broad pattern of labeled peptides confirmed that puromycylation indeed takes place in both 293T cells and neurons. However, although preincubation with PatA or HHT reduces puromycylation in 293T cells to near-background levels, these inhibitors failed to significantly reduce puromycylation in neurons (Fig. 3E). Together, these observations point to a distinct ribosomal landscape in neurons, exemplified by the presence of polyribosomes stalled at the level of elongation/termination.
Fig. 3.
Neuronal polyribosomes are stalled downstream of the first round of translation elongation. (A) Treatment with drugs that induce ribosome runoff reveals the persistence of RPM puncta (arrows). (B) Magnified images of soma and straightened neurites from confocal images in A. (C) Quantified RPM signal in soma and neurites from images represented in B. (D) Quantified number of RPM puncta in neurites found more than 50 μm from the nucleus (refer to Materials and Methods for parameters). (E) Comparison of puromycylated peptides between nonneuronal HEK293T cells and primary rat hippocampal neurons by Western blot analysis following RPM (Upper). Total protein levels were assessed by Ponceau staining (Lower). Blot is representative of three independent experiments. All bars represent mean ± SEM and n = 9 cells from three independent experiments.
mGluR Activation Reduces the Number of Stalled Polyribosomes and Induces Initiation-Independent Peptide Synthesis in Neurites.
mGluR activation by 3,5-dihydrophenylglycine (DHPG) induces a form of plasticity called mGluR-LTD that depends on local (i.e., synaptic) translation. Because two mRNA-binding proteins implicated in mGluR-LTD, FMRP and Staufen 2, are localized to RPM puncta (Fig. 1 E and F), we hypothesized that DHPG could reactivate stalled polyribosomes, thereby synthesizing proteins necessary for mGluR-LTD. Contrary to a model where DHPG would increase the number of RPM puncta (i.e., polyribosomes) through translation initiation and elongation, our stalled polyribosome hypothesis would predict that DHPG would instead cause a decrease in the number of RPM puncta as elongation and termination proceed. To test this, we applied DHPG to hippocampal neuronal cultures for 0, 1, 5, 10, and 20 min followed by RPM. We found a progressive decrease in the number of RPM-positive puncta per micrometer in neurites >50 μm from the cell soma (Fig. 4A; quantified in Fig. 4B). As expected, this decrease was also seen in the presence of HHT, suggesting that it is independent of initiation events (Fig. 4B, open circles). A similar decrease in the number of puncta was also seen in proximal dendrites (Fig. S6).
Fig. 4.
Stalled neuronal polyribosomes can be reanimated upon the activation of metabotropic glutamate receptors. (A) DHPG treatment reduces the number of RPM puncta. Representative confocal images of polyribosome labeling by RPM in neurites following bath application of 50 μM DHPG for 0–20 min. (B) Quantification of RPM puncta per micrometer in distal neurites (>50 μm from soma; green box in A represents the region quantified) following DHPG treatment over time in the absence (closed circles) or presence (open circles) of HHT. n = 14–17 neurites from three independent experiments. P value was determined by a two-tailed Student t test. (C) Experimental schema for assessing DHPG-mediated changes in bulk protein synthesis in the presence or absence of HHT. (D) Inhibition of the first round of elongation by HHT does not impair DHPG-mediated increases in protein synthesis as detected by AHA labeling. n = 46–54 neurites from three independent experiments. All bars represent mean ± SEM, and the P value was calculated using a one-way ANOVA with Bonferroni post hoc tests. ns, P > 0.05.
If DHPG induces release of stalled polyribosomes and, moreover, if the latter represent a sizeable proportion of the ribosomes present in neurites, one might expect to see an initiation-independent increase in protein synthesis in the presence of DHPG. We used AHA labeling in hippocampal neurites to determine whether DHPG could increase peptide synthesis in the presence of HHT (see Fig. 4C for experimental schema). Despite the pronounced reduction of AHA incorporation upon treatment of neurons with HHT alone, we observed that DHPG significantly increased AHA labeling in the presence of HHT (Fig. 4D). The loss of RPM puncta and the initiation-independent increase in bulk protein synthesis following DHPG treatment, is consistent with a stimulus-dependent reactivation of stalled polyribosomes mediated through mGluRs.
A Concurrent Block in Translation Initiation Does Not Impair mGluR-Dependent MAP1b Protein Synthesis.
MAP1b mRNA is one of several transcripts whose local translation is required for mGluR-LTD (11, 25). We have previously shown that mGluR activation by DHPG induces release of MAP1b reporter mRNA from neurite RNA granules (11), consistent with storage of this mRNA in a stalled polyribosome. Therefore, we predicted that rapid MAP1b protein synthesis in distal dendrites would occur independently of translation initiation. Indeed, Pat A and HHT failed to impede DHPG-induced MAP1b protein expression in distal neurites as revealed by immunocytochemistry (Fig. 5A; quantified in Fig. 5B). Importantly, blocking elongation with emetine prevented the DHPG-mediated increase in MAP1b expression. These data demonstrate that mGluR-mediated translation of a known LTD target mRNA, although still requiring elongation and termination, can occur independently of the initiation step of protein synthesis.
Fig. 5.
Transient block in translation initiation or the first round of elongation does not impair mGluR-dependent MAP1b protein synthesis or LTD in hippocampal neurons. (A) Confocal images of MAP1b protein expression in hippocampal neurons assessed by immunocytochemistry. Neurons were incubated with PatA, HHT, or emetine for 10 min before coincubation with DHPG for an additional 10 min. (B) Quantification of MAP1b protein expression from experiments in A. Intensity of MAP1b signal was assessed at least 50 μm from the soma (delineated by the yellow circles in A; boxed areas indicate the neurite regions quantified). (C) Time plots of normalized field excitatory postsynaptic potential (fEPSP) slope evoked by Schaffer collateral stimulation in hippocampal slices incubated with vehicle only (No DHPG; n = 8) or DHPG (100 μΜ) alone for 5 min to induce mGluR-mediated long-term depression (DHPG; n = 9), or in the presence of emetine (DHPG+Emetine; n = 9) or HHT (n = 9). (D) Summary bar graph showing mGluR-LTD (at 40–70 min postinduction) was prevented by emetine but not HHT. (E) Time plots of normalized fEPSP slope showing that L-LTP induced by theta-burst stimulation (TBS) is impaired by HHT. (F) Summary bar graph showing differences in L-LTP between the control and HHT group (at 150–180 min postinduction). (C and E) (Insets) Average fEPSPs (n = 5 traces) taken during the baseline period (1; dotted trace) and after DHPG or TBS (2; solid trace). (Scale bars, 0.2 mV, 5 ms.) Results in B are means ± SEM; n = 72 neurites from three independent experiments. *P < 0.05, ***P < 0.001 were calculated using a one-way ANOVA with Dunnett’s post hoc tests. Results in D and F are means of fEPSP slopes normalized to baseline ± SEM. Statistical significances were calculated using a one-way ANOVA with Bonferroni post hoc tests (D) and Student t test (F).
Blocking the First Round of Translation Elongation Does Not Impair mGluR-Dependent LTD in Hippocampal Neurons.
If reanimation of translation stalled at the level of elongation/termination is a mechanism used by all or most of the mRNA targets up-regulated during mGluR-LTD, one would predict that protein synthesis-dependent LTD would be sensitive to general elongation inhibitors but not inhibitors that target initiation or only the first round of elongation. Indeed, it is well-established in the field that elongation inhibitors such as emetine, anisomycin, and cycloheximide block mGluR-LTD. To determine whether this form of plasticity is mainly mediated by reactivation of stalled polyribosomes, we induced mGluR-LTD in rat hippocampal slices using DHPG in the presence or absence of emetine or HHT. Strikingly, although emetine impaired mGluR-LTD, HHT failed to do so (Fig. 5 C and D). HHT was indeed effective in slices, as it inhibited late-LTP (L-LTP), a distinct form of translation-dependent plasticity (Fig. 5 E and F), and was found to reduce protein synthesis in slices by more than 90% (Fig. S7). Earlier studies had shown that blocking initiation with an excess of RNA cap analog was effective in occluding hippocampal mGluR-LTD (9). We thus examined whether longer incubations with HHT would be more effective. Neither a 30-min preincubation nor continuous perfusion of HHT during the entire course of the experiment was effective in blocking LTD (Fig. S8). However, preincubating with HHT for 1 h did block mGluR-LTD (Fig. S8), suggesting that in the prolonged absence of initiation, factors important for mGluR-LTD fail to be synthesized. The insensitivity of mGluR-LTD, but not L-LTP, to a de facto translation initiation inhibitor is consistent with distinct mRNA storage mechanisms for at least some of the mRNAs that underlie these distinct forms of plasticity.
Discussion
A recent study reported that polyribosomes in lysates from a mouse neuroblastoma cell line (N2A) are insensitive to runoff mediated by hippuristanol (an inhibitor of initiation) or puromycin (12). Specific mGluR-LTD mRNA targets, including MAP1b, were also retained on polyribosomes in the presence of hippuristanol.
Here we extend these in vitro data revealing stalled translation as a predominant feature in primary neurons, specifically targeting MAP1b translation and a protein synthesis-dependent form of synaptic plasticity, mGluR-LTD. Our findings strongly suggest elongation and termination rather than initiation as the critical steps of translation during mGluR-LTD (Fig. S5). It should be noted that other studies have indicated a role for initiation in mGluR-LTD (9). While we observed that pre-incubation with HHT for 1 h blocks mGluR-LTD (Fig. S8), blocking translation initiation at least 30 min before and during stimulation of mGluRs has no effect, and suggests that DHPG-mediated increases in initiation are not required for mGluR-LTD. Alternatively, differences in experimental conditions may alter the rate-limiting step that underlies this form of plasticity (11, 26, 27). Nevertheless, under the conditions used in this study, mGluR-LTD is resistant to an inhibitor that blocks the translation of newly initiated mRNAs, demonstrating that release of stalled polyribosomes is an important and sufficient step in producing the proteins necessary for this form of plasticity. Moreover, it highlights an important mechanistic difference between mGluR-LTD and L-LTP; although both types of plasticity are translation-dependent, the latter shows a stricter requirement for translation of newly initiated mRNAs. Our results are consistent with recent studies using the initiation inhibitor 4EGI-1, which report a block in L-LTP (28) but not striatal mGluR-dependent LTD (28, 29).
How does the puromycin reaction take place on stalled polyribosomes (i.e., engaged but not translocating ribosomes)? One might predict that stalled polyribosomes would be resistant to puromycin as it competes with aminoacyl-tRNAs that cannot undergo A- to P-site translocation. We speculate that the high concentration of puromycin used in this study allows it to displace the charged tRNA present at the A site. Another possibility is that the stalling involves occupation of the A site by other proteins that could be competed out with the addition of puromycin. Alternatively, a single 3′ proximal ribosome (perhaps at the stop codon) is stalled, causing the remaining upstream ribosomes to slow but perhaps not stall completely, thus allowing puromycylation.
If most neuronal ribosomes are occupied in stalled polyribosomes, what constitutes the large amount of basal translation in dendrites that is blocked by HHT and PatA (Fig. 2 E–G)? We suggest that most of this translation is mediated by monosomes that fall below the detection limit of the RPM technique. In this context, a slow rate of initiation relative to elongation would favor a small number of ribosomes processing an entire mRNA before another is initiated. It should also be noted that our AHA labeling measures translation products accumulated over a period of 60 min (Fig. 2G), whereas RPM offers a snapshot of polyribosomes. Therefore, even a small percentage of ribosomes occupied in active translation could mediate the initiation-dependent basal translation observed in this study.
Although the colocalization of RPM puncta in neurites with FMRP and Staufen 2 establishes the puncta as RNA granules, we also observed that many of the Staufen 2 and FMRP puncta in neurites do not label with RPM (Fig. S2). These data demonstrate the heterogeneity of these neuronal structures (30). Indeed, these two RNA-binding proteins play important roles in other ribonucleoprotein structures, including RNA interference complexes (31) and RNA transport complexes blocked at initiation (32). What determines whether a particular RNA, such as MAP1b, is stored in a stalled polyribosome is probably not defined by binding to a single RNA-binding protein, and may require some combinatorial code (33).
There is now mounting evidence that dysregulated translation affecting mGluR-LTD is responsible for the overt autistic-like behaviors found in mice and humans lacking FMRP, although the exact mechanism(s) by which this protein negatively regulates translation is unknown (34, 35). We therefore anticipate that elucidating the upstream pathways regulating elongation of mGluR-LTD target proteins will offer a tractable strategy for the design of new therapies targeting neurodevelopmental disorders such as fragile X syndrome and autism spectrum disorders in general.
Reactivation of translation stalled at elongation/termination is an elegant solution to the general problem of inducing protein synthesis with maximal alacrity when needed in special circumstances. It seems likely that this mechanism is widely applicable to vertebrate cell biology and beyond, and our findings demonstrate that the combined use of RPM with initiation inhibitors provides a robust strategy for examining this phenomenon.
Materials and Methods
Detailed descriptions can be found in SI Materials and Methods.
Animals and Cell Culture.
Sprague–Dawley rats were obtained from Charles River Canada. All experiments were approved by the Animal Ethics Committee of the Montreal Neurological Institute, and abided by the guidelines of the Canadian Council on Animal Care. Rat primary hippocampal neurons were dissected from embryonic day 18 Sprague–Dawley embryos and cultured as previously described for 8–10 d before experimentation (11). HEK293T cells were cultured in DMEM (Life Technologies) supplemented with 10% (vol/vol) FBS, sodium pyruvate, penicillin, and streptomycin.
AHA Labeling.
To assess active protein synthesis in primary rat hippocampal neurons at 8–10 d in vitro (DIV) or HEK293T cells, the methionine analog AHA (Life Technologies) was incubated with methionine-starved cells for 60 min before conjugation of AHA peptides to a fluorophore using the Click-IT Cell Labeling Kit (Life Technologies). Cells were treated with inhibitors of translation 10 min before and during the entire AHA incubation period. See SI Materials and Methods for details.
Ribopuromycylation.
RPM was performed in HEK293T cells as previously described (14) and in 8–10 DIV primary rat hippocampal neurons using 0.0003% digitonin for the extraction step. The cells were preincubated with inhibitors of translation for 10 min before and during the 5-min RPM procedure. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
Pateamine A was a generous gift of Dr. Jerry Pelletier. The GFP-FMRP expression plasmid was a kind gift of Dr. Keith Murai. T.E.G. is supported by a Jeanne Timmins-Costello Fellowship from the Montreal Neurological Institute and a Postdoctoral Fellowship from the Fonds de recherche du Québec-Santé. S.H.-S. is supported by a Graduate Studentship from Université de Montréal. This work was funded by the Canadian Institutes of Health Research (J.-C.L. and W.S.S.) and Fonds de recherche du Québec-Santé (J.-C.L. and W.S.S.). J.-C.L. is the recipient of the Canada Research Chair in Cellular and Molecular Neurophysiology.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307747110/-/DCSupplemental.
References
- 1.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61(1):10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32(4):683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 3.Elvira G, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5(4):635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci USA. 2012;109(12):4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boström K, et al. Pulse-chase studies of the synthesis and intracellular transport of apolipoprotein B-100 in Hep G2 cells. J Biol Chem. 1986;261(29):13800–13806. [PubMed] [Google Scholar]
- 7.Villareal G, Li Q, Cai D, Glanzman DL. The role of rapid, local, postsynaptic protein synthesis in learning-related synaptic facilitation in aplysia. Curr Biol. 2007;17(23):2073–2080. doi: 10.1016/j.cub.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mameli M, Balland B, Luján R, Lüscher C. Rapid synthesis and synaptic insertion of GluR2 for mGluR-LTD in the ventral tegmental area. Science. 2007;317(5837):530–533. doi: 10.1126/science.1142365. [DOI] [PubMed] [Google Scholar]
- 9.Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288(5469):1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 10.Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7(12):1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 11.Lebeau G, et al. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn Mem. 2011;18(5):314–326. doi: 10.1101/lm.2100611. [DOI] [PubMed] [Google Scholar]
- 12.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David A, et al. RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J Biol Chem. 2011;286(23):20688–20700. doi: 10.1074/jbc.M110.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David A, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197(1):45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willett M, Brocard M, Davide A, Morley SJ. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem J. 2011;438(1):217–227. doi: 10.1042/BJ20110435. [DOI] [PubMed] [Google Scholar]
- 16.Knowles RB, et al. Translocation of RNA granules in living neurons. J Neurosci. 1996;16(24):7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Primerano B, et al. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8(12):1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 18.Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4(6):350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 19.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134(6):1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Barbarese E, et al. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108(Pt 8):2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- 21.Bordeleau M-E, et al. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem Biol. 2006;13(12):1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Huang MT. Harringtonine, an inhibitor of initiation of protein biosynthesis. Mol Pharmacol. 1975;11(5):511–519. [PubMed] [Google Scholar]
- 23.Fresno M, Jiménez A, Vázquez D. Inhibition of translation in eukaryotic systems by harringtonine. Eur J Biochem. 1977;72(2):323–330. doi: 10.1111/j.1432-1033.1977.tb11256.x. [DOI] [PubMed] [Google Scholar]
- 24.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13(7):897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J Neurosci. 2007;27(48):13273–13278. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480(7375):63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24(28):6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoeffer CA, et al. Multiple components of eIF4F are required for protein synthesis-dependent hippocampal long-term potentiation. J Neurophysiol. 2013;109(1):68–76. doi: 10.1152/jn.00342.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santini E, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493(7432):411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller LC, et al. Combinations of DEAD box proteins distinguish distinct types of RNA:protein complexes in neurons. Mol Cell Neurosci. 2009;40(4):485–495. doi: 10.1016/j.mcn.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Jin P, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7(2):113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, et al. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell. 2008;19(1):105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8(7):533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 34.Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelleher RJ, III, Bear MF. The autistic neuron: Troubled translation? Cell. 2008;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.